Ancient role of sulfakinin/cholecystokinin-type signalling in inhibitory regulation of feeding processes revealed in an echinoderm
Figures
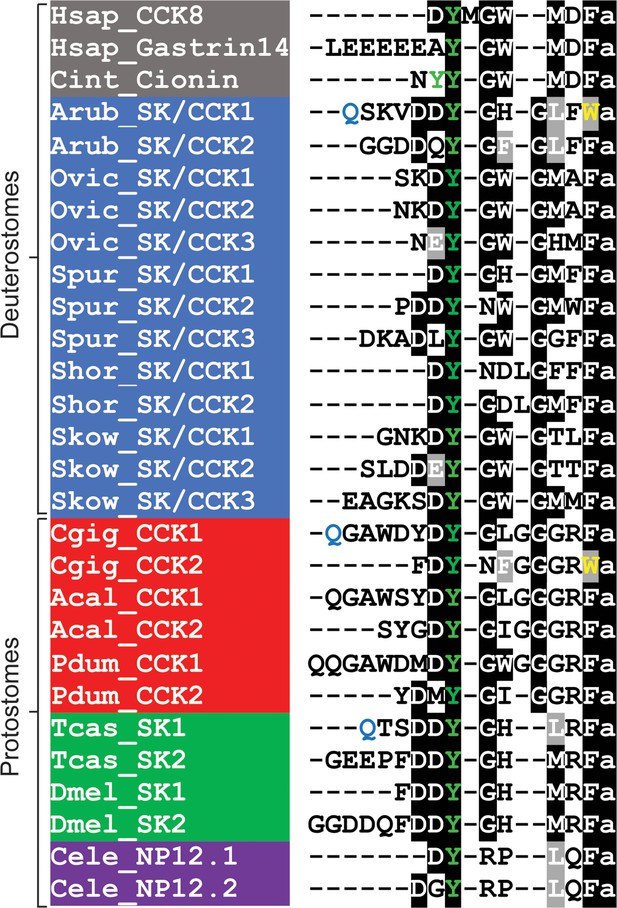
Comparison of the Asterias rubens sulfakinin/cholecystokinin (SK/CCK)-type neuropeptides ArSK/CCK1 and ArSK/CCK2 with SK/CCK-type neuropeptides from other taxa.
Conserved residues are highlighted, with conservation in more than 70% of sequences highlighted in black and conservative substitutions highlighted in grey. Experimentally verified conversion of an N-terminal glutamine residue (Q) to pyroglutamate in the mature peptide is indicated by the letter Q being shown in light blue. Tyrosine (Y) residues that are known or predicted to be subject to post-translational sulphation are shown in green. The C-terminal tryptophan (W) in ArSK/CCK1 and in a Crassostrea gigas SK/CCK-type peptide are shown in yellow to highlight that this feature is atypical of SK/CCK-type peptides. Predicted or experimentally verified C-terminal amides are shown as the letter ‘a’ in lowercase. Species names are highlighted in taxon-specific colours: grey (Chordata), blue (Ambulacraria), red (Lophotrochozoa), green (Arthropoda),and purple (Nematoda). Abbreviations are as follows: Acal (Aplysia californica), Arub (Asterias rubens), Cele (Caenorhabditis elegans), Cgig (Crassostrea gigas), Cint (Ciona intestinalis), Dmel (Drosophila melanogaster), Hsap (Homo sapiens), Ovic (Ophionotus victoriae), Pdum (Platynereis dumerilii), Shor (Stichopus horrens), Skow (Saccoglossus kowalevskii), Spur (Strongylocentrotus purpuratus), Tcas (Tribolium castaneum). The accession numbers of the sequences are listed in Figure 1—source data 1. The nucleotide sequence of a cloned cDNA encoding the A. rubens SK/CCK-type precursor is shown in Figure 1—figure supplement 1. Mass spectroscopic analysis of the structures of the peptides derived from the A. rubens SK/CCK-type precursor is presented in Figure 1—figure supplement 2. The raw data for the results shown in Figure 1—figure supplement 2 can be found in Figure 1—figure supplement 2—source data 1.
-
Figure 1—source data 1
Accession numbers for precursors of the neuropeptides shown in the sequence alignment in Figure 1.
- https://cdn.elifesciences.org/articles/65667/elife-65667-fig1-data1-v1.docx

The Asterias rubens sulfakinin/cholecystokinin (SK/CCK)-type precursor (ArSK/CCKP).
The nucleotide sequence of contig 1124413 derived from radial nerve cord transcriptome data (Semmens et al., 2016; GenBank accession number KT601716) is shown in lowercase (3434 bases) and the encoded precursor protein sequence is shown in uppercase (163 residues). The predicted signal peptide is shown in dark-blue, two putative SK/CCK-type peptides are shown in red but with an N-terminal glutamine residue in first peptide shown light blue to indicate that it is a potential substrate for pyroglutamination. Putative dibasic cleavage sites are shown in green. The asterisk shows the position of the stop codon. The sequences of primers used for PCR cloning of a cDNA encoding ArSK/CCKP are highlighted in yellow. The sequence of the cloned cDNA was found to be identical to the corresponding sequence of contig 1124413.
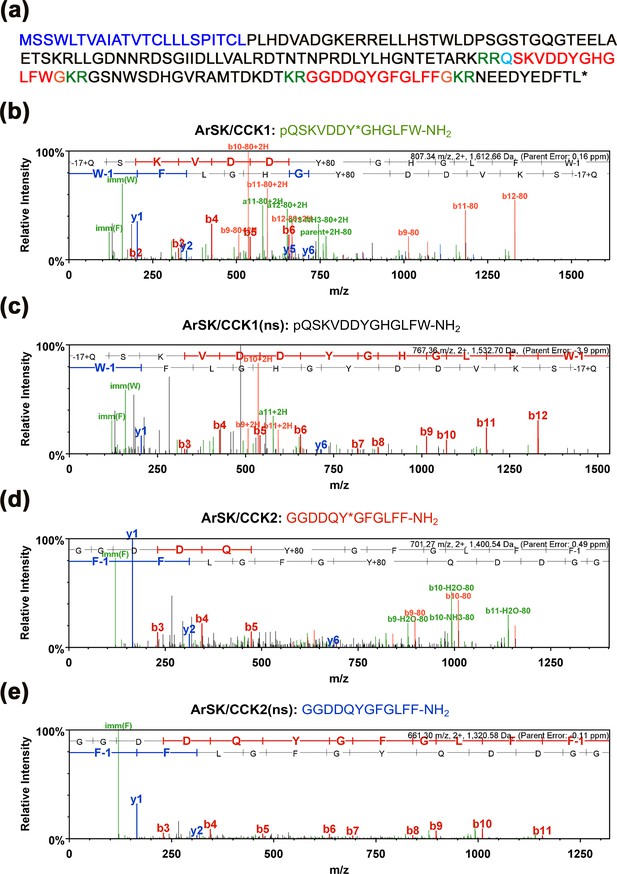
Determination of the structures of peptides derived from ArSK/CCKP by mass spectrometric (LC-MS-MS) analysis of Asterias rubens radial nerve cord extract.
