Neuronal regulated ire-1-dependent mRNA decay controls germline differentiation in Caenorhabditis elegans
Figures
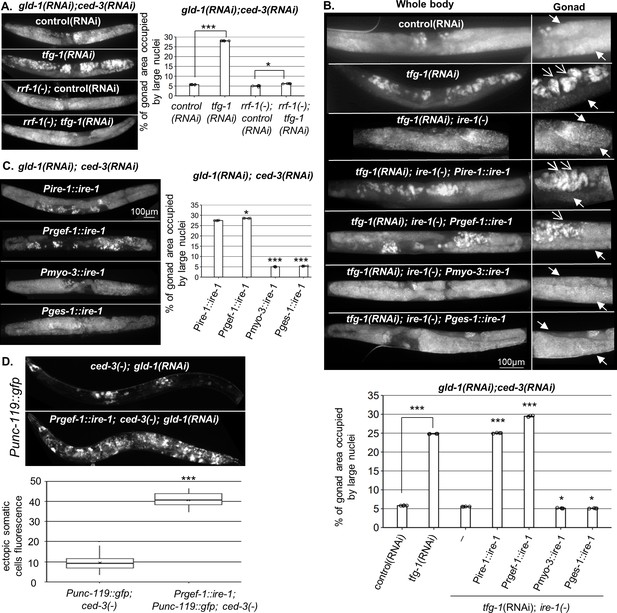
Endoplasmic reticulum (ER) stress in the soma regulates germline ectopic differentiation (GED).
Percent of gonad area occupied by aberrant somatic-like cells determined by DAPI staining of day 4 gld-1(RNAi); ced-3(RNAi) animals. (A) tfg-1 RNAi treatment resulted in increased levels of aberrant somatic-like cells in the gonads of wild-type animals but not in rrf-1 mutants (n = 320 gonads per genotype, N = 6). Asterisks mark one-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001 compared to the same genetic background treated with control RNAi. (B) Rescue of ire-1 expression in the soma (Pire-1::ire-1) and in the neurons (Prgef-1::ire-1) increased levels of aberrant somatic-like cells in the gonads upon tfg-1 RNAi treatment, whereas expression of ire-1 in the muscles (Pmyo-3::ire-1) and in the intestine (Pges-1::ire-1) did not. Full arrows indicate mitotic germ cells. Open arrows indicate aberrant nuclei (n = 210 gonads per genotype, N = 4). Asterisks mark one-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001 compared to ire-1(-) animals unless indicated otherwise. (C) Overexpression of ire-1 in the soma (Pire-1::ire-1) and in the neurons (Prgef-1::ire-1) of animals with wild-type ire-1 resulted in high levels of aberrant somatic-like cells in the gonads even in the absence of ER stress, whereas overexpression of ire-1 in the muscles (Pmyo-3::ire-1) and in the intestine (Pges-1::ire-1) did not (n = 250 gonads per genotype, N = 4). Asterisks mark one-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001 compared to Pges-1::ire-1 animals. (D) Overexpression of ire-1 in the neurons (Prgef-1::ire-1) of animals expressing the neuronal reporter Punc-119::gfp resulted in high GED levels in gld-1(RNAi); ced-3(-) genetic background, even in the absence of an ER stress-inducing treatment. Asterisks mark nested one-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001. Means represented by ‘X’ (n = 70 gonads per genotype, N = 2). Triple asterisks mark significant results of at least a twofold change.
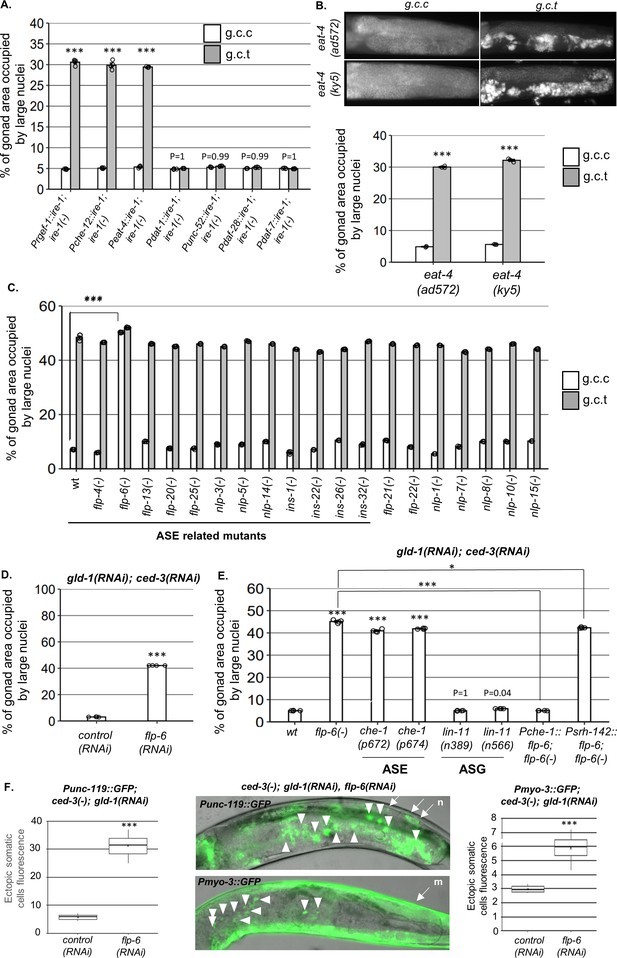
ASE-secreted FLP-6 regulates germline ectopic differentiation (GED).
Percent of gonad area occupied by ectopic somatic cells determined by DAPI-staining of day 4 animals. g.c.c represents treatment with gld-1, ced-3, and control RNAi mixture. g.c.t represents treatment with gld-1, ced-3, and tfg-1 RNAi mixture. (A) Rescue of ire-1 expression in all neurons (Prgef-1::ire-1), in sensory neurons (Pche-12::ire-1 (or in glutamatergic neurons [Peat-4::ire-1])) resulted in high levels of ectopic somatic cells in the gonads upon tfg-1 RNAi treatment, whereas expression of ire-1 in the dopaminergic (Pdat-1::ire-1), GABAergic (Punc-25::ire-1), and ASI/ASJ neurons (Pdaf-28::ire-1and Pdaf-7::ire-1) did not (n = 210 gonads per genotype, N = 4). Asterisks mark two-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001 relative to the same animals treated with g.c.c RNAi. For representative animals, see Figure 2—figure supplement 1. (B) eat-4 mutants displayed high levels of ectopic somatic cells in the gonads only upon tfg-1 RNAi treatment (n = 180 gonads per genotype, N = 3). Asterisks mark two-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001 relative to the same animals treated with g.c.c RNAi. (C, D) flp-6 mutants (C) and flp-6 RNAi-treated animals (D) displayed high levels of ectopic somatic cells in the gonads in the absence of endoplasmic reticulum (ER) stress (n = gonads per genotype, N = 4). Asterisks mark two-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 relative to wild-type animals treated with the same RNAi treatment (C) and Student’s t-test of p<0.001 (D). (E) che-1 mutants (with defective ASE) displayed high levels of ectopic somatic cells in the gonads, whereas lin-11 mutants (with defective ASG) did not. Rescue of flp-6 expression in ASE (Pche-1::flp-6) suppressed GED in flp-6(-) mutants, whereas its expression in ADF (Psrh-142::flp-6) did not. (n = 195 gonads per genotype, N = 4). Asterisks mark one-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001 relative to wild-type animals, unless indicated otherwise. All animals were treated with gld-1 and ced-3 RNAi. (F) gld-1(RNAi); ced-3(-) animals expressing the neuronal reporter Punc-119::gfp or the muscle reporter Pmyo-3::gfp displayed high GED levels upon treatment with flp-6 RNAi (n = 70 gonads per genotype, N = 2). Full arrows point at the labeling of somatic neurons (n) or muscles (m) within the soma. Arrowheads point at the labeling of somatic neuron or muscle cells within the gonad. Asterisks mark nested one-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001. Means represented by ‘X.’ Triple asterisks mark significant results of at least a twofold change.
