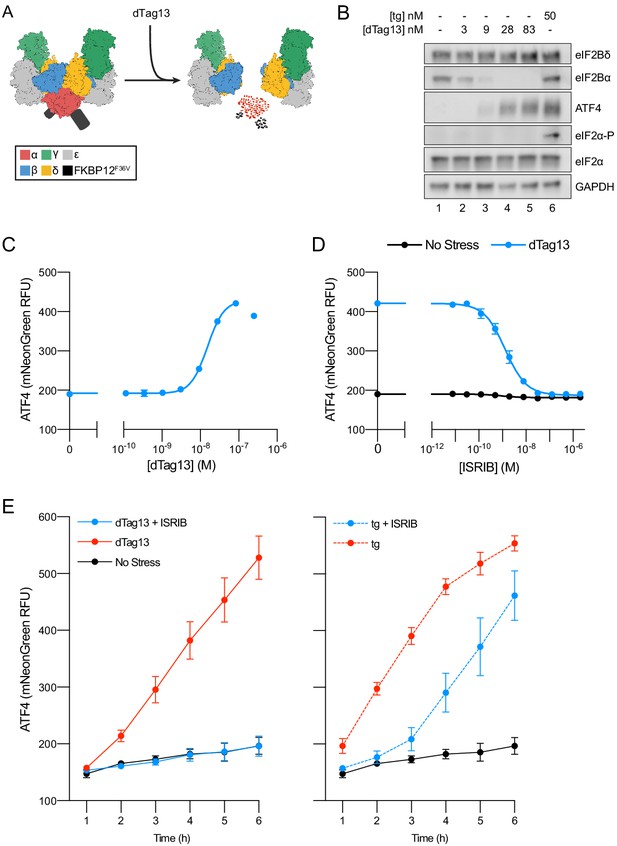
eIF2B conformation and assembly state regulate the integrated stress response
Figures
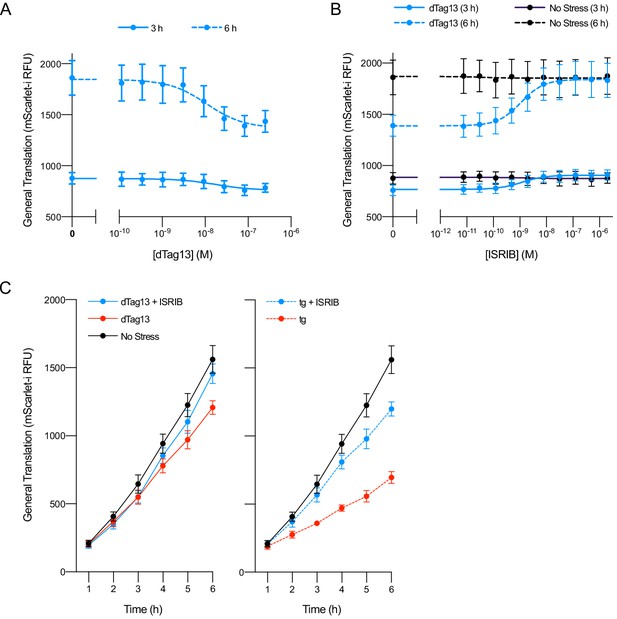
Cellular eIF2B assembly state in cells modulates the integrated stress response (ISR).
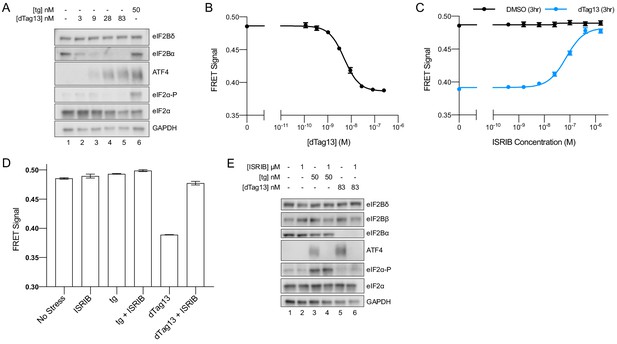
(A) Schematic of eIF2B assembly state modulation via the FKBP12F36V/dTag13 system used to induce degradation of eIF2Bα. (B) Western blot of K562 cell extracts after treatment with thapsigargin (tg) or dTag13 for 3 hr as indicated. Thapsigargin induces the ISR by depleting Ca2+ levels in the endoplasmic reticulum. Loading of all lanes was normalized to total protein. (C–E) ATF4 reporter levels as monitored by flow cytometry. Trimethoprim was at 20 μM. (C) Samples after 3 hr of dTag13 treatment (EC50 = 15 nM; s.e.m. = 1 nM). (D) Samples after 3 hr of ISRIB treatment ± 83 nM dTag13 (EC50 = 1.4 nM; s.e.m. = 0.3 nM). (E) Timecourse of tg treatment (dTag13 = 83 nM, tg = 100 nM, ISRIB = 2 μM). For (B), eIF2Bδ, eIF2Bα, and GAPDH blots, and the ATF4 and eIF2α blots are from the same gels, respectively; the eIF2α-P blot is from its own gel. For (C–E), biological replicates: n = 3. All error bars represent s.e.m.