(a) Amino acid sequence of ArSK/CCKP, with the predicted signal peptide shown in dark blue, predicted dibasic cleavage sites shown in green, and predicted sulfakinin/cholecystokinin (SK/CCK)-type neuropeptides shown in red. C-terminal glycine residues that are predicted substrates for amidation are shown in orange and an N-terminal glutamine residue that is a potential substrate for pyroglutamination (pQ) is shown in light blue The nucleotide sequence of a cDNA encoding ArSK/CCKP is shown in Figure 1—figure supplement 1. (b–d) MS/MS data for four SK/CCK-type peptides derived from ArSK/CCKP detected in A. rubens radial nerve cord extracts: (b) pQSKVDDY(SO3H)GHGLFW-NH2 (ArSK/CCK1, which has a sulphated tyrosine), (c) pQSKVDDYGHGLFW-NH2 (ArSK/CCK1(ns), which is not sulphated), (d) GGDDQY(SO3H)GFGLFF-NH2 (ArSK/CCK2, which has a sulphated tyrosine), and (e) GGDDQYGFGLFF-NH2 (ArSK/CCK2(ns), which is not sulphated). The b series of peptide fragment ions are shown in red, the y series are shown in blue, and additional identified peptide fragment ions are shown in green. The amino acid sequence identified in each mass spectrum is shown above it, with –17 + Q representing an N-terminal pyroglutamate residue (pQ), W-1 representing an amidated C-terminal tryptophan residue (W–NH2), F-1 representing an amidated C-terminal phenylalanine residue (F–NH2), and Y + 80 representing a sulphated tyrosine residue (Y*). The observed m/z of the precursor ion for each peptide is with a charge state of +2 and with errors between the experimentally determined and predicted values ranging from –3.9 to +0.49 ppm.
-
Figure 1—figure supplement 2—source data 1
Data for the mass spectra shown in Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/65667/elife-65667-fig1-figsupp2-data1-v1.xlsx
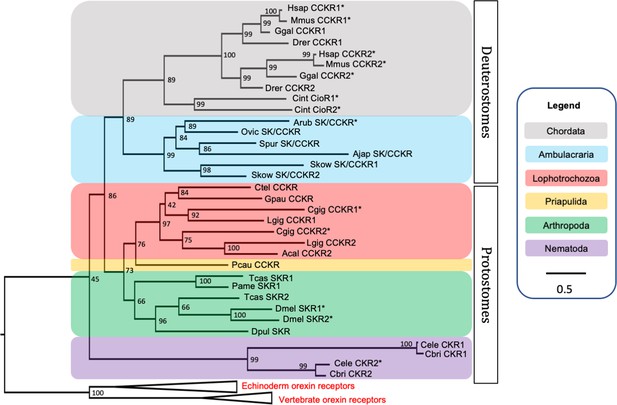
Phylogenetic tree showing that the predicted Asterias rubens sulfakinin/cholecystokinin (SK/CCK)-type receptor (ArSK/CCKR) is an ortholog of SK/CCK-type receptors in other taxa, which include receptors (*) for which the peptide ligands have been identified experimentally.
The tree was generated using the maximum likelihood method (Guindon et al., 2009), with the percentage of replicate trees in which two or more sequences form a clade in a bootstrap test (1000 replicates) shown at the node of each clade (Zhaxybayeva and Gogarten, 2002). Orexin receptors were included as an outgroup. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Evolutionary analyses were conducted using the IQ-tree server (Trifinopoulos et al., 2016). Taxa are colour-coded as explained in the key. Abbreviations of species names are as follows: Acal (Aplysia californica), Ajap (Apostichopus japonicus), Arub (Asterias rubens), Cbri (Caenorhabditis briggsae), Cele (Caenorhabditis elegans), Cgig (Crassostrea gigas), Cint (Ciona intestinalis), Ctel (Capitella teleta), Dmel (Drosophila melanogaster), Dpul (Daphnia pulex), Drer (Danio rerio), Ggal (Gallus gallus), Gpau (Glossoscolex paulistus), Hsap (Homo sapiens), Lgig (Lottia gigantea), Mmus (Mus musculus), Ovic (Ophionotus victoriae), Pame (Periplaneta americana), Pcau (Priapulus caudatus), Skow (Saccoglossus kowalevskii), Spur (Strongylocentrotus purpuratus), Tcas (Tribolium castaneum). The accession numbers of the sequences used for this phylogenetic tree are listed in Figure 2—source data 1. The nucleotide sequence and the predicted topology of ArSK/CCKR are shown in Figure 2—figure supplement 1 and Figure 2—figure supplement 2, respectively.
-
Figure 2—source data 1
Accession numbers for the receptor sequences used for the phylogenetic tree in Figure 2.
- https://cdn.elifesciences.org/articles/65667/elife-65667-fig2-data1-v1.docx

The Asterias rubens sulfakinin/cholecystokinin (SK/CCK)-type receptor (ArSK/CCKR).
The nucleotide sequence of contig 1110296 derived from radial nerve cord transcriptome data (GenBank accession number MW261740) is shown in lowercase (1530 bases; numbering on the left) and the encoded ArSK/CCKR protein sequence is shown in uppercase (434 residues). The start and stop codons are highlighted in yellow. The coding sequence was synthesized (GenScript) to enable testing of the ArSK/CCKP-derived SK/CCK-type peptides as ligands for ArSK/CCKR (see Figure 3).
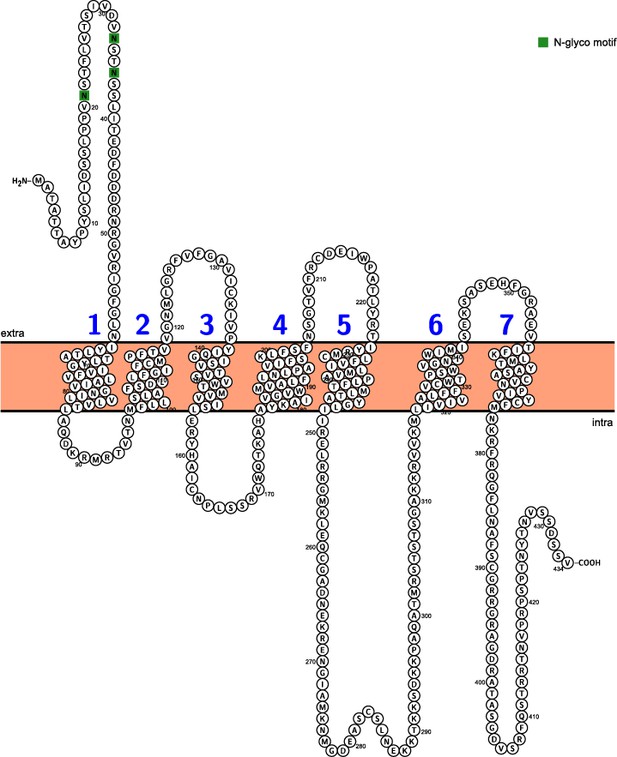
Topology of ArSK/CCKR.