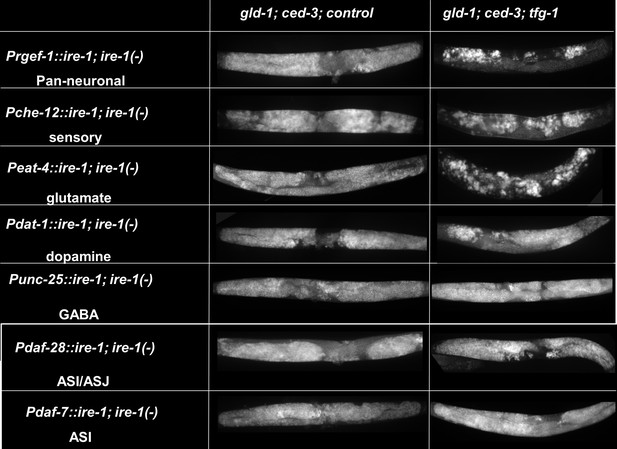
Endoplasmic reticulum (ER) stress-induced germline ectopic differentiation (GED) is controlled by sensory neuronal IRE-1.
Representative micrographs of whole-body (×100) DAPI stained day 4 ire-1(-) transgenic animals, each expressing an ire-1-rescuing transgene driven by the indicated promoters, treated with either a mixture of control, gld-1, and ced-3 RNAi or with a mixture of tfg-1, gld-1, and ced-3 RNAi (n = 210 gonads per genotype, N = 4). See Figure 2A for quantification.
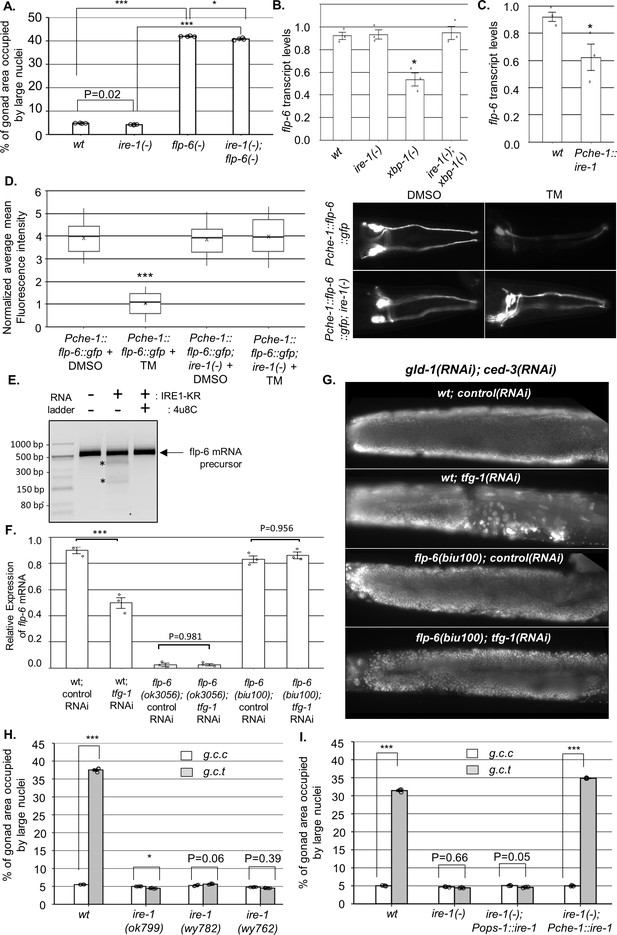
IRE-1 acts upstream of flp-6 and controls its transcript stability.
(A) flp-6 ok3056 mutation increased the levels of ectopic somatic cells in the gonads upon treatment with gld-1 and ced-3 RNAi independently of ire-1 (n = 200 gonads per genotype, N = 4). The ire-1 ok799 deletion mutant was examined. Asterisks mark one-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001 as indicated. Germline ectopic differentiation (GED) levels were assessed by DAPI staining of day 4 animals. Consistent with the lack of dependency on ire-1 for GED induction, flp-6 deficiency did not induce expression of the Phsp-4::gfp endoplasmic reticulum (ER) stress reporter throughout the body or within the ASE neuron (see Figure 3—figure supplement 1). (B, C) flp-6 transcript levels were assessed by qRT-PCR and normalized to actin transcript levels. flp-6 transcript levels were significantly reduced in xbp-1 mutants compared to wild-type animals but not in ire-1 mutants. flp-6 transcript levels were also did not significantly change in ire-1; xbp-1 mutants compared to wild-type animals. Asterisks mark one-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001 compared to wild-type animals (B). flp-6 transcript levels were also reduced upon treatment with tfg-1 RNAi (see Figure 3—figure supplement 2A). flp-6 transcript levels were significantly reduced in Pche-1::ire-1 animals, overexpressing ire-1 in the ASE neurons compared to wild-type animals. Asterisks indicate Student’s t-test p-value of <0.05 (C). (D) L4-staged animals were treated with either 25 µg/ml tunicamycin (TM) or with DMSO till day 2 of adulthood and then scored. TM treatment significantly reduced the fluorescence levels of the translation reporter Pche-1::flp-6::gfp compared to DMSO treatment, whereas it did not significantly change the fluorescence levels of Pche-1::flp-6::gfp; ire-1(-) animals compared to DMSO. n = 65 gonads per genotype, N = 2. p<0.001 of one-way nested ANOVA followed by Tukey’s post hoc analysis is indicated. Means represented by ‘X.’ tfg-1 RNAi treatment also reduced the fluorescence levels of the translation reporter Pche-1::flp-6::gfp (see Figure 3—figure supplement 2C). In contrast, the fluorescence levels of the Pflp-6::gfp and Pche-1::mcherry transcriptional reporters were not significantly altered by tfg-1 RNAi or TM treatment (see Figure 3—figure supplement 2B,D,E). (E) Purified recombinant human IRE-1α comprising the kinase and RNase domains (IRE1-KR) was incubated with in vitro-transcribed flp-6 RNA fragment in the presence of vehicle or 4µ8c (5 µM). Cleavage products of the flp-6 RNA fragment (marked by asterisks) were observed upon its incubation with IRE1-KR, but not in the presence of the specific IRE-1 ribonuclease inhibitor 4μ8C. Cleavage products of the flp-6 RNA fragment were not observed upon scrambling the predicted hairpin sequence (see Figure 3—figure supplement 3B). (F) flp-6 transcript levels were assessed by qRT-PCR and normalized to actin transcript levels (N = 3). flp-6 transcript levels were significantly reduced by tfg-1 RNAi treatment, but not in flp-6(biu100) mutants, in which the putative stem-loop structure has been disrupted while preserving the coding sequence. Negligible levels of flp-6 transcript were detected in flp-6(ok3056) deletion mutants. Asterisks mark nested one-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001 compared to the corresponding control RNAi treatment. (G) Representative micrographs of whole-body DAPI stained day 4 animals treated with either a mixture of control, gld-1, and ced-3 RNAi or with a mixture of tfg-1, gld-1, and ced-3 RNAi. tfg-1 RNAi treatment increased the levels of ectopic somatic cells in the gonads of animals with a wild-type flp-6 transcript (wt) but not in animals with a stem-loop disrupted flp-6 transcript (biu100) (see Figure 3—figure supplement 3C for quantifications). (H) Treatment with a mixture of gld-1; ced-3; tfg-1 RNAi (g.c.t) failed to increase the levels of ectopic somatic cells in the gonads in the absence of functional ire-1 (n = 210 gonads per genotype, N = 5). ok799 is an ire-1 deletion mutation. wy762 is an IRE-1 endoribonuclease missense mutation. wy782 is an IRE-1 kinase missense mutation. Asterisks mark two-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 as indicated. (I) Rescue of ire-1 expression in the ASE neuron (Pche-1::ire-1) restored ER stress-induced GED upon treatment with a mixture of gld-1; ced-3; tfg-1 RNAi (g.c.t), whereas expression of ire-1 in the ASG neuron (Pops-1::ire-1) did not (n = 110 gonads per genotype, N = 3). g.c.c indicates treatment with a mixture of gld-1; ced-3; control RNAi. Asterisks mark two-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 relative to the same animals treated with gld-1; ced-3; control RNAi (g.c.c). In all panels, triple asterisks mark significant results resulting in a twofold change or more.