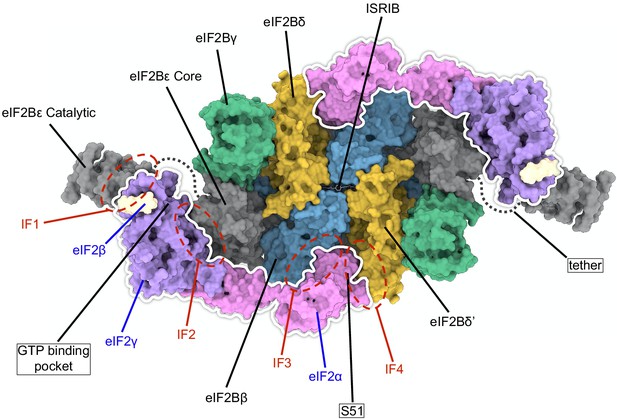
Overview of key eIF2 and eIF2B interaction surfaces.
A surface representation of a model of two eIF2 heterotrimers and ISRIB bound to an eIF2B decamer is shown (PDB ID: 6O85). Individual subunits of eIF2 and eIF2B are indicated. The eIF2 heterotrimers are outlined in white and the locations of interfaces IF1–IF4 are indicated, as are the positions of eIF2α S51, the GTP binding pocket (empty in the structure), and ISRIB (shown in stick representation). The eIF2Bα2 dimer is hidden in this orientation. eIF2Bε contains two domains linked by a flexible tether that was not resolved in the structure.
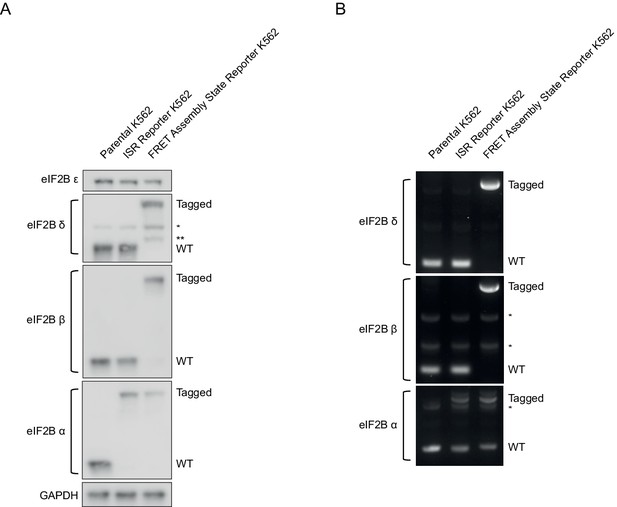
Tagging of eIF2B subunits in K562 cells.
(A) Western blot of eIF2B subunits in parental and edited K562 cells. Integrated stress response (ISR) reporter cells and assembly state reporter cells were edited at the EIF2B1 locus (eIF2Bα-FKBP12F36V N-terminal fusion). No evidence of WT protein is observed in either cell line. Assembly state reporter cells were edited at the EIF2B2 locus (eIF2Bβ-mNeonGreen C-terminal fusion) and the EIF2B4 locus (eIF2Bδ-mScarlet-i C-terminal fusion). No evidence of WT protein is observed in these cells. The asterisk denotes a non-specific band. The double asterisk denotes a minor eIF2Bδ species likely resulting from mScarlet-i/G/S linker proteolysis during sample preparation. eIF2Bδ and eIF2Bα blots and eIF2Bε and GAPDH blots are from the same gel, respectively; eIF2Bβ is from its own blot. (B) One percent agarose gel of PCR amplified eIF2Bα-, eIF2Bβ-, and eIF2Bδ-encoding loci from parental and edited cell line gDNA preps. The lengths of the eIF2Bβ and eIF2Bδ products demonstrate that no unedited alleles are present in the assembly state reporter cells. The length of the eIF2Bα product demonstrates that some tagged as well as some untagged alleles are present in both cell lines. Based on the lack of WT length protein, the remaining untagged alleles likely harbor deletions or frameshift mutations that prevent synthesis or destroy the protein product. The asterisk denotes a non-specific band.
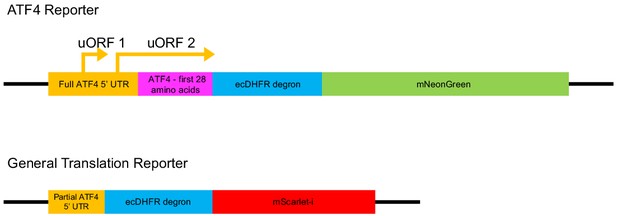
Integrated stress response (ISR) reporter design.
A schematic of the ATF4 translation and general translation reporters used to read out ISR activation.
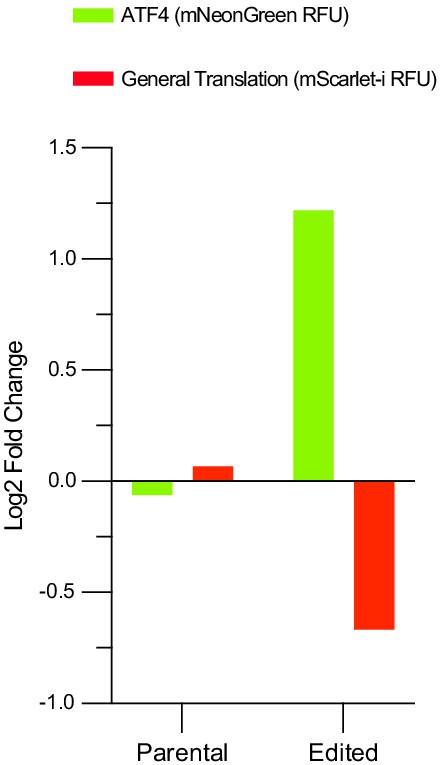
dTag13 treatment alone does not activate the integrated stress response (ISR).