Predicted topology of ArSK/CCKR inferred by the Protter tool (Omasits et al., 2014) with seven transmembrane domains numbered in blue and predicted N-glycosylation sites shown in green.
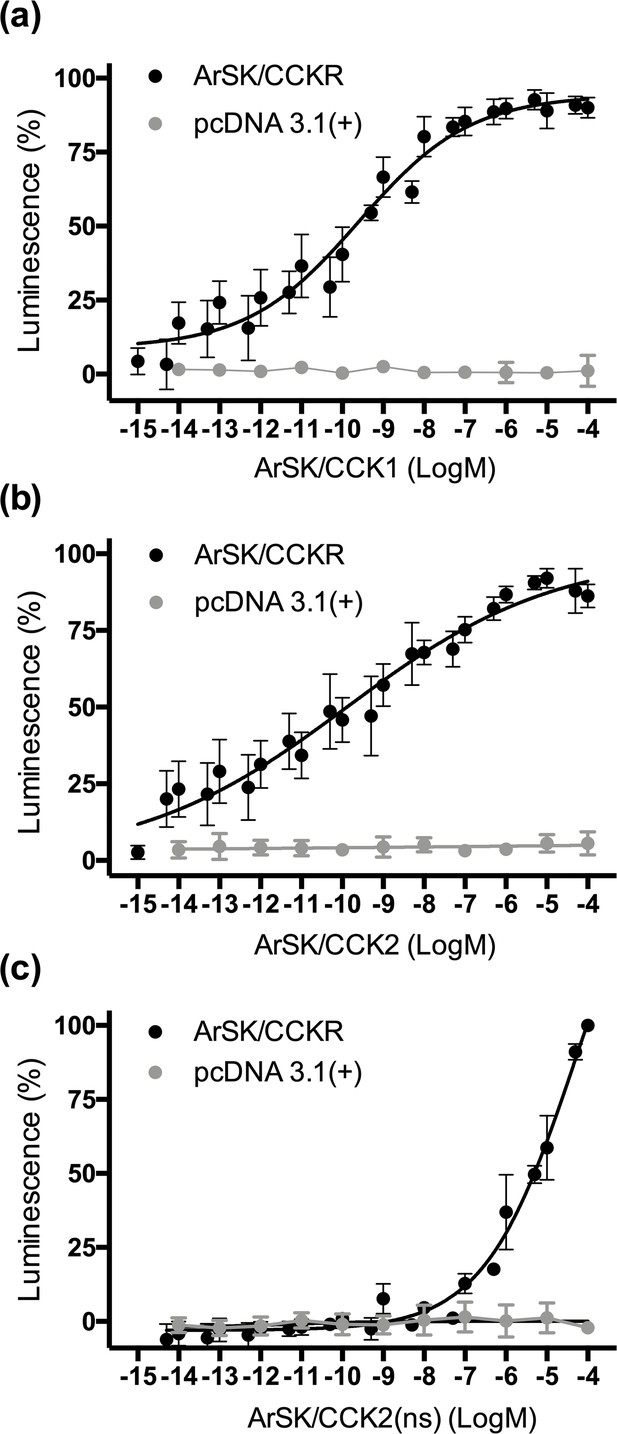
Experimental demonstration that the Asterias rubens SK/CCK-type peptides ArSK/CCK1 and ArSK/CCK2 act as ligands for the A. rubens SK/CCK-type receptor ArSK/CCKR.
The sulphated peptides ArSK/CCK1 (a), ArSK/CCK2 (b), and the non-sulphated peptide ArSK/CCK2(ns) (c) trigger dose-dependent luminescence in Chinese hamster ovary (CHO)-K1 cells stably expressing mitochondrial targeted apoaequorin (G5A) that were co-transfected with plasmids encoding the promiscuous human G-protein Gα16 and ArSK/CCKR (black). Control experiments where cells were transfected with an empty pcDNA 3.1(+) vector are shown in grey. Each point represents mean values (± s.e.m.) from at least four independent experiments performed in triplicate. Luminescence is expressed as a percentage of the maximal response observed in each experiment. The EC50 values for ArSK/CCK1 (a) and ArSK/CCK2 (b) are 0.25 and 0.12 nM, respectively. In comparison, the absence of tyrosine (Y) sulphation in ArSK/CCK2(ns) (c) causes a massive loss of potency (EC50 = 48 µM), indicating that the sulphated peptides act as ligands for ArSK/CCKR physiologically. A graph showing the selectivity of ArSK/CCKR as a receptor for ArSK/CCK-type peptides is presented in Figure 3—figure supplement 1. See Figure 3—source data 1 for source data.
-
Figure 3—source data 1
Data for the graphs shown in Figure 3 and Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/65667/elife-65667-fig3-data1-v1.xlsx
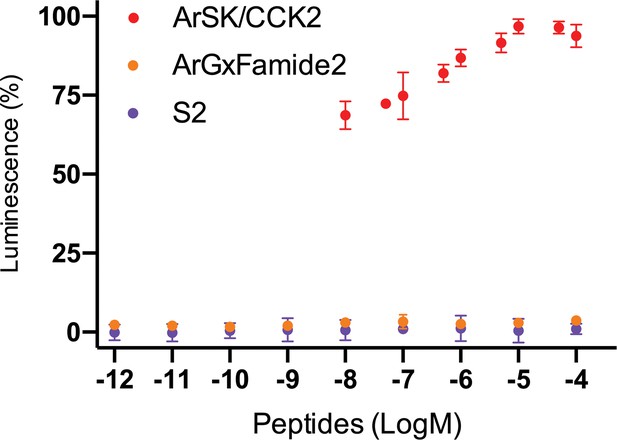
Graph showing the selectivity of ArSK/CCKR as a receptor for sulfakinin/cholecystokinin (SK/CCK)-type peptides.
The Asterias rubens SK/CCK-type peptide ArSK/CCK2 (red) triggers luminescence in Chinese hamster ovary (CHO)-K1 cells expressing the receptor ArSK/CCKR, the promiscuous G-protein Gα16, and the calcium-sensitive luminescent GFP-apoaequorin fusion protein G5A. In these experiments ArSK/CCK2 was tested at concentrations between 10–8 and 10–4 M; see Figure 3b for experiments showing complete concentration-response curves. The receptor is not activated by the SALMFamide-type neuropeptide S2 (SGPYSFNSGLTF-NH2; purple) or by the A. rubens tachykinin-like peptide ArGxFamide2 (GGGVPHVFQSGGIF-NH2; orange). Each point represents mean values (± s.e.m) from at least three independent experiments done in triplicate.