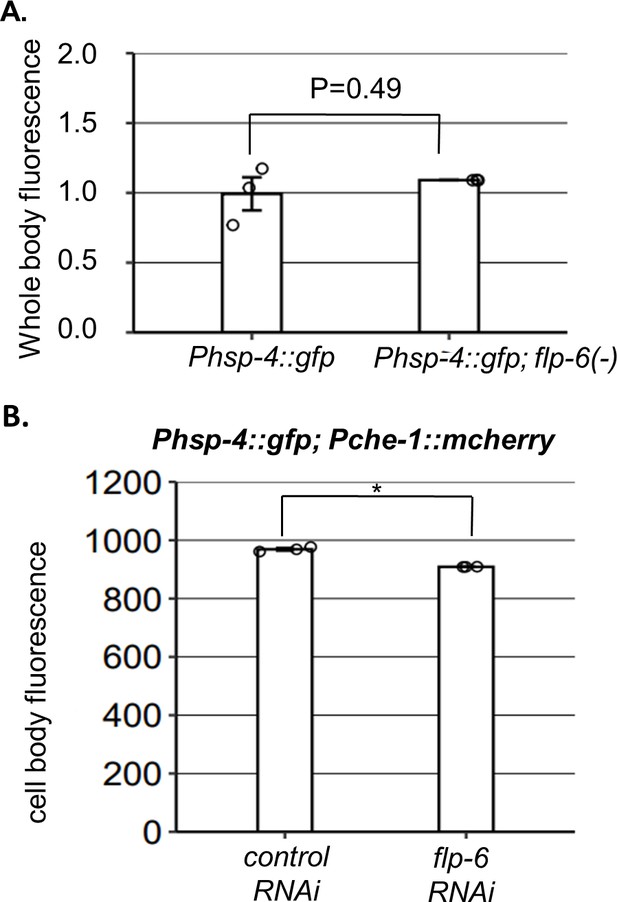
Phsp-4 expression is not increased by flp-6 deficiency.
(A) The whole-body fluorescence levels of the Phsp-4::gfp IRE-1 reporter were comparable in flp-6(ok3056) and wild-type flp-6(+) day 1 animals (Student’s t-test p=0.49, n = 130 gonads per genotype, N = 3). (B) The fluorescence levels of the Phsp-4::GFP endoplasmic reticulum (ER) stress reporter in the ASE neuron were not increased and were even slightly reduced by flp-6 RNAi treatment (Student’s t-test p=0.0056, n = 95 animals per genotype, N = 3). To measure the reporter’s fluorescence specifically in ASE, the selected areas for fluorescence measurements were manually selected based on their overlap with the Pche-1::mcherry marker.
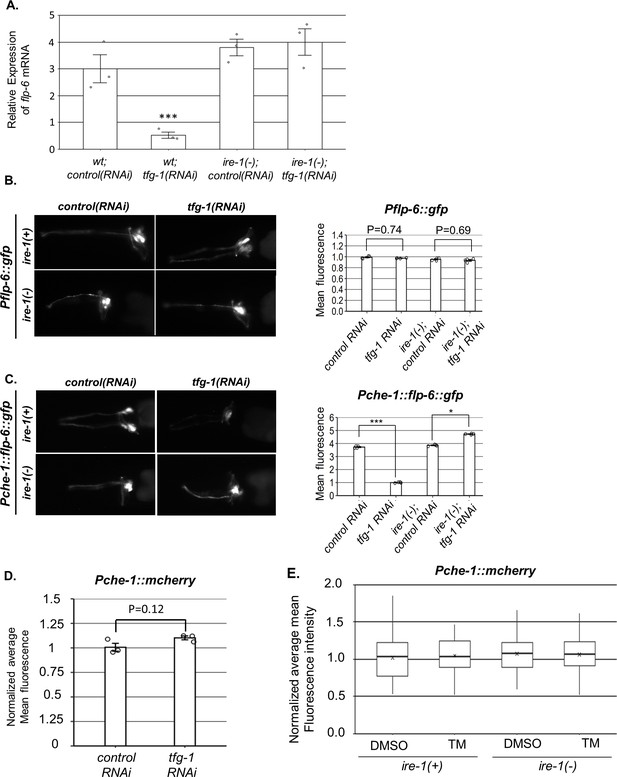
ire-1 regulates flp-6 transcript levels post-transcriptionally.
(A) flp-6 transcript levels were assessed by qRT-PCR and normalized to actin transcript levels (N = 3). Endoplasmic reticulum (ER) stress induced by tfg-1 RNAi reduced flp-6 transcript levels in wild-type animals but not in ire-1 mutants. Triple asterisks mark one-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 relative to the corresponding animals treated with control RNAi. (B) tfg-1 RNAi treatment did not change the fluorescence levels of the Pflp-6::gfp transcriptional reporter (n = 205 gonads per genotype, N = 4). p-Values of one-way ANOVA followed by Tukey’s post hoc analysis are indicated. (C) tfg-1 RNAi treatment reduced the levels of the Pche-1::flp-6::gfp translational reporter in ire-1(+) animals, but not in ire-1(-) animals. Asterisks mark one-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 as indicated (n = 205 gonads per genotype, N = 4). (D) The fluorescence levels of the Pche-1::mcherry transcriptional reporter were not significantly altered by tfg-1 RNAi treatment (Student’s t-test p=0.1258, n = 120 animals per genotype, N = 3). (E) L4-staged animals were treated with either 25 µg/ml tunicamycin (TM) or with DMSO till day 2 of adulthood and then scored. Normalized average mean fluorescence levels of the Pche-1::mcherry transcriptional reporter were not significantly altered by TM treatment compared to DMSO. ire-1 mutated background did not significantly change on either TM or DMSO. Nested one-way ANOVA followed by Tukey’s post hoc analysis indicated no significant difference between groups. Means represented by ‘X.’ n = 60 animals per genotype, N = 2.