Parental cells containing the ATF4 and general translation reporters as well as the edited cells where eIF2Bα was tagged with an FKBP12F36V degron were treated with 500 nM dTag13 or untreated (0.1% dimethyl sulfoxide (DMSO)) for 24 hr and then 20 μM trimethoprim for 3 hr. ATF4 and general translation reporter levels were monitored by flow cytometry, and the change in reporter signal between dTag13 treated and untreated conditions is shown. dTag13 only activates the ISR when eIF2Bα is endogenously tagged with the FKBP12F36V degron.
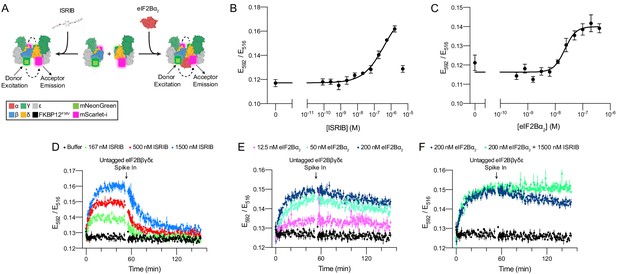
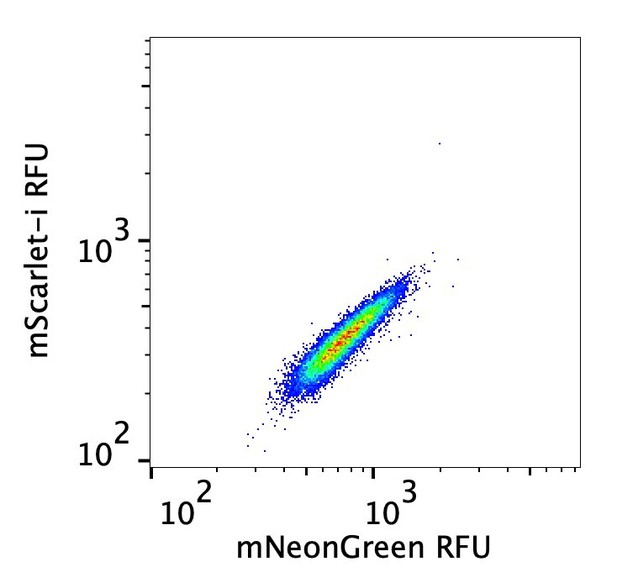
Förster resonance energy transfer (FRET) system monitors eIF2B assembly state.
(A) Schematic depicting the principle of eIF2B assembly state modulation by ISRIB and eIF2Bα2 and FRET readout. (B, C) FRET signal (E592/E516) measured after 1 hr of incubation with (B) ISRIB (EC50 = 250 nM; s.e.m. = 80 nM) or (C) eIF2Bα2 (EC50 = 20 nM; s.e.m. = 4 nM) at 50 nM eIF2Bβδγε-F. (D–F) Timecourse monitoring of FRET signal (E592/E516) after addition of (D) ISRIB (association t1/2 = 5.1 min, s.e.m. = 0.5 min; dissociation t1/2 = 15 min, s.e.m. = 1 min), (E) eIF2Bα2 (association t1/2 = 7.3 min, s.e.m. = 0.6 min; dissociation t1/2 = 180 min, s.e.m. = 10 min), or (F) ISRIB + eIF2Bα2 (association t1/2 = 7 min, s.e.m. = 1 min; dissociation t1/2 = N/A) at 50 nM eIF2Bβδγε-F. At t = 52 min, unlabeled eIF2Bβδγε was added to a final concentration of 1 μM. For (B, C), representative replicate averaging four technical replicates is shown. For (D–F), representative replicate averaging three technical replicates is shown. For (B–F), biological replicates: n = 3. All error bars represent s.e.m.
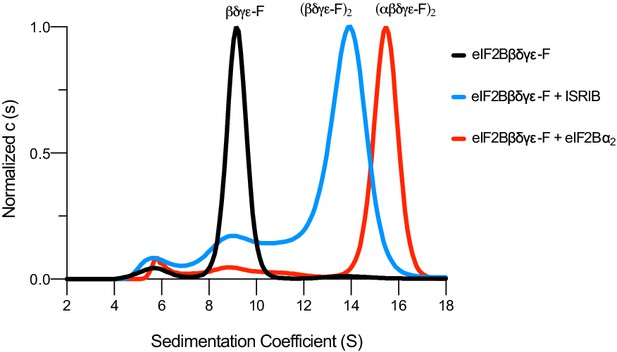
eIF2Bβδγε-F can octamerize and decamerize.
Analytical ultracentrifugation (sedimentation velocity) was used to determine eIF2B complex assembly state. Treatment with ISRIB induces octamerization of eIF2Bβδγε-F. Treatment with eIF2Bα2 induces decamerization. Also, 1 μM ISRIB, 1 μM eIF2Bβδγε-F, and 500 nM eIF2Bα2 were used.
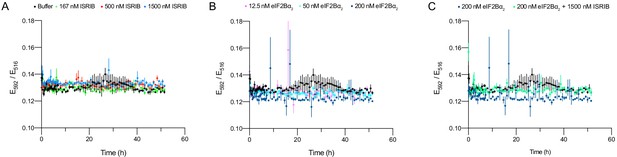
Validation of eIF2Bβδγε-F kinetics.