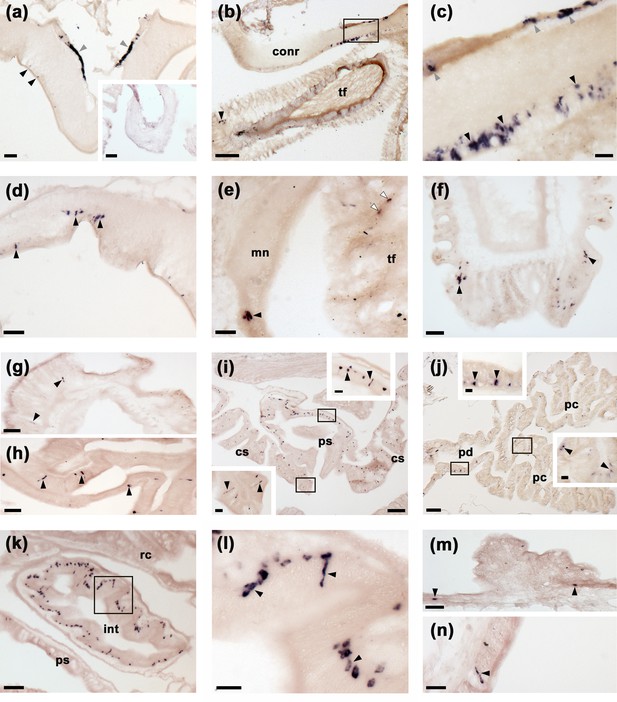
Localisation of ArSK/CCKP expression in Asterias rubens using mRNA in situ hybridization.
(a) Using antisense probes, ArSK/CCKP-expressing cells are revealed in the ectoneural (black arrowheads) and hyponeural (grey arrowheads) regions of a radial nerve cord. The specificity of staining with antisense probes is demonstrated by the absence of staining in a radial nerve cord section incubated with sense probes (see inset). (b) ArSK/CCKP-expressing cells in the circumoral nerve ring (see boxed area) and in the disk region of a peri-oral tube foot (black arrowhead). (c) High magnification image of the boxed area in (b), showing stained cells in the ectoneural (black arrowheads) and hyponeural (grey arrowheads) regions of the circumoral nerve ring. (d) ArSK/CCKP-expressing cells in a lateral branch of the radial nerve cord (black arrowheads). (e) ArSK/CCKP-expressing cells adjacent to the marginal nerve (black arrowhead) and in the stem of a tube foot (white arrowheads). (f) ArSK/CCKP-expressing cells (black arrowheads) adjacent to the basal nerve ring in the disk region of a tube foot. (g) ArSK/CCKP-expressing cells (black arrowheads) in the mucosal layer of the oesophagus. (h) ArSK/CCKP-expressing cells (black arrowheads) in the mucosal layer of the cardiac stomach. (i) ArSK/CCKP-expressing cells (black arrowheads) in the cardiac stomach and pyloric stomach, with the boxed regions shown at higher magnification in the insets. (j) ArSK/CCKP-expressing cells (black arrowheads) in the pyloric duct and pyloric caeca, with the boxed regions shown at higher magnification in the insets. (k,l) ArSK/CCKP-expressing cells (black arrowheads) in an oblique section of the intestine; the boxed region in (k) is shown at higher magnification in (l). (m,n) ArSK/CCKP-expressing cells (black arrowheads) in the external epithelium of the body wall. Abbreviations: conr, circumoral nerve ring; int, intestine; mn, marginal nerve; pc, pyloric caecum; pd, pyloric duct; ps, pyloric stomach; rc, rectal caeca; tf, tube foot. Scale bars: (b), (i), (j) = 120 μm; (a), (a-inset), (k) = 60 μm; (d), (e), (f), (g), (h), (m) = 32 μm; (c), (i-insets), (j-insets), (l), (n) = 16 μm.
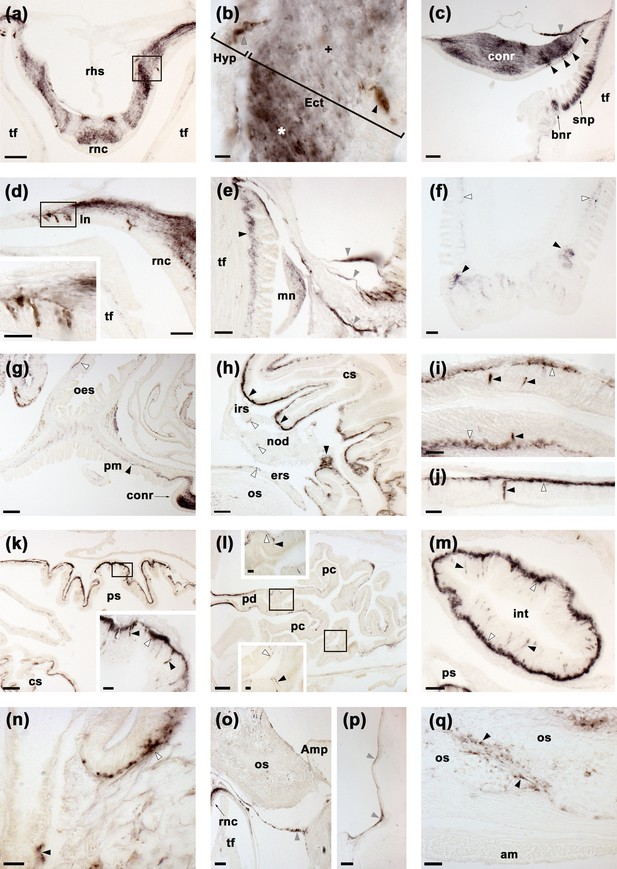
Localisation of ArSK/CCK1 expression in Asterias rubens using immunohistochemistry.