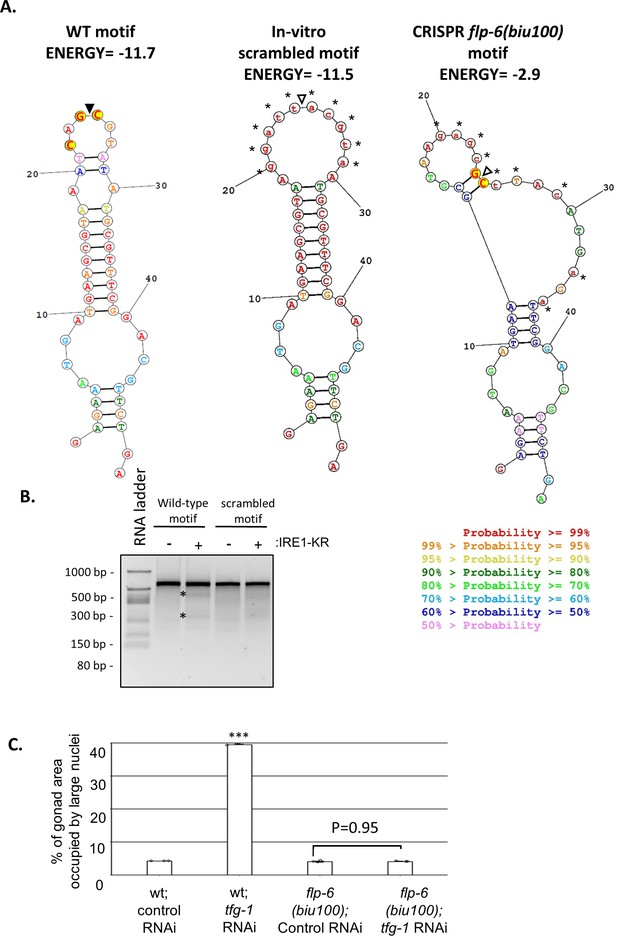
The flp-6 hairpin motif is required for ER stress-induced GED in gld-1; ced-3 deficient animals.
(A) The lowest free energy predicted secondary RNA structure for the given fragment of the flp-6 transcript sequence as calculated by the RNA structure web server (Reuter and Mathews, 2010), before and after scrambling. The conserved residues in the nucleotide loop of putative RIDD substrates (Moore and Hollien, 2015) are highlighted in yellow. Black triangle marks putative cleavage site by IRE1 based on the wild-type transcript sequence. In the scrambled in vitro cleavage substrate, the loop structure has been extended and the conserved nucleotides within the loop have been altered. In the scrambled CRISPR cleavage substrate, corresponding to the biu100 mutation, the entire stem-loop structure has been disrupted using silent mutations to preserve the coding sequence. (B) Purified recombinant human IRE-1α comprising the kinase and RNase domains (IRE1-KR) was incubated with wild-type or scrambled in vitro-transcribed flp-6 RNA fragments. Cleavage products of the flp-6 RNA fragment (marked by asterisks) were observed upon the incubation of the wild-type transcript with IRE1-KR. These fragments were not observed upon incubation of the in vitro-scrambled flp-6 transcript with IRE1-KR. (C) Treatment with a mixture of gld-1; ced-3; tfg-1 RNAi (g.c.t) failed to increase the levels of ectopic somatic cells in the gonads of hairpin-mutated flp-6(biu100) animals (n = 80 gonads per genotype, N = 3). See (A) for the description of the biu100 mutation. Asterisks mark one-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 compared to the corresponding control RNAi treatment. Triple asterisks mark significant results resulting in a twofold change or more.
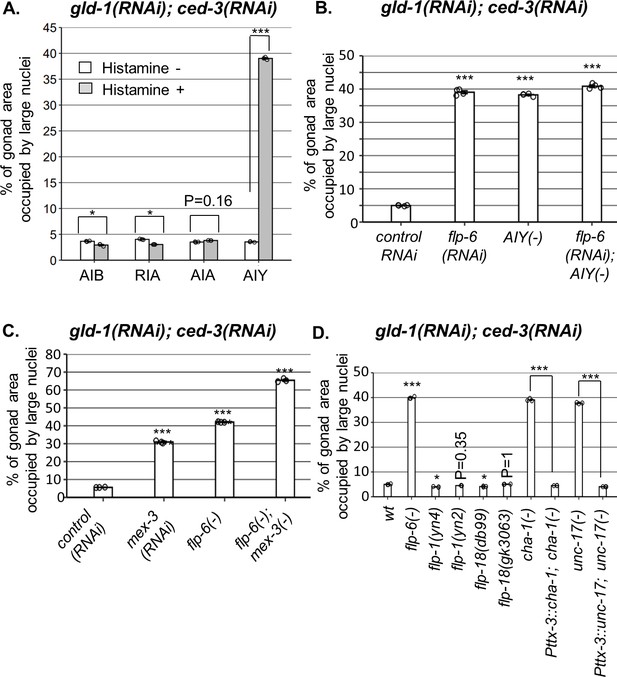
AIY produces acetylcholine (ACh) to prevent germline ectopic differentiation (GED).
Percent of gonad area occupied by ectopic somatic cells determined by DAPI staining of day 4 animals treated with a mixture of gld-1 and ced-3 RNAi. (A) Four histamine-inducible interneurons silencing mutants (AIA, AIB, AIY, RIA) were examined (n = 170 gonads per genotype, N = 3). While AIA, AIB, and RIA histamine-treated animals exhibited low levels of ectopic somatic cells in the gonads, AIY histamine-treated animals had high GED levels. Asterisks mark two-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 relative to the same animals without histamine treatment. (B) flp-6 RNAi did not additively increase GED levels in AIY histamine-treated animals (n = 180 gonads per genotype, N = 4). p-Values were determined by one-way ANOVA followed by Tukey’s post hoc analysis. All strains were grown in the presence of histamine. (C) Animals deficient in both flp-6(ok3056) and mex-3 additively increased GED (n = 215 gonads per genotype, N = 3). Asterisks mark one-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 relative to animals treated with the gld-1, ced-3, control RNAi mix. (D) cha-1(p1152) and unc-17(e113) mutants had high levels of ectopic somatic cells upon gld-1 and ced-3 RNAi treatment (n = 190 gonads per genotype, N = 3). These were suppressed upon expression of the corresponding transgenes in the AIY neuron (Pttx-3::cha-1 and Pttx-3::unc-17). Asterisks mark one-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 relative to wild-type animals unless indicated otherwise. In all panels, triple asterisks mark significant results resulting in a twofold change or more.
