Latrophilin GPCR signaling mediates synapse formation
Figures
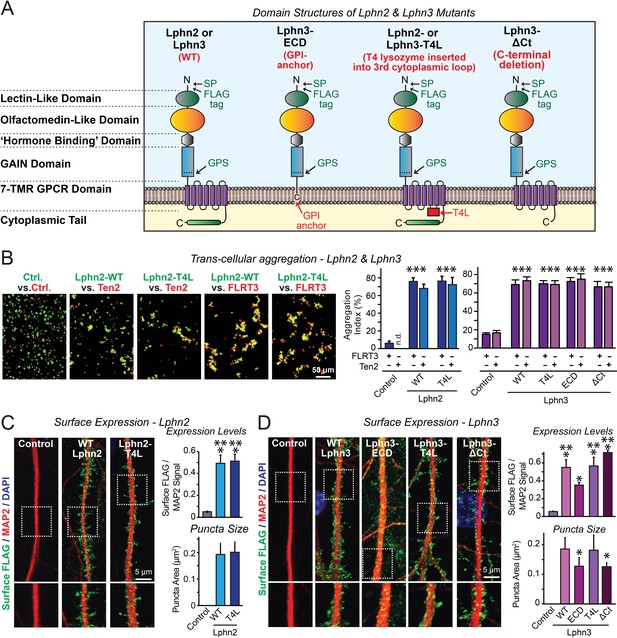
Latrophilin-2 (Lphn2) and Latrophilin-3 (Lphn3) signal transduction mutants engage in teneurin- and FLRT-mediated intercellular interactions on the cell surface.
(A) Domain structures of wild-type and mutant Lphn2 and Lphn3. Mutants contain either only the extracellular Lphn3 domains attached to the membrane via a GPI-anchor (Lphn3-ECD), block G-protein coupling (Lphn2-T4L and Lphn3-T4L), or truncate the intracellular C-terminal tail of Lphn3 (Lphn3-ΔCt). (B) Wild-type and mutant Lphn2 and Lphn3 proteins are robustly expressed on the cell surface of HEK293 cells and efficiently mediate trans-cellular adhesion by binding to FLRT3 and Ten2 ligands (left, representative images of transcellular aggregation assays as indicated (Ctrl = empty plasmid); right, quantification of the aggregation efficiency for Lphn2 and Lphn3 constructs). (C and D) Wild-type and mutant Lphn2 (C) and Lphn3 proteins (D) are robustly expressed on the cell surface of hippocampal neurons (left, representative images of dendritic segments of surface-labeled cultured hippocampal neurons infected with lentiviruses expressing the indicated proteins); right, summary graphs of the surface levels of FLAG-tagged Lphn3 forms relative to MAP2 signal, as well as the FLAG puncta area. Numerical data are means ± SEM. Statistical significance was assessed by two-tailed t-test (For C, puncta area) or one-way ANOVA with post-hoc Tukey tests for multiple comparisons (For B, C and D) (*** denotes p<0.001; * denotes p<0.05). For additional images and control experiments, see Figure 1—figure supplement 1.
-
Figure 1—source data 1
Latrophilin-2 (Lphn2) and Latrophilin-3 (Lphn3) signal transduction mutants engage in teneurin- and FLRT-mediated intercellular interactions on the cell surface.