(a) ArSK/CCK1-immunoreactivity (ArSK/CCK1-ir) in a transverse section of the V-shaped radial nerve cord, with bilaterally symmetrical regional variation in the density of immunostaining in the ectoneural neuropile. (b) High magnification image of the boxed region in (a), showing stained cell bodies in the hyponeural (grey arrowhead) and ectoneural (black arrowhead) regions of the radial nerve cord. Regions of the ectoneural neuropile containing a higher (*) and lower (+) densities of immunostained fibres can be seen here. (c) ArSK/CCK1-ir in the circumoral nerve ring, with stained cells present in the hyponeural region (grey arrowheads) and in the ectoneural epithelium (black arrowheads); in the ectoneural neuropile there is regional variation in the density of immunostained fibres. Immunostaining can also be seen here in the sub-epithelial nerve plexus and basal nerve ring of an adjacent peri-oral tube foot. (d) ArSK/CCK1-ir in cells and fibres in a lateral branch of the radial nerve cord; the inset shows immunostained cells in the boxed region at higher magnification. (e) ArSK/CCK1-ir in the marginal nerve, the sub-epithelial nerve plexus of an adjacent tube foot (black arrowhead) and in branches of the lateral motor nerve (grey arrowheads). (f) ArSK/CCK1-ir in the sub-epithelial nerve plexus (white arrowheads) and basal nerve ring (black arrowheads) of a tube foot. (g) ArSK/CCK1-ir in the basi-epithelial nerve plexus of the peristomial membrane (black arrowhead) and the oesophagus (white arrowhead); immunostaining in the circumoral nerve can also be seen here. (h) ArSK/CCK1-ir in the lateral pouches of the cardiac stomach; note that the density of immunostained fibres is highest (black arrowheads) in regions of the mucosa adjacent to the intrinsic retractor strand; immunostaining in the intrinsic retractor strand, nodule and extrinsic retractor strand can also be seen here (white arrowheads). (i,j) High magnification images of cardiac stomach tissue showing ArSK/CCK1-ir in cell bodies (black arrowheads) and their processes in the basi-epithelial nerve plexus (white arrowheads); note that in (j) a process emanating from an immunostained cell body can be seen projecting into the plexus. (k) ArSK/CCK1-ir in the cardiac stomach and pyloric stomach; the boxed region is shown at higher magnification in the inset, showing immunostaining in cells (black arrowheads) and the basi-epithelial nerve plexus (white arrowhead). (l) ArSK/CCK1-ir in the pyloric duct and pyloric caeca; the boxed regions are shown at higher magnification in the insets, where immunostained cells (black arrowheads) and fibres (white arrowheads) can be seen. (m) ArSK/CCK1-ir in an oblique section of the intestine, with immunostained cells in the mucosa (black arrowheads) and intense immunostaining in the basi-epithelial nerve plexus (white arrowheads). (n) ArSK/CCK1-ir in the basi-epithelial nerve plexus of the body wall external epithelium (white arrowhead) and in the lining of a papula (black arrowhead). (o) ArSK/CCK1-ir in nerve fibres projecting around the base of a tube foot at its junction with the neck of its ampulla. (p) ArSK/CCK1-ir in nerve fibres located in the coelomic lining of the lateral region of the body wall. (q) ArSK/CCK1-ir in inter-ossicular tissue of the body wall. Abbreviations: am, apical muscle; Amp, ampulla; bnr, basal nerve ring; conr, circumoral nerve ring; cs, cardiac stomach; Ect, ectoneural; ers, extrinsic retractor strand; Hyp, hyponeural; int, intestine; irs, intrinsic retractor strand; ln, lateral nerve; mn, marginal nerve; nod, nodule; oes, oesophagus; os, ossicle; pm, peristomial membrane; pc, pyloric caecum; pd, pyloric duct; ps, pyloric stomach; rhs, radial hemal strand; rnc, radial nerve cord; snp, sub-epithelial nerve plexus; tf, tube foot. Scale bars: (g), (k), (l) = 120 μm; (a), (c), (f), (h), (o), (p) = 60 μm; (d), (e), (m), (q) = 32 μm; (d-inset), (i), (j), (k-inset), (l-insets), (n) = 16 μm; (b) = 6 μm. Graphs showing enzyme-linked immunosorbent assay (ELISA)-based characterisation of the antibodies to ArSK/CCK1 used here for immunohistochemistry are presented in Figure 5—figure supplement 1.
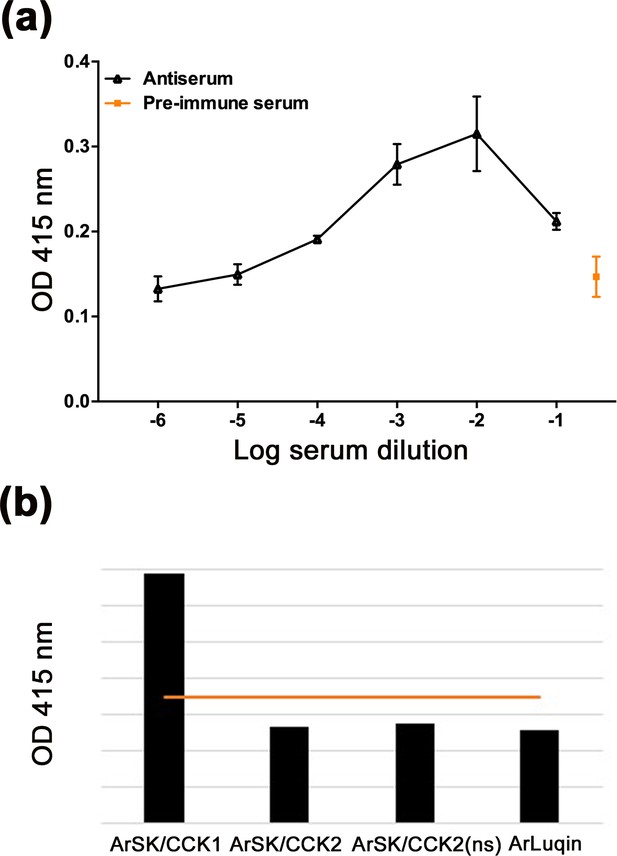
Characterisation of a rabbit antibodies to ArSK/CCK1 using an enzyme-linked immunosorbent assay (ELISA).
(a) By comparison with pre-immune serum (orange), 0.1 nmol of the ArSK/CCK1 antigen peptide is detected by the ArSK/CCK1 antiserum (black line) at dilutions between 1:10 and 1:10,000. All data points are mean values from two replicates. (b) Graph showing the results of ELISA tests using the trimethylamine (TEA) fraction of affinity-purified antibodies to ArSK/CCK1 (dilution 1:10). The red line indicates the mean optical density (OD) value of negative control experiments without peptides. The four peptides tested were applied at a concentration of 0.1 nmol but only the mean OD value for ArSK/CCK1 is above the OD value for the negative control. All data points are mean values from six replicates. These experiments demonstrate that specific antibodies to ArSK/CCK1 were successfully generated.
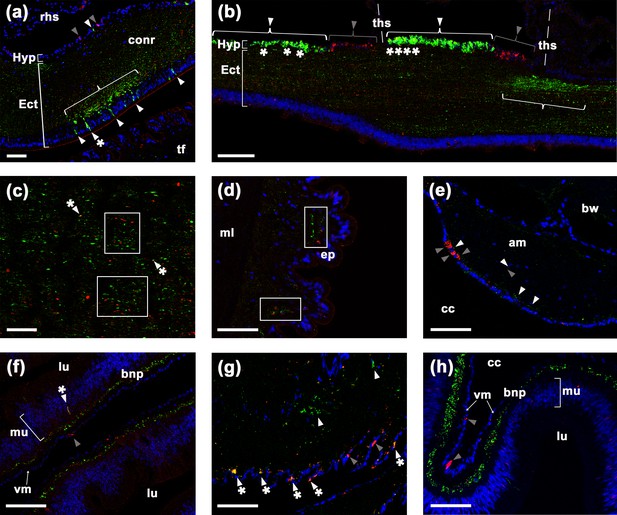
Comparison of ArSK/CCK1 and ArSK/CCK receptor (ArSK/CCKR) expression in Asterias rubens using double immunofluorescence labelling.