(A–C) Treatment of 50 nM eIF2Bβδγε-F with ISRIB or eIF2Bα2 led to no changes in Förster resonance energy transfer signal when simultaneously treated with excess of untagged eIF2Bβδγε (1 μM). For (A–C), representative replicate averaging three technical replicates is shown. Biological replicates: n = 2. All error bars represent s.e.m.
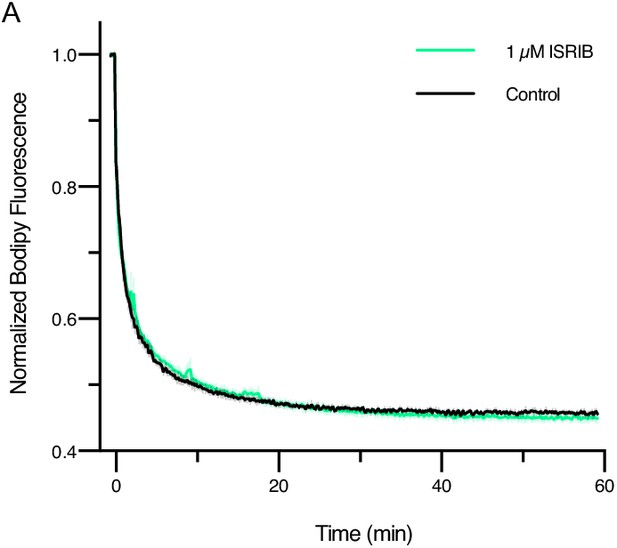
ISRIB treatment does not impact guanine nucleotide exchange factor (GEF) activity when eIF2Bα2 is saturating.
GEF activity of eIF2B as assessed by BODIPY-FL-GDP exchange. BODIPY-FL-GDP fluorescence decreases when free in solution. t1/2 = 1.6 min (Control) and 1.9 min (1 μM ISRIB). Biological replicates: n = 3.
eIF2B is a decamer in both unstressed and stressed cells, and ISRIB blocks integrated stress response (ISR) activation.
(A) Western blot of K562 ISR reporter cell extracts after treatment with thapsigargin (tg) or dTag13 for 3 hr as indicated. (B–D) Förster resonance energy transfer signal as monitored by flow cytometry after 3 hr treatment with (B) dTag13 (EC50 = 5.1 nM; s.e.m. = 0.2 nM), (C) ISRIB ± 83 nM dTag13 (EC50 = 80 nM; s.e.m. = 10 nM), and (D) various stressors (83 nM dTag13, 50 nM tg, ± 1.6 μM ISRIB). The ratio of mScarlet-i/mNeonGreen emission is presented. (E) Western blot of K562 ISR reporter cell extracts treated for 3 hr with ISRIB, tg, and/or dTag13 as indicated. All lanes across gels were loaded with equal total protein. For (A), eIF2Bδ, eIF2Bα, and GAPDH blots, and the ATF4 and eIF2α blots are from the same gels, respectively; the eIF2α-P blot is from its own gel. For (E), eIF2Bδ, eIF2Bβ, and GAPDH blots, ATF4 and eIF2α blots, and eIF2Bα and eIF2α-P blots are from the same gels, respectively. For (B–D), biological replicates: n = 3. All error bars represent s.e.m.
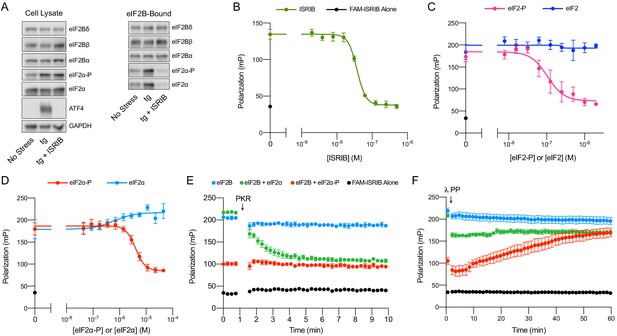
ISRIB and eIF2-P compete for eIF2B binding.
(A) Western blot of K562 integrated stress response (ISR) reporter cell extracts after treatment with tg ± ISRIB as indicated (left panel) or of eIF2B-bound fraction isolated by anti-FLAG immunoprecipitation of the eIF2B-mNeonGreen-FLAG tagged subunit under native conditions (right panel). (B–D) Plot of fluorescence polarization signal after incubation of FAM-ISRIB (2.5 nM) with 100 nM eIF2B(αβδγε)2 and varying concentrations of (B) ISRIB (IC50 = 37 nM; s.e.m. = 1 nM), (C) eIF2 or eIF2-P (IC50 = 210 nM; s.e.m. = 120 nM), and (D) eIF2α or eIF2α-P (IC50 = 4000 nM; s.e.m. = 200 nM). (E–F) Timecourse of fluorescence polarization signal after addition of (E) eIF2α kinase PKR and ATP or (F) λ phosphatase. FAM-ISRIB was at 2.5 nM. eIF2B(αβδγε)2 was at 100 nM. eIF2α and eIF2α-P were at 5.6 μM. In (A), eIF2Bδ, eIF2Bα, and eIF2α blots, eIF2Bβ and eIF2α-P blots, and ATF4 and GAPDH blots are from the same gels, respectively. All cell lysate or eIF2B-bound lanes across all gels were loaded with equal total protein. Biological replicates: (B) n = 3; (C) n = 5 (n = 4 at 2 μM); and (D–F) n = 3. All error bars represent s.e.m.