- https://cdn.elifesciences.org/articles/65717/elife-65717-fig1-data1-v2.xlsx
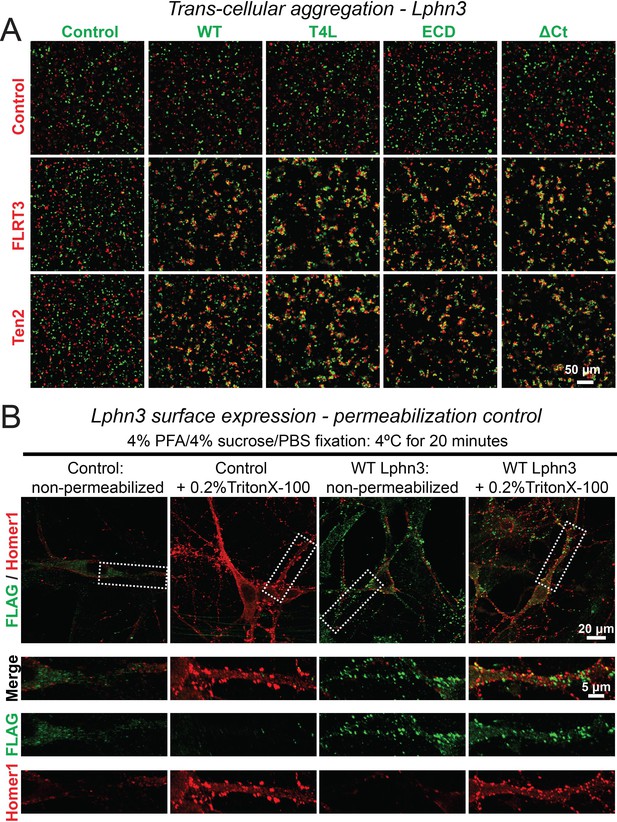
Representative images of aggregation assays for wild-type and mutant Lphn3 constructs (A) and cell-surface labeling control (B).
(A) Wild-type and mutant Lphn3 proteins used in rescue experiments are robustly expressed on the cell surface of transfected HEK293 cells and efficiently mediate trans-cellular adhesion by binding to FLRT3 and Ten2 ligands. Representative images show trans-cellular aggregation assays using the indicated Lphn3 proteins and either FLRT3 or Ten2 as ligands; for summary graph, see Figure 1B. (B) Cell fixation with 4% PFA/4% sucrose/PBS alone produced minor membrane permeability and exposure of intracellular antigens. Primary hippocampal cultures infected with lentiviral FLAG-WT Lphn3 or control cells were fixed with 4% PFA/4% sucrose/PBS for 20 min at 4°C and either un-permeabilized or treated with 0.2% Triton X-100/PBS for 5 min. Cells were subsequently labeled for FLAG together with the intracellular synaptic protein Homer1. Permeabilization exposed a substantial amount of Homer1 signal, thereby supporting that non-permeabilized conditions sample predominately surface protein.
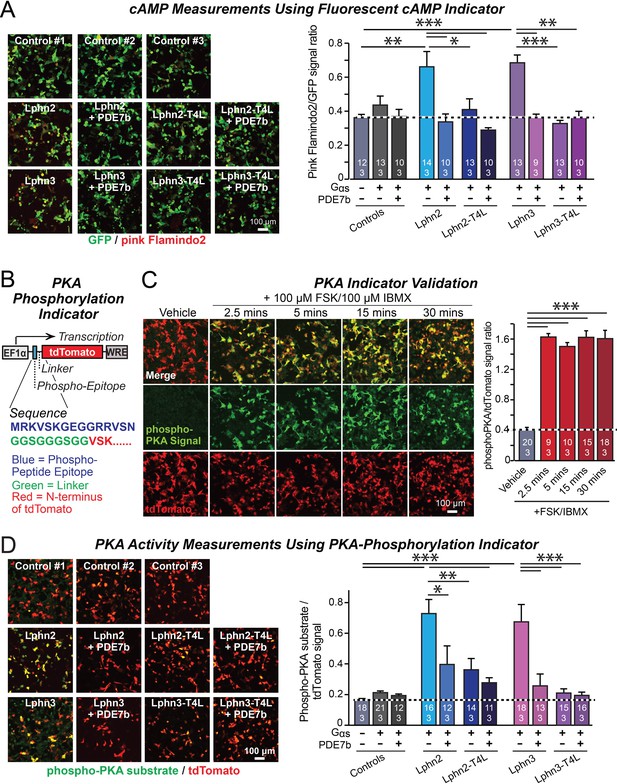
Lphn2- and Lphn3-induced cAMP signaling is blocked by insertion of T4-lysozyme into the third cytoplasmic loop.