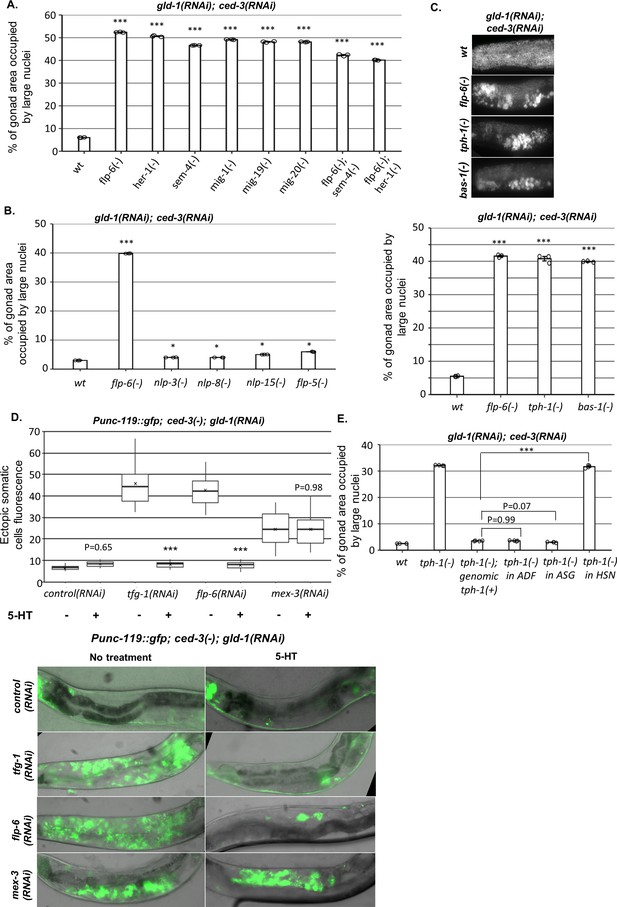
HSN produces serotonin to prevent germline ectopic differentiation (GED).
(A–C) Gonad area occupied by ectopic somatic cells determined by DAPI staining of day 4 animals treated with a mixture of gld-1 and ced-3 RNAi. All HSN deficiency and migration mutants had high levels of ectopic somatic cells in their gonads. flp-6 deletion did not further increase GED levels in HSN-defective animals (n = 175 gonads per genotype, N = 3) (A). HSN-related neuropeptide mutants had low levels of GED, similar to wild-type animals (n = 200 gonads per genotype, N = 3) (B). Both tph-1(mg280) and bas-1(tm351) serotonin mutants had high levels of ectopic somatic cells in their gonads, similarly to flp-6 mutants (n = 185 gonads per genotype, N = 3) (C). See Figure 5—figure supplement 1 for the analysis of serotonin receptor mutants. (D) Treatment with 20 mM serotonin (5-HT) suppressed the high ectopic somatic fluorescence of the neuronal reporter Punc-119::gfp in the gonads of gld-1(RNAi); tfg-1(RNAi) or gld-1(RNAi); flp-6(RNAi)-treated ced-3 mutants to the same extent as in the control RNAi group. However, treatment with serotonin (5-HT) did not suppress the ectopic somatic fluorescence of the neuronal reporter in gld-1(RNAi); mex-3(RNAi)-treated animals. Representative fluorescent images are shown. n = 90 gonads per genotype, N = 2. Asterisks mark nested one-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 relative to the same animals without serotonin supplementation. Means represented by ‘X.’ (E) tph-1 deficiency in HSN, but not in other serotonin-producing neurons, resulted in high levels of ectopic somatic-like cells in the gonads (n = 195 gonads per genotype, N = 4). In (A–C) and (E), asterisks mark one-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 relative to wild-type animals treated with gld-1 and ced-3 RNAi, unless indicated otherwise. In all panels, triple asterisks mark significant results resulting in a twofold change or more.
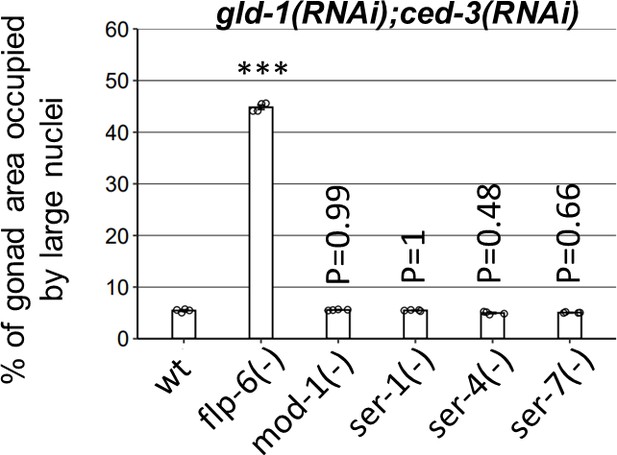
Four major serotonin receptor mutants are not required for germline ectopic differentiation (GED) in gld-1; ced-3-deficient animals.
Percentage of gonad area occupied by DAPI-labeled ectopic cells in day 4 animals treated with gld-1 and ced-3 RNAi (n = 140 gonads per genotype, N = 4). Asterisks mark one-way ANOVA values followed by Tukey’s post hoc analysis of p<0.001 relative to wild-type animals treated with gld-1 and ced-3 RNAi. mod-1, ser-1, ser-4, and ser-7 deficiencies were obtained by ok103, ok345, ok512, ok1944 mutations, respectively. Triple asterisks mark significant results resulting in a twofold change or more.
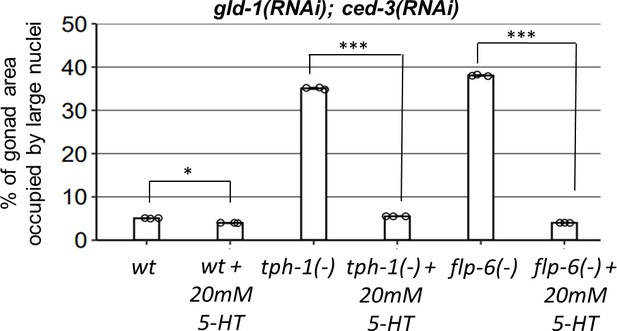
Treatment with 20 mM serotonin (5-HT) decreased the levels of abnormal nuclei in the gonads of gld-1 and ced-3 RNAi-treated tph-1(mg280) and flp-6(ok3056) mutants (n = 165 gonads per genotype, N = 3).
Asterisks mark one-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 relative to the same animals without serotonin supplementation.