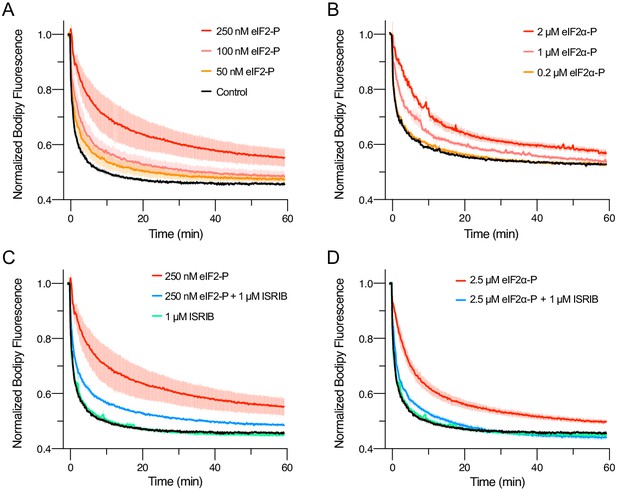
eIFα-P is the minimal unit needed to inhibit nucleotide exchange by eIF2B.
(A–D) Guanine nucleotide exchange factor (GEF) activity of eIF2B as assessed by BODIPY-FL-GDP exchange. eIF2B(αβδγε)2 was at 10 nM throughout. For (A) t1/2 = 1.6 min (Control), 2.5 min (50 nM eIF2-P), 3.5 min (100 nM eIF2-P), and 7.2 min (250 nM eIF2-P). For (B) t1/2 = 2.4 min (Control), 3.0 min (0.2 μM eIF2α-P), 5.0 min (1 μM eIF2α-P), and 6.7 min (2 μM eIF2α-P). For (C) t1/2 = 1.6 min (Control), 1.9 min (1 μM ISRIB), 3.1 min (250 nM eIF2-P + 1 μM ISRIB), and 7.2 min (250 nM eIF2-P). For (D) t1/2 = 1.6 min (Control), 1.9 min (1 μM ISRIB), 3.1 min (2.5 μM eIF2α-P + 1 μM ISRIB), and 5.3 min (2.5 μM eIF2α-P). All error bars represent s.e.m. Biological replicates: (A–D) n = 3 except for the 100 and 50 nM eIF2-P conditions in (A), where n = 2.
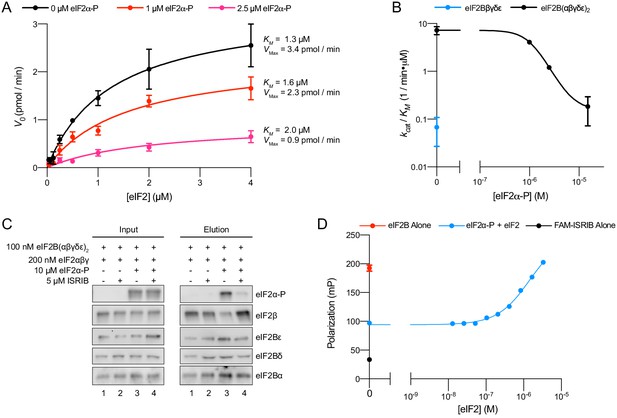
eIFα-P reduces eIF2B’s catalytic activity and antagonizes eIF2 binding.
(A) Initial velocity of eIF2B-catalyzed nucleotide exchange as a function of eIF2 concentration. eIF2B(αβδγε)2 concentration was 10 nM. (B) Plot of kcat/KM for tetramer and decamer at varying eIF2α-P concentrations, obtained by fitting the linear portion of the Michaelis–Menten saturation curve. Keeping the number of eIF2 binding sites constant, the eIF2B(αβδγε)2 concentration was 10 nM while eIF2Bβδγε was 20 nM. (C) Western blot of purified protein recovered after incubation with eIF2B(αβδγε)2 immobilized on anti-protein C antibody conjugated resin. eIF2Bα was protein C tagged. (D) Plot of fluorescence polarization signal before (black) and after incubation of FAM-ISRIB (2.5 nM) with 100 nM eIF2B(αβδγε)2 (red) or 100 nM eIF2B(αβδγε)2 + 6.0 μM eIF2α-P and varying concentrations of eIF2 (blue). For elution samples in (C), eIF2β, eIF2Bε, and eIF2Bα, and the eIF2Bδ and eIF2α-P blots are from the same gels, respectively. For input samples eIF2β and eIF2Bα, and the eIF2Bδ and eIF2α-P blots are from the same gels, respectively; eIF2Bε is from its own gel. Biological replicates: (A, B) n = 2; (D) n = 3. All error bars represent s.e.m.
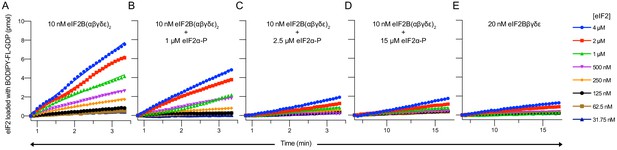
eIF2α-P decreases the initial velocity of eIF2B’s guanine nucleotide exchange factor (GEF) activity.
(A–E) Initial velocity of the eIF2B GEF reaction under varying conditions. Initial velocity was determined by a linear fit to timepoints acquired from 50 to 200 s (A–C) or 400–1000 s (D–E) after addition of eIF2B. For (A–E), representative replicates of n = 2 biological replicates are shown.