(a) ArSK/CCK1-immunoreactivity (ArSK/CCK1-ir; green) and ArSK/CCKR immunoreactivity (ArSK/CCK-R-ir; red) in a transverse section of the circumoral nerve ring, with cell nuclei stained using DAPI (blue). Fibres exhibiting ArSK/CCK1-ir or ArSK/CCKR-ir can be seen throughout the neuropile of the ectoneural region. However, there is variation in the density of immunostained fibres/cells and a bracketed neuropile region containing a higher density of ArSK/CCK1-ir fibres is located proximal to a cluster of cells in the ectoneural epithelium that exhibit ArSK/CCK1-ir (green; white arrowheads) or ArSK/CCK1-ir and ArSK/CCKR-ir (green and yellow; white arrowheads with asterisk). Cells exhibiting ArSK/CCK1-ir (white arrowhead) or ArSK/CCKR-ir (grey arrowhead) can be seen in the hyponeural region. (b) Longitudinal section of a radial nerve cord showing segmentally repeated clusters of labelled cells in the hyponeural region, with each segment bounded by transverse hemal strands. Note the distinct clusters of cells exhibiting ArSK/CCKR-ir (grey arrowhead) and cells exhibiting ArSK/CCK1-ir (white arrowhead). However, the presence of yellow regions in some of the cells exhibiting ArSK/CCK1-ir (asterisks) indicates that ArSK/CCKR may be co-expressed in these cells. As in the circumoral nerve (a), there is variation in the density of immunostained fibres in the ectoneural neuropile and a region with a high density of fibres exhibiting ArSK/CCK1-ir is bracketed. (c) High magnification image of a longitudinal section of a radial nerve cord showing a ‘salt and pepper’ pattern of labelling in the ectoneural neuropile, consistent with expression of ArSK/CCK1 and ArSK/CCKR in different but often adjacent populations of fibres (see white rectangles). However, the presence of yellow labelling (white arrowhead with asterisk) may be indicative of co-expression in a few fibres. (d) Immunostained processes exhibiting ArSK/CCK1-ir or ArSK/CCK-R-ir can be seen here in close proximity (white rectangles) within the sub-epithelial nerve plexus of a tube foot podium. (e) Immunostained cells/processes exhibiting ArSK/CCK1-ir (white arrowheads) or ArSK/CCKR-ir (grey arrowheads) can be seen here in the coelomic epithelial layer, sub-epithelial nerve plexus, or muscle fibre layer of an apical muscle. (f) High magnification image of the lateral pouches of cardiac stomach showing fibres exhibiting ArSK/CCK1-ir or ArSK/CCKR-ir in the basiepithelial nerve plexus. Cells exhibiting ArSK/CCKR-ir can be seen in the visceral muscle layer (grey arrowhead) and a cell that exhibits both ArSK/CCK1-ir and ArSK/CCKR-ir can be seen in the mucosal layer (white arrowhead with asterisk). (g) Immunostained cells/processes exhibiting ArSK/CCK1-ir (white arrowhead) or ArSK/CCK-R-ir (grey arrowhead) or both ArSK/CCK1-ir and ArSK/CCK-R-ir (white arrowhead with asterisks) in a nodule that links the cardiac stomach to extrinsic retractor strands. (h) High magnification image of pyloric stomach showing fibres exhibiting ArSK/CCK1-ir or ArSK/CCKR-ir in the basiepithelial nerve plexus. Cells exhibiting ArSK/CCKR-ir can be seen in the visceral muscle layer (grey arrowhead). Abbreviations: am, apical muscle; bnp, basiepithelial nerve plexus; bw, body wall; cc, coelomic cavity; conr, circumoral nerve ring; Ect, ectoneural; ep, epithelium; Hyp, hyponeural; ml, muscle layer; mu, mucosa; rhs, radial hemal strand; tf, tube foot; ths, transverse hemal strand; vm, visceral muscle layer. Scale bars: (b) = 70 μm; (a), (d), (e), (f), (g), (h) = 40 μm; (c) = 20 μm.
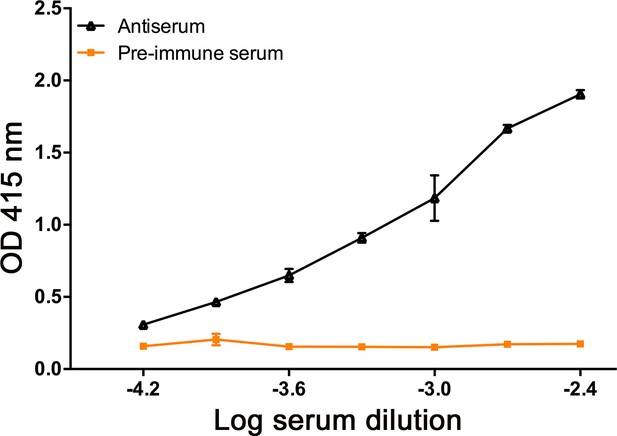
Characterisation of rabbit antibodies to ArSK/CCKR using an enzyme-linked immunosorbent assay (ELISA).
By comparison with pre-immune serum (orange), 0.1 nmol of the ArSK/CCKR antigen peptide is detected by the ArSK/CCKR antiserum (black line) at dilutions between 1:250 and 1:16,000. All data points are mean values from three replicates.
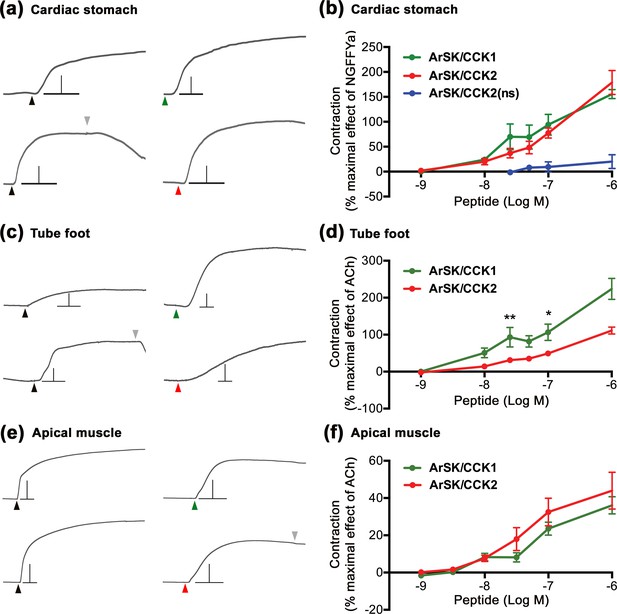
ArSK/CCK1 and ArSK/CCK2 cause concentration-dependent contraction of in vitro preparations of cardiac stomach, tube foot, and apical muscle from Asterias rubens.