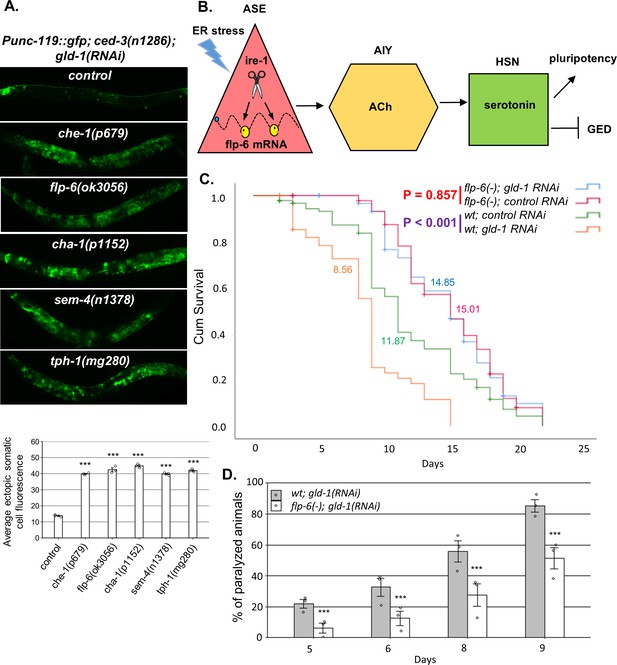
A neuronal circuit actively prevents germline ectopic differentiation (GED).
(A) Abnormal expression of the neuronal reporter Punc-119::gfp within the tumorous gonads of gld-1 RNAi-treated animals with a perturbed ASE-AIY-HSN-germline circuit compared to animals with an intact circuit. che-1 and sem-4 mutants are ASE and HSN-deficient, respectively. flp-6 mutants are deficient in the FLP-6 neuropeptide. cha-1 and tph-1 mutants are compromised in acetylcholine (Ach) and serotonin production. Asterisks mark one-way ANOVA followed by Tukey’s post hoc analysis of p<0.001 relative to control ced-3(-) animals treated with gld-1 RNAi. Triple asterisks mark significant results resulting in a twofold change or more. (B) Summarizing model of ASE-AIY-HSN-germline circuit which actively maintains germline pluripotency and suppresses ectopic germline differentiation in gld-1 tumorous animals. Implicated neurons and signaling molecules are indicated. Endoplasmic reticulum (ER) stress suppresses this circuit by regulated Ire1-dependent decay (RIDD)-mediated destabilization of the flp-6 transcript in the ASE neuron. This releases the inhibition from the tumorous germ cells that acquire somatic fate by default. (C) Lifespan analysis of wild-type animals and flp-6 mutants treated with either gld-1 RNAi to induce tumor formation or with control RNAi. Lifespan shortening was significantly suppressed in flp-6 mutants treated with gld-1 RNAi. Mean lifespan is indicated within each graph. Log rank (Mantel–Cox) p<0.001 between wild-type animals treated with either gld-1 RNAi or with control RNAi is indicated in purple. Log rank (Mantel–Cox) p=0.857 between flp-6 mutants treated with either gld-1 RNAi or with control RNAi is indicated in red. See Supplementary file 2 for statistical data of two additional replicates. (D) Paralysis assay in wild-type animals and flp-6 mutants treated with gld-1 RNAi. 90 synchronized adult animals per genotype were placed on gld-1 RNAi plates and their paralysis was scored on days 5, 6, 8, and 9. Bar graphs present percentage of paralyzed animals. At all timepoints, the flp-6 deficiency resulted in a significant decrease in the paralysis of the tumorous animals in comparison to wild-type animals. Proportions of paralyzed animals were compared between different genotypes in three replicates using the Cochran–Mantel–Haenszel test. Triple asterisks mark significant reduction of at least twofold change in the paralysis relative to wild-type animals treated with gld-1 RNAi. Note that in the lifespan and paralysis, experiments shown in (C) and (D) were done in ced-3(+) background to allow the apoptosis-mediated clearance of the ER stress-induced aberrant somatic cells in the gonad.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Caenorhabditiselegans) | N2 | Caenorhabditis Genetics Center | Wild type | |
| Strain, strain background (C. elegans) | SHK124 | This paper | rrf-1(pk1417) I | x4 outcrosses, strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | CF2473 | Cynthia Kenyon lab | ire-1(ok799) II | x3 outcrosses, strain created in C. Kenyon lab |
| Strain, strain background (C. elegans) | SHK27 | Levi-Ferber et al., 2015 PMID:25340700 | ire-1(ok799) II; biuEx2[pRF4(rol-6(su1006)); Pire-1::ire-1(cDNA)]; svls69[Pdaf-28::daf-28::gfp] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK185 | Levi-Ferber et al., 2014 PMID:25340700 | ire-1(ok799) II; biuEx49[pRF4(rol-6(su1006)); Prgef-1::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK14 | Levi-Ferber et al., 2014 PMID:25340700 | ire-1(ok799) II; biuEx5[pRF4(rol-6(su1006)); Pmyo-3::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK703 | This paper | ire-1(ok799) II wdIs52 [F49H12.4::GFP + unc-119(+)]; biuEx50[pRF4(rol-6(su1006)); Pges-1::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK4 | Levi-Ferber et al., 2014 PMID:25340700 | biuEx2[pRF4(rol-6(su1006)); Pire-1::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK182 | Levi-Ferber et al., 2014 PMID:25340700 | biuEx49[pRF4(rol-6(su1006)); Prgef-1::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK8 | Levi-Ferber et al., 2014 PMID:25340700 | biuEx5[pRF4(rol-6(su1006)); Pmyo-3::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK282 | This paper | biuEx50[pRF4(rol-6(su1006)); Pges-1::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK268 | This paper | ire-1(ok799) II; biuEx11[Pgrd-10::gfp; Pche-12::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK271 | This paper | ire-1(ok799) II; biuEx14[Pgrd-10::gfp; Peat-4::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK270 | This paper | ire-1(ok799) II; biuEx13[Pgrd-10::gfp; Pdat-1::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK266 | This paper | ire-1(ok799) II; biuEx10[Pgrd-10::gfp; Punc-25::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK15 | Levi-Ferber et al., 2014 PMID:25340700 | ire-1(ok799) II; biuEx4[pRF4(rol-6(su1006)); Pdaf-28::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK256 | This paper | ire-1(ok799) II; biuEx51[pRF4(rol-6(su1006)); Pdaf-7::gfp; Pdaf-7::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK592 | This paper | ire-1(ok799) II; biuEx57[pRF4(rol-6(su1006)); Pche-1::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK593 | This paper | ire-1(ok799) II; biuEx58[pRF4(rol-6(su1006)); Pops-1::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | OH10434 | Caenorhabditis Genetics Center | otls232 [che-1p::mcherry(C. elegans-optimized)::che-1 3'UTR + rol-6(su1006)] | |
| Strain, strain background (C. elegans) | DA572 | Caenorhabditis Genetics Center | eat-4(ad572) | |
| Strain, strain background (C. elegans) | MT6308 | Caenorhabditis Genetics Center | eat-4(ky5) | |
| Strain, strain background (C. elegans) | NY119 | Chris Li lab | flp-4(yn35) II | |
| Strain, strain background (C. elegans) | VC2324 | Caenorhabditis Genetics Center | flp-6(ok3056) V | |