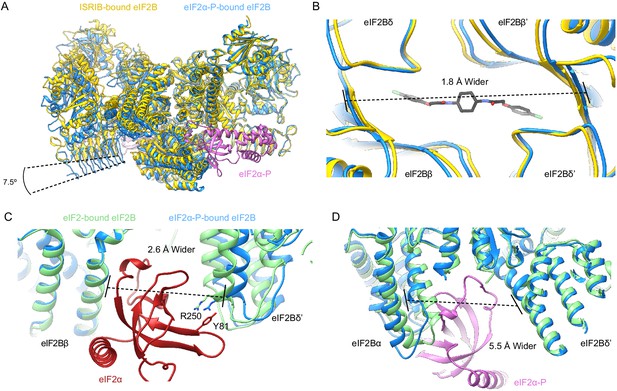
eIF2α-P binding conformationally inactivates eIF2B.
(A) Overlay of the ISRIB-bound eIF2B structure (PDB ID: 6CAJ) to the eIF2α-P-bound eIF2B structure (PDB ID: 6O9Z). The 7.5° hinge movement between the two eIF2B halves was measured between the lines connecting eIF2Bε H352 and P439 in the ISRIB-bound versus eIF2α-P-bound structures. (B) Zoom-in view of the ISRIB binding pocket upon eIF2α-P binding. The ~2 Å pocket lengthening was measured between eIF2Bδ and eIF2Bδ′ L482; the ‘prime’ to indicate the subunit of the opposing tetramer. ISRIB is shown in stick representation. (C) Overlay of eIF2-bound eIF2B (PDB ID: 6O85) and eIF2α-P-bound eIF2B. The 2.6 Å widening of the eIF2 binding site induced by eIF2α-P binding was measured between E139 and R250 of eIF2Bβ and eIF2Bδ′, respectively. The side chains involved in the key cation–π interaction between R250 in eIF2Bδ and Y81 in eIF2α that is lost due to pocket expansion are shown. (D) Overlay of the eIF2-bound eIF2B to the eIF2α-P-bound eIF2B. The 5.5 Å narrowing of the eIF2α-P binding pocket causing a steric clash between eIF2Bα and eIF2α-P in the eIF2-bound state was measured between eIF2Bα S77 and eIF2Bδ L314. ISRIB-bound eIF2B is colored in gold, eIF2α-P-bound eIF2B in blue, and eIF2-bound eIF2B in light green. eIF2α-P is shown in pink and eIF2α in red. ISRIB is colored in CPK.
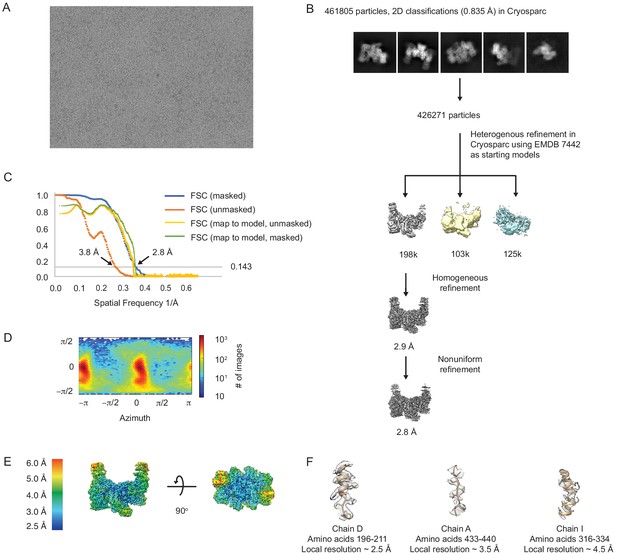
Cryo-electron microscopy workflow for apo-eIF2B decamer.
(A) Representative micrograph showing the quality of data used for the final reconstruction of the apo eIF2B structure. (B) Data processing scheme of the apo eIF2B. (C) Fourier shell correlation (FSC) plots of the 3D reconstructions of the apo eIF2B masked (dark blue), unmasked (orange), and map to model (yellow). (D) Orientation angle distribution of the apo eIF2B reconstruction. (E) Local resolution map of the apo eIF2B showing that the peripheral regions of the γ and α subunits are dynamic. (F) Electron microscopy maps of different regions of the apo eIF2B structure showing the quality of the data and the fit of the model. Regions close to the core (chain D, on the left) are well-resolved and have clear density for most side chains; regions close to the periphery of the molecule (chains A and I, middle and right) are less well-resolved due to higher flexibility.
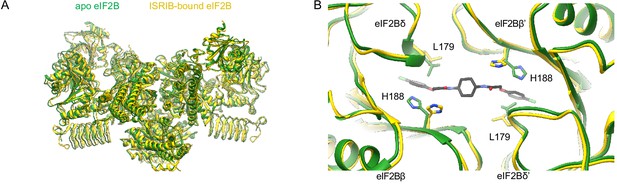
ISRIB binding induces local pocket changes.
(A) Overlay of ISRIB-bound eIF2B (PDB ID: 6CAJ) to the apo eIF2B (PDB ID: 7L70) showing both structures share a similar global conformation. (B) Zoom-in view of the ISRIB binding pocket showing that in the apostate L179 occupies a position in the ISRIB binding pocket that would clash with ISRIB binding. H188 changes its rotameric conformation upon ISRIB binding. The apo eIF2B is shown in green, and the ISRIB-bound eIF2B in gold. ISRIB is shown in stick representation, colored in CPK.