(a) Representative recordings of the effects ArSK/CCK1 (1 µM; green arrowhead) and ArSK/CCK2 (1 µM; red arrowhead) in causing contraction of cardiac stomach preparations and compared with the effect of NGFFYamide (100 nM; black arrowhead) on the same preparation. The downward pointing grey arrowhead shows when a preparation was washed with seawater. Scale bar: vertical 0.5 mV; horizontal 1 min. (b) Concentration-response curves comparing the effects of ArSK/CCK1, ArSK/CCK2, and ArSK/CCK2(ns) on cardiac stomach preparations. The effects of peptides (means ± s.e.m.; n = 5–9) were normalized to the effect of 100 nM NGFFYamide (NGFFYa). (c) Representative recordings of the effects ArSK/CCK1 (1 µM; green arrowhead) and ArSK/CCK2 (1 µM; red arrowhead) in causing contraction of tube foot preparations and compared with the effect of acetylcholine (10 µM; black arrowhead) on the same preparation. The downward pointing grey arrowhead shows when a preparation was washed with seawater. Scale bar: vertical 0.08 mV; horizontal 1 min. (d) Concentration-response curves comparing the effects of ArSK/CCK1 and ArSK/CCK2 on tube foot preparations. The effects of peptides (means ± s.e.m.; n = 8–10) were normalized to the effect of 10 µM acetylcholine (ACh). * indicates statistically significant differences between ArSK/CCK1 and ArSK/CCK2 when tested at concentrations of 25 and 100 nM (p < 0.01 and p < 0.05, respectively) as determined by two-way ANOVA and Bonferroni’s multiple comparison test. (e) Representative recordings of the effects ArSK/CCK1 (1 µM; green arrowhead) and ArSK/CCK2 (1 µM; red arrowhead) in causing contraction of apical muscle preparations and compared to the effect of acetylcholine (10 µM; black arrowhead) on the same preparation. The downward pointing grey arrowhead shows when a preparation was washed with seawater. Scale bar: vertical 0.4 mV; horizontal 1 min. (f) Concentration-response curves comparing the effects of ArSK/CCK1 and ArSK/CCK2 on apical muscle preparations. The effects of peptides (means ± s.e.m; n = 20–23) were normalized to the effect of 10 µM acetylcholine (ACh).
-
Figure 7—source data 1
Data for graphs shown in Figure 7b, d and f.
- https://cdn.elifesciences.org/articles/65667/elife-65667-fig7-data1-v1.xlsx
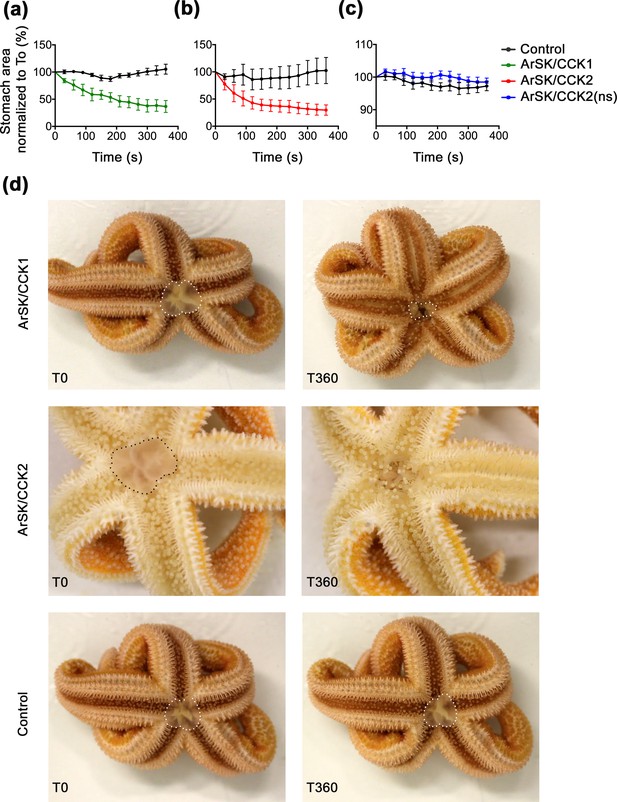
ArSK/CCK1 and ArSK/CCK2 trigger cardiac stomach retraction in Asterias rubens.
The graphs compare experiments where starfish were first injected with vehicle (black line; 10 µl of distilled water) and then injected with (a) ArSK/CCK1 (green line; 10 mol; 10 µl 1 mM), or (b) ArSK/CCK2 (red line; 10 mol; 10 µl 1 mM), or (c) ArSK/CCK2(ns) (blue line; 10 mol; 10 µl 1 mM). Stomach eversion was induced by placing starfish in seawater containing 2% MgCl2 and then the area of cardiac stomach everted (in 2D) at 30 s intervals (0–360 s) following injection of water (control) or peptide was measured, normalizing to the area of cardiac stomach everted at the time of injection (T0). Data (means ± s.e.m.) were obtained from 6 (ArSK/CCK1), 6 (ArSK/CCK2), or 8 (ArSK/CCK2(ns)) experiments. Both ArSK/CCK1 and ArSK/CCK2 cause retraction of the cardiac stomach, with >50% reduction in the area of cardiac stomach everted within 360 s (Figure 8—videos 1; 2), whereas ArSK/CCK2(ns) has no effect. (d) Photographs from representative experiments showing that injection of ArSK/CCK1 (10 mol; 10 µl 1 mM at T0) or ArSK/CCK2 (10 mol; 10 µl 1 mM at T0) causes retraction of the everted cardiac stomach (marked with white or black dots), which is reflected in a reduction in the area everted after 360 s (T360). By way of comparison, in a control experiment injection with vehicle (10 µl of distilled water at T0) does not trigger cardiac stomach retraction. See Figure 8—source data 1 for source data.
-
Figure 8—source data 1
Data for graphs shown in Figure 8a, b and c,.
- https://cdn.elifesciences.org/articles/65667/elife-65667-fig8-data1-v1.xlsx
ArSK/CCK1 (10 mol; 10 µl 1 mM) induced retraction of the cardiac stomach in the starfish Asterias rubens.
ArSK/CCK2 (10 mol; 10 µl 1 mM) induced retraction of the cardiac stomach in the starfish Asterias rubens.
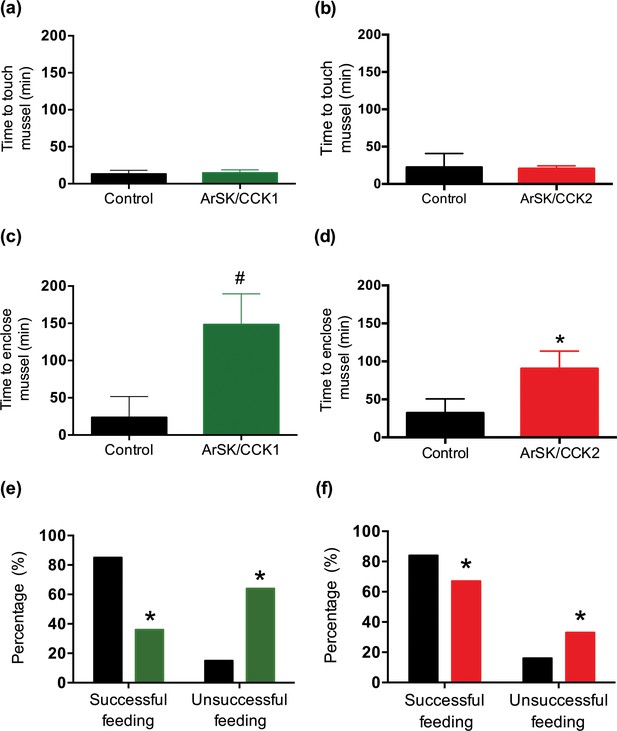
Effects of ArSK/CCK1 and ArSK/CCK2 on feeding behaviour in Asterias rubens.