| Strain, strain background (C. elegans) | FX02427 | Dr. Shohei Mitani, National Bioresource Project for the nematode, Japan | flp-13(tm2427) IV | |
| Strain, strain background (C. elegans) | PT505 | Caenorhabditis Genetics Center | flp-20(pk1596) X | |
| Strain, strain background (C. elegans) | VC1982 | Caenorhabditis Genetics Center | flp-25(gk1016) III | |
| Strain, strain background (C. elegans) | FX03023 | Dr. Shohei Mitani, National Bioresource Project for the nematode, Japan | nlp-3(tm3023) X | |
| Strain, strain background (C. elegans) | RB1609 | Caenorhabditis Genetics Center | nlp-5(ok1981) II | |
| Strain, strain background (C. elegans) | FX1880 | Dr. Shohei Mitani, National Bioresource Project for the nematode, Japan | nlp-14(tm1880) X | |
| Strain, strain background (C. elegans) | FX1888 | Caenorhabditis Genetics Center | ins-1(tm1888) IV | |
| Strain, strain background (C. elegans) | RB2594 | Caenorhabditis Genetics Center | ins-22(ok3616) III | |
| Strain, strain background (C. elegans) | FX14756 | Dr. Shohei Mitani, National Bioresource Project for the nematode, Japan | ins-26(tm1983) I | |
| Strain, strain background (C. elegans) | FX06109 | Caenorhabditis Genetics Center | ins-32(tm6109) II | |
| Strain, strain background (C. elegans) | RB982 | Caenorhabditis Genetics Center | flp-21(ok889) V | |
| Strain, strain background (C. elegans) | RB1340 | Caenorhabditis Genetics Center | nlp-1(ok1469) X | |
| Strain, strain background (C. elegans) | FX02984 | Dr. Shohei Mitani, National Bioresource Project for the nematode, Japan | nlp-7(tm2984) X | |
| Strain, strain background (C. elegans) | VC1309 | Caenorhabditis Genetics Center | nlp-8(ok1799) I | |
| Strain, strain background (C. elegans) | FX06232 | Dr. Shohei Mitani, National Bioresource Project for the nematode, Japan | nlp-10(tm6232) III | |
| Strain, strain background (C. elegans) | VC1063 | Caenorhabditis Genetics Center | nlp-15(ok1512) I | |
| Strain, strain background (C. elegans) | SHK497 | This paper | biuEx52[pRF4(rol-6(su1006)); Pche-1::flp-6(cDNA)]; flp-6(ok3056) V | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK498 | This paper | biuEx53[pRF4(rol-6(su1006)); Psrh-142::flp-6]; flp-6(ok3056) V | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | PR672 | Caenorhabditis Genetics Center | che-1(p672) I | |
| Strain, strain background (C. elegans) | PR674 | Caenorhabditis Genetics Center | che-1(p674) I | |
| Strain, strain background (C. elegans) | MT633 | Caenorhabditis Genetics Center | lin-11(n389) I; him-5(e1467)V | |
| Strain, strain background (C. elegans) | MT1196 | Caenorhabditis Genetics Center | lin-11(n566) I | |
| Strain, strain background (C. elegans) | CF2260 | Cynthia Kenyon lab | Zcls4[Phsp-4::gfp] V | |
| Strain, strain background (C. elegans) | SHK314 | This paper | Zcls4[Phsp-4::gfp] V; flp-6(ok3056) V | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK315 | This paper | ire-1(ok799) II, flp-6(ok3056) V | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | NY2067 | Caenorhabditis Genetics Center | ynIs67[Pflp-6::gfp] III; him-5(e1490) V | |
| Strain, strain background (C. elegans) | SHK403 | This paper | ynIs67[Pflp-6::gfp] III; ire-1(ok799) II | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK474 | This paper | biuEx54[pRF4(rol-6(su1006)); Pche-1::flp-6(cDNA)::gfp] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK491 | This paper | biuEx52[pRF4(rol-6(su1006)); Pche-1::flp-6(cDNA)]; ire-1(ok799) II | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | TV13656 | Kang Shen lab | ire-1(wy782) II | |
| Strain, strain background (C. elegans) | TV13763 | Kang Shen lab | ire-1(wy762) II | |
| Strain, strain background (C. elegans) | NY7 | Caenorhabditis Genetics Center | flp-1(yn2) IV | |
| Strain, strain background (C. elegans) | NY16 | Caenorhabditis Genetics Center | flp-1(yn4) IV | |
| Strain, strain background (C. elegans) | AX1410 | Caenorhabditis Genetics Center | flp-8(db99) X | |
| Strain, strain background (C. elegans) | VC2016 | Caenorhabditis Genetics Center | flp-18(gk3063) X | |
| Strain, strain background (C. elegans) | PR1152 | Caenorhabditis Genetics Center | cha-1(p1152) IV | |
| Strain, strain background (C. elegans) | SHK605 | This paper | biuEx55[pRF4(rol-6(su1006)); Pttx-3::cha-1]; cha-1(p1152) IV | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | GG201 | Caenorhabditis Genetics Center | ace-2(g72) I; ace-1(p1000) X | |
| Strain, strain background (C. elegans) | PR1300 | Caenorhabditis Genetics Center | ace-3(dc2) II | |
| Strain, strain background (C. elegans) | CB113 | Caenorhabditis Genetics Center | unc-17(e113) IV | |
| Strain, strain background (C. elegans) | SHK586 | This paper | biuEx56[pRF4(rol-6(su1006)); Pttx-3::unc-17]; unc-17(e113) IV | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | MT9668 | Caenorhabditis Genetics Center | mod-1(ok103) V | |
| Strain, strain background (C. elegans) | DA1814 | Caenorhabditis Genetics Center | ser-1(ok345) X | |
| Strain, strain background (C. elegans) | AQ866 | Caenorhabditis Genetics Center | ser-4 (ok512) III | |
| Strain, strain background (C. elegans) | RB1585 | Caenorhabditis Genetics Center | ser-7(ok1944) X | |
| Strain, strain background (C. elegans) | MT1446 | Caenorhabditis Genetics Center | her-1(n695) V | |
| Strain, strain background (C. elegans) | MT5825 | Caenorhabditis Genetics Center | sem-4(n1378) I | |
| Strain, strain background (C. elegans) | MT3969 | Caenorhabditis Genetics Center | mig-1(n1652) I | |
| Strain, strain background (C. elegans) | NF69 | Caenorhabditis Genetics Center | mig-19(k142) II | |
| Strain, strain background (C. elegans) | NF78 | Caenorhabditis Genetics Center | mig-20(k148) X | |