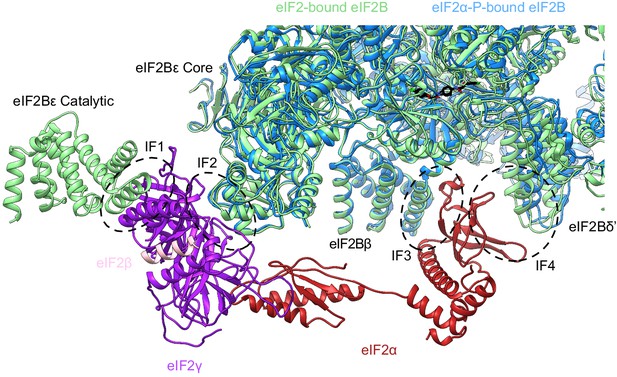
eIF2-P binding pulls IF4 away but leaves IF1–IF3.
Overlay of eIF2-bound eIF2B (PDB ID: 6O85) and eIF2α-P-bound eIF2B (PDB ID: 6O9Z). IF4 is pulled away from IF3 by 2.6 Å but IF1 (eIF2Bε Catalytic and eIF2γ), IF2 (eIF2Bε Core and eIF2γ), and IF3 (eIF2Bβ and eIF2α) remain available for eIF2 binding. eIF2α-P-bound eIF2B is shown in blue and eIF2-bound eIF2B in light green. eIF2γ is shown in purple, eIF2β in pink, and eIF2α in red. ISRIB is colored in CPK.
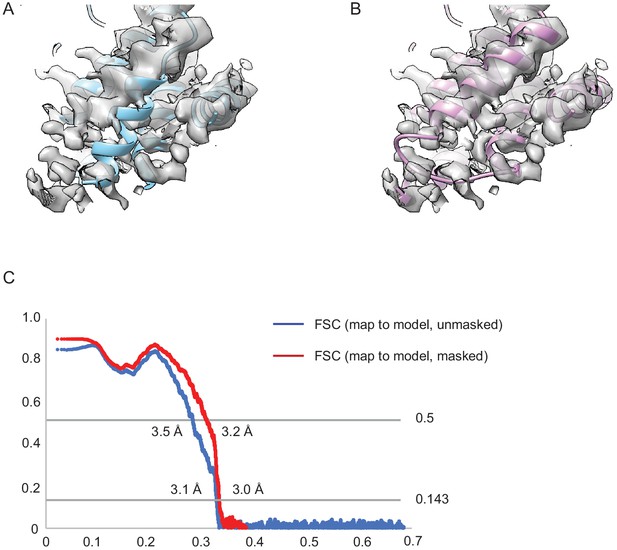
Re-refinement of the ISRIB-bound eIF2B decamer.
(A) The distal portion of the original model eIF2Bα from the ISRIB-bound eIF2B decamer placed within EMDB:7443 after lowpass filtering to 3.0 Å resolution. There is a helix (amino acids 44–56) out of place. The average CC value for the chains belonging to eIF2Bα from this model is ~0.74. (B) After manual adjustments in Coot and re-refinement in phenix.real_space_refine, this short helix is placed inside the cryo-electron microscopy (EM) density with an average CC value for the chains belonging to eIF2Bα of ~0.77. (C) The map-to-model Fourier shell correlation plots of the updated model. EM validation report.
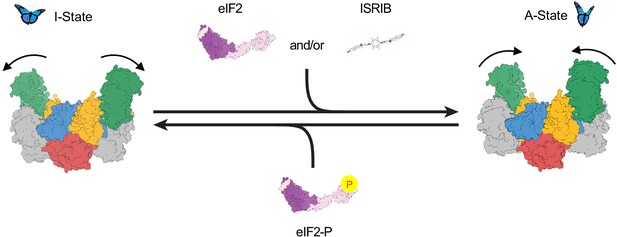
Model for modulation of eIF2B activity.
ISRIB and eIF2 binding to eIF2B stabilize the active, ‘wings up’ conformation of eIF2B (A-State) while both eIF2-P (as well as eIF2α-P alone; not shown) stabilize the inactive ‘wings down’ conformation of eIF2B (I-State), which cannot engage ISRIB and exhibits reduced enzymatic activity and eIF2 binding (akin to an eIF2Bβδγε tetramer). As indicated by the structure of the apo eIF2B decamer, the conformational equilibrium in the absence of ligand likely favors the A-State, which is further stabilized by substrate eIF2 and/or ISRIB binding but antagonized by eIF2-P binding.
Tables
Data collection, reconstruction, and model refinement statistics for the apo eIF2B decamer.
| Structure | Apo eIF2B decamer (PDB ID: 7L70; EMD-23209) |
|---|---|
| Data collection | |
| Microscope | Titan Krios |
| Voltage (keV) | 300 |
| Nominal magnification | ×105,000 |
| Exposure navigation | Image shift |
| Electron dose (e−Å−2) | 67 |
| Dose rate (e−/pixel/s) | 8 |
| Detector | K3 summit |
| Pixel size (Å) | 0.835 |
| Defocus range (μm) | 0.6–2.0 |
| Micrographs | 1699 |
| Reconstruction | |
| Total extracted particles (no.) | 461,805 |
| Final particles (no.) | 198,362 |
| Symmetry imposed | C1 |
| FSC average resolution, masked (Å) | 3.8 |
| FSC average resolution, unmasked (Å) | 2.8 |
| Applied B-factor (Å) | 92.4 |
| Reconstruction package | Cryosparc 2.15 |
| Refinement | |
| Protein residues | 3156 |
| Ligands | 0 |
| RMSD bond lengths (Å) | 0.004 |
| RMSD bond angles (o) | 0.978 |
| Ramachandran outliers (%) | 0.06 |
| Ramachandran allowed (%) | 3.81 |
| Ramachandran favored (%) | 96.13 |
| Poor rotamers (%) | 2.61 |
| CaBLAM outliers (%) | 2.00 |
| Molprobity score | 1.83 |
| Clash score (all atoms) | 4.77 |
| B-factors (protein) | 88.43 |
| B-factors (ligands) | N/A |
| EMRinger score | 2.68 |
| Refinement package | Phenix 1.17.1-3660-000 |
| FSC: Fourier shell correlation. | |
Oligos and sgRNAs.