To investigate if injection of ArSK/CCK1 and/or ArSK/CCK2 affects feeding behaviour, starved animals were presented with a mussel as prey and then behaviour was observed. By comparison with vehicle-injected animals (control; 10 µl of distilled water; shown in black), injection of ArSK/CCK1 (a; 10 mol or 10 µl of 1 mM; shown in green) or ArSK/CCK2 (b; 10 mol or 10 µl of 1 mM; shown in red) had no effect on the time taken for starfish to make first contact with the mussel. However, by comparison with vehicle-injected animals (control; 10 µl of distilled water; shown in black) injection of ArSK/CCK1 (c; 10 mol or 10 µl of 1 mM; shown in green) or ArSK/CCK2 (d; 10 mol or 10 µl of 1 mM; shown in red) causes an increase in the time elapsed before starfish enclose a mussel. Data are expressed as means ± s.e.m. (n = 13 for control- and 11 for ArSK/CCK1-treated groups; n = 19 for control- and 19 for ArSK/CCK2-treated groups). # indicates a nearly statistically significant difference (p = 0.0523) between vehicle-injected and ArSK/CCK1-injected groups, as determined by two-tailed Mann-Whitney U-test. * indicates statistically significant differences (p < 0.05) between vehicle-injected and ArSK/CCK2-injected groups, as determined by two-tailed Welch’s unequal variances t-test. Furthermore, injection of ArSK/CCK1 (e; 10 mol or 10 µl of 1 mM; shown in green) or ArSK/CCK2 (f; 10 mol or 10 µl of 1 mM; shown in red) causes a significant decrease in the percentage of starfish that initiate feeding after the mussel is touched for the first time, by comparison with vehicle-injected animals (control; 10 µl of distilled water; shown in black). * indicates statistically significant differences (p < 0.0001 and p < 0.01 for ArSK/CCK1- and ArSK/CCK2-treated groups, respectively) between vehicle-injected and ArSK/CCK1- or ArSK/CCK2-injected groups, as determined by Fisher’s exact test. See Figure 9—source data 1 for source data.
-
Figure 9—source data 1
Data for graphs shown in Figure 9.
- https://cdn.elifesciences.org/articles/65667/elife-65667-fig9-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Sequence-based reagent | 5’-TCGCTACTGTTTCTCTCGCA-3’5’-AAAGGCGTCAACAACTGCTT-3’ | Custom synthesized by Sigma-Aldrich | Oligonucleotide primers for cloning of ArSK/CCKP cDNA (Asterias rubens) | |
| Recombinant DNA reagent | Zero Blunt Topo | Thermo Fisher Scientific | Cat. no. 450,159 | |
| Recombinant DNA reagent | pBluescript SKII (+) | Agilent Technologies | Cat. no. 212,205 | |
| Recombinant DNA reagent | pcDNA 3.1+ vector with neomycin selectable marker (mammalian expression vector) | Invitrogen | Cat. no. V790-20 | |
| Transfected construct (Asterias rubens) | Asterias rubens ArSK/CCKR cDNA cloned in expression vector pcDNA 3.1+ | This paper | GenBank: MW261740 | See Materials and methods section ‘Cell lines and pharmacological characterization of ArSK/CCKR’ |
| Cell line (Cricetus griseus) | Chinese hamster ovary cells (CHO-K1) | Sigma-Aldrich | RRID: CVCL_0214 | Cat. no. 85051005 |
| Antibody | (Rabbit polyclonal) antibodies to ArSK/CCK1 | This paper | RRID: AB_2877176 | Antibodies generated by Elphick group at QMUL (m.r.elphick@qmul.ac.uk) – see Materials and methods section ‘Generation and characterisation of antibodies to ArSK/CCK1 and ArSK/CCKR’. Affinity-purified antibodies were used for immunohistochemistry (1:10 or 1:15). |
| Antibody | (Guinea pig polyclonal) antibodies to ArSK/CCKR | This paper | RRID: AB_2891354 | Antibodies generated by Elphick group at QMUL (m.r.elphick@qmul.ac.uk) – see Materials and methods section ‘Generation and characterisation of antibodies to ArSK/CCK1 and ArSK/CCKR’. Affinity-purified antibodies were used for immunohistochemistry (1:3.3). |
| Antibody | (Sheep polyclonal)Sheep alkaline phosphatase (AP)-conjugated anti-DIG antibody | Roche | Cat. no. 11093274910 | Used for mRNA in situ hybridisation (1:3000). |
| Antibody | (Goat polyclonal)Peroxidase-AffiniPure Goat Anti-Rabbit IgG (H + L) Horseradish Peroxidase conjugated | Jackson ImmunoResearch | RRID: AB_2313567Cat. no. 111-035-003 | Used for immunohistochemistry (1:1000). |
| Antibody | (Goat polyclonal)Alexa Fluor 488-conjugated AffiniPure Goat Anti-Rabbit IgG (H + L) | Jackson ImmunoResearch | RRID: AB_2338046Cat. no. 111-545-003 | Used for immunohistochemistry (1:500). |
| Antibody | (Goat polyclonal)Cy3-conjugated AffiniPure Goat Anti-Guinea Pig IgG (H + L) | Jackson ImmunoResearch | RRID: AB_2337423Cat. no. 106-165-003 | Used for immunohistochemistry (1:300). |
| Commercial assay, kit | AminoLink Plus Immobilization Kit | Thermo Fisher Scientific | Cat. no. 44,894 | |
| Commercial assay, kit | MycoAlert Mycoplasma Detection Kit | Lonza | LT07-118 | |
| Software, algorithm | MSConvert | ProteoWizard Toolkit | Version 3.0.5759 | doi: 10.1093/bioinformatics/btn323 |
| Software, algorithm | Scaffold | Proteome Software Inc | Version 4.6.1RRID:SCR_014345 | |
| Software, algorithm | MAFFT | MAFFT | Version 7RRID:SCR_011811 | http://mafft.cbrc.jp/alignment/server/ |
| Software, algorithm | IQ-tree web server | http://iqtree.cibiv.univie.ac.at | ||
| Software, algorithm | MUSCLE | https://www.ebi.ac.uk/Tools/msa/muscle/ | RRID:SCR_011812 | |
| Software, algorithm | Volocity | PerkinElmer | Version 6.3.1 | |
| Software, algorithm | Adobe Photoshop CC | Adobe | Version 19.1.4, ×64RRID:SCR_014199 | |
| Software, algorithm | Leica Application Suite Advanced Fluorescence (LAS AF) | Leica | Version 2.6.0.7266 | |
| Software, algorithm | LabChart | ADInstruments | Version 8.0.7RRID:SCR_017551 | |
| Software, algorithm | ImageJ | http://rsb.info.nih.gov/ij | Version 1.0RRID:SCR_003070 | |
| Software, algorithm | Prism | GraphPad | Version 6.0RRID:SCR_002798 |