| Strain, strain background (C. elegans) | SHK492 | This paper | flp-6(ok3056) V; sem-4(n1378) I | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK490 | This paper | flp-6(ok3056) V; her-1(n695) V | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | FX30280 | Dr. Shohei Mitani, National Bioresource Project for the nematode, Japan | flp-5(tm10075) X | |
| Strain, strain background (C. elegans) | LC33 | Dr. Shohei Mitani, National Bioresource Project for the nematode, Japan | bas-1(tm351) III | |
| Strain, strain background (C. elegans) | CX13228 | Cori Bargman Lab | tph-1(mg280) II; kySi56[ tph-1 genomic rescue] IV | Strain created in C. Bargman Lab |
| Strain, strain background (C. elegans) | CX13576 | Cori Bargman Lab | tph-1(mg280) II; kySi56[ tph-1 genomic rescue] IV; kyEx4107[egl6::nCre] | Strain created in C. Bargman Lab |
| Strain, strain background (C. elegans) | CX13571 | Cori Bargman Lab | tph-1(mg280) II; kySi56[ tph-1 genomic rescue] IV; kyEx4077[srh142::nCre] | Strain created in C. Bargman Lab |
| Strain, strain background (C. elegans) | CX13574 | Cori Bargman Lab | tph-1(mg280) II; kySi56[ tph-1 genomic rescue] IV; kyEx4081[ops1::nCre] | Strain created in C. Bargman Lab |
| Strain, strain background (C. elegans) | CX14909 | Cori Bargman Lab | kyEx4925 [ttx-3::hisCl1*::sl2::GFP; myo-3::mCherry] | Strain created in C. Bargman Lab |
| Strain, strain background (C. elegans) | CX14849 | Cori Bargman Lab | kyEx4867 [ins-1::HisCl1::sl2mCherry; unc-122::GFP] | Strain created in C. Bargman Lab |
| Strain, strain background (C. elegans) | CX14908 | Cori Bargman Lab | kyEx4924 [inx1::hisCl1*::sl2::GFP; myo-3::mCherry] | Strain created in C. Bargman Lab |
| Strain, strain background (C. elegans) | CX16069 | Cori Bargman Lab | kyEx5493 [pNP443 (glr3::HisCl1::SL2::mCherry); elt-2:mCherry] | Strain created in C. Bargman Lab |
| Strain, strain background (C. elegans) | DP132 | Caenorhabditis Genetics Center | edIs6 (punc-119::GFP) IV | |
| Strain, strain background (C. elegans) | SHK659 | This paper | che-1(p679) I; edIs6 (punc-119::GFP) IV | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK361 | This paper | edIs6 (punc-119::GFP) IV; flp-6(ok3056) V | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK660 | This paper | cha-1(p1152) edIs6 (punc-119::GFP) IV | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK661 | This paper | sem-4(n1378) I; edIs6 (punc-119::GFP) IV | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK662 | This paper | tph-1(mg280) II; edls6 (punc-119::GFP) IV | Strain created in S. Henis-Korenblit lab |
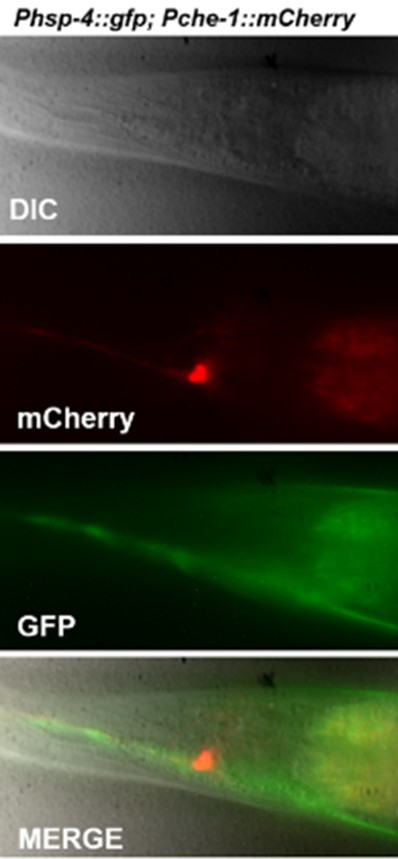
| Strain, strain background (C. elegans) | SHK663 | This paper | Zcls4[Phsp-4::gfp] V; otls232 [che-1p::mcherry(C. elegans-optimized)::che-1 3'UTR + rol-6(su1006)] | |
| Strain, strain background (C. elegans) | CF3208 | Cynthia Kenyon lab | xbp-1(tm2457) III | X4 outcrosses |
| Strain, strain background (C. elegans) | SHK62 | Levi-Ferber et al., 2014 PMID:25340700 | xbp-1(tm2457) III; ire-1(ok799) II | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK152 | Levi-Ferber et al., 2015 PMID:26192965 | edIs6 (punc-119::GFP) IV; ced-3(n1286) IV | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK86 | Levi-Ferber et al., 2015 PMID:26192965 | Pmyo-3::GFP; ced-3(n1286) | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK697 | This paper | flp-6(biu100) | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK698 | This paper | ire-1(ok799) II; otls232 [che-1p::mcherry(C. elegans-optimized)::che-1 3'UTR + rol-6(su1006)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK699 | This paper | edIs6 (punc-119::GFP) IV; ced-3(n1286) IV; biuEx49[pRF4(rol-6(su1006)); Prgef-1::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Strain, strain background (C. elegans) | SHK584 | This paper | biuEx57[pRF4(rol-6(su1006)); Pche-1::ire-1(cDNA)] | Strain created in S. Henis-Korenblit lab |
| Sequence-based reagent | act-1 Fw | Cohen-Berkman et al., 2020 PMID:32213289 | qPCR primer | CCAATCCAAGAGAGGTATCCTTAC |
| Sequence-based reagent | act-1 Bw | Cohen-Berkman et al., 2020 PMID:32213289 | qPCR primer | CATTGTAGAAGGTGTGATGCCAG |
| Sequence-based reagent | flp-6 Fw | This paper | qPCR primer | GTGAAGTGGAGAGAGAAATGATGA |
| Sequence-based reagent | flp-6 Bw | This paper | qPCR primer | CCGCTACTTCTCTTTCCAAAACG |
| Chemical compound, drug | TRIzol | Ambion | 15596026 | |
| Chemical compound, drug | Maxima SYBR GREEN | Thermo Scientific | K0221 | |
| Chemical compound, drug | IPTG | Gold Bio | I2481C | |
| Chemical compound, drug | DAPI | Sigma | D9542 | |
| Chemical compound, drug | Serotonine creatinine sulfate monohydrate | Sigma | H7752 | |
| Chemical compound, drug | Histamine dihydrochloride | Sigma | H7250 | |
| Chemical compound, drug | Levamisol hydrochloride | Sigma | 31742 | |
| Chemical compound, drug | Tunicamycin | Cayman | 11445 | |
| Other | CFX-96 real time system | Bio-Rad | ||
| Software, algorithm | SPSS | SPSS | ||
| Software, algorithm | R statistical environment | R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. | RRID:SCR_001905 | |
| Sequence-based reagent | tracrRNA | IDT | ||
| Sequence-based reagent | cas9 enzyme | IDT | ||
| Sequence-based reagent | dpy-10 crRNA | IDT | GCTACCATAGGCACCACGAG | |
| Sequence-based reagent | flp-6 crRNA | IDT | AAATCAGCGTATATGCGTTT | |
| Sequence-based reagent | dpy-10 ssODN | IDT | CACTTGAACTTCAATACGGCAAGATGAGAATGACTGGAAACCGTACCGCATGCGGTGCCTATGGTAGCGGAGCTTCACATGGCTTCAGACCAACAGCCTAT | |
| Sequence-based reagent | flp-6 (biu100) ssODN | IDT | tcaaaaatatgttttgcagAAATGATGAAGCGTAAgagcGCtTAcATGaGaTTCGGACGTTCTGACGGTGGAAACCCAATGGAAATGGAAA |
Additional files
-
Supplementary file 1
List of sensory neurons included under the che-1 promoter.
- https://cdn.elifesciences.org/articles/65644/elife-65644-supp1-v1.xlsx
-
Supplementary file 2
Statistical data.
- https://cdn.elifesciences.org/articles/65644/elife-65644-supp2-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65644/elife-65644-transrepform1-v1.pdf