| Oligo | Sequence | Use |
|---|---|---|
| oMS266 | /5InvddT/G*G*G*A*A*CCTCTTCTGTAACTCCTTAGC | Amplify HDR template |
| oMS267 | /5InvddT/C*C*T*G*A*G*GGCAAACAAGTGAGCAGG | Amplify HDR template |
| oMS269 | TCGTGCCAGCCCCCTAATCT | Validate eIF2Bα tagging |
| oMS270 | CTGAACGGCGCTGCTGTAGC | Validate eIF2Bα tagging |
| oMS256 | AGTGAACTCTACCATCCTGA | Validate eIF2Bβ tagging |
| oMS258 | TTAGGTGGACTCCTGTGC | Validate eIF2Bβ tagging |
| oMS096 | CTGGCTAACTGGCAGAACC | Validate eIF2Bδ tagging |
| oMS268 | AGAAACAAAGGCAGCAGAGT | Validate eIF2Bδ tagging |
| sgMS001 | CAATCTGCTTAGGACACGTG | Target Cas9 to eIF2BβC-terminus |
| sgMS004 | AGAGCAGTGACCAGTGACGG | Target Cas9 to eIF2Bδ C-terminus |
| sgMS006 | GAGGACGCCATGGACGACAA | Target Cas9 to eIF2Bα N-terminus |
| HDR: homology-directed recombination. | ||
Antibodies for western blotting.
| Antibody target | Host | Dilution | Manufacturer | Blocking conditions |
|---|---|---|---|---|
| GAPDH | Rabbit | 1/2000 | Abcam | TBS-T + 3% BSA |
| eIF2Bα | Rabbit | 1/1000 | ProteinTech | TBS-T + 3% milk |
| eIF2Bβ | Rabbit | 1/1000 | ProteinTech | TBS-T + 3% milk |
| eIF2Bδ | Rabbit | 1/1000 | ProteinTech | TBS-T + 3% milk |
| eIF2Bε | Mouse | 1/1000 | Santa Cruz Biotechnology | PBS-T + 3% milk |
| ATF4 | Rabbit | 1/1000 | Cell Signaling | PBS-T + 3% milk |
| eIF2α-P | Rabbit | 1/1000 | Cell Signaling | PBS-T + 1% BSA |
| eIF2α | Rabbit | 1/1000 | Cell Signaling | PBS-T + 3% milk |
| eIF2β | Rabbit | 1/1000 | ProteinTech | PBS-T + 3% milk |
| BSA: bovine serum albumin. | ||||
Data collection, reconstruction and refinement statistics for the ISRIB-bound eIF2B decamer.
| Structure | ISRIB-bound eIF2B decamer from Janelia (PDB ID: 6CAJ) (Tsai et al., 2018) | ISRIB-bound eIF2B decamer from Berkeley (PDB ID: 6CAJ) (Tsai et al., 2018) |
|---|---|---|
| Data collection | ||
| Voltage (keV) | 300 | 300 |
| Nominal magnification | ×29,000 | ×29,000 |
| Per frame electron dose (e−Å−2) | 1.19 | 1.63 |
| Spherical aberration (mm) | 2.7 | 2.62 |
| Number of frames | 67 | 27 |
| Detector | K2 summit | K2 summit |
| Pixel size (Å) | 1.02 | 0.838 |
| Defocus range (μm) | −0.3 to −3.9 | −0.3 to −3.9 |
| Micrographs | 1780 | 1515 |
| Frame length (s) | 0.15 | 0.18 |
| Detector pixel size (μm) | 5.0 | 5.0 |
| Reconstruction using particles from both data sets after magnification rescaling | ||
| Particles following 2D classification | 202,125 | |
| FSC average resolution unmasked (Å) | 3.4 | |
| FSC average resolution masked (Å) | 3.0 | |
| Map sharpening B-factor | −60 | |
| Refinement PDB ID: 7L7G (update to 6CAJ); EMD-7443 | ||
| Protein residues | 3198 | |
| Ligands | 1 | |
| RMSD bond lengths (Å) | 0.004 | |
| RMSD bond angles (o) | 0.967 | |
| Ramachandran outliers (%) | 0.00 | |
| Ramachandran allowed (%) | 5.40 | |
| Ramachandran favored (%) | 94.60 | |
| Poor rotamers (%) | 1.00 | |
| Molprobity score | 1.81 | |
| Clash score (all atoms) | 7.95 | |
| B-factors (protein) | 65.93 | |
| B-factors (ligands) | 52.57 | |
| EMRinger score | 2.37 | |
| Refinement package | Phenix 1.17.1-3660-000 | |
| FSC: Fourier shell correlation. | ||