(A) Expression of wild-type Lphn2 or Lphn3 in HEK293T cells together with Gαs and Gβγ constitutively induces cAMP accumulation. The T4L mutation and co-expression of the cAMP-specific phosphodiesterase PDE7b prevent cAMP increases by Lphn2 or Lphn3. The Pink Flamindo cAMP reporter (Odaka et al., 2014; Harada et al., 2017) was used to measure cAMP levels in live cells, and was quantified relative to the GFP signal as an internal standard. (B) Design of a new phospho-PKA indicator to measure PKA activation (top, plasmid design; bottom, sequences of the N-terminal PKA phospho-peptide substrate (blue) that was fused to the N-terminus of tdTomato (red) via a glycine-rich linker (green)). (C) Characterization of the phospho-PKA indicator in HEK293T cells. Treatment of cells with 100 µM Forskolin (FSK)/100 µM IBMX induces rapid phosphorylation of the phospho-PKA substrate (left, representative images; right, quantification of the immunocytochemical phospho-PKA signal relative to the tdTomato signal as an internal standard as a function of FSK/IBMX). (D) Lphn2 and Lphn3 constitutively activate PKA signaling in a manner blocked by the T4L-mutation. Experiments were performed as in A, but with the PKA-indicator described in B and C (left, representative images; right, summary graph). Numerical data are means ± SEM (number of analyzed ROIs/experiments in bars). Statistical significance was assessed by one-way ANOVA with post-hoc Tukey tests for multiple comparisons (For A, C and D) (*** denotes p<0.001; ** denotes p<0.01; * denotes p<0.05). For more representative images, see Figure 2—figure supplement 1.
-
Figure 2—source data 1
Lphn2- and Lphn3-induced cAMP signaling is blocked by insertion of T4-lysozyme into the third cytoplasmic loop.
- https://cdn.elifesciences.org/articles/65717/elife-65717-fig2-data1-v2.xlsx
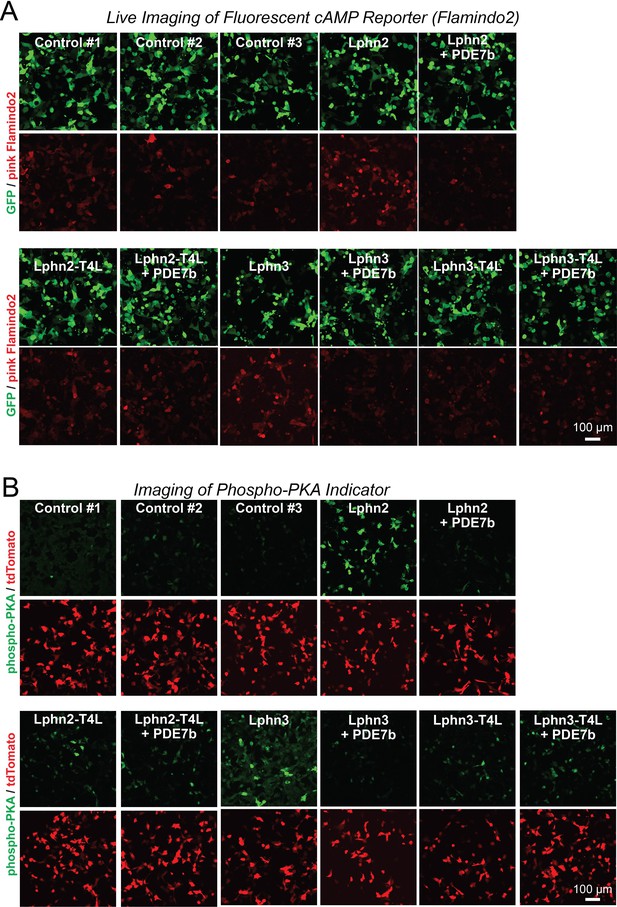
Representative images of cAMP measurements using Pink Flamindo (A) and of PKA activity measurements using a new reporter protein (B) in HEK293T cells expressing wild-type and mutant Lphn2 and Lphn3 constructs.
(A) Further representative images for the green GFP used as an internal control and the red pink Flamindo signal during the cAMP measurements shown in Figure 2A. (B) Further representative images for the green phospho-peptide and the red tdTomato signal used as an internal control during the PKA measurements shown in Figure 2D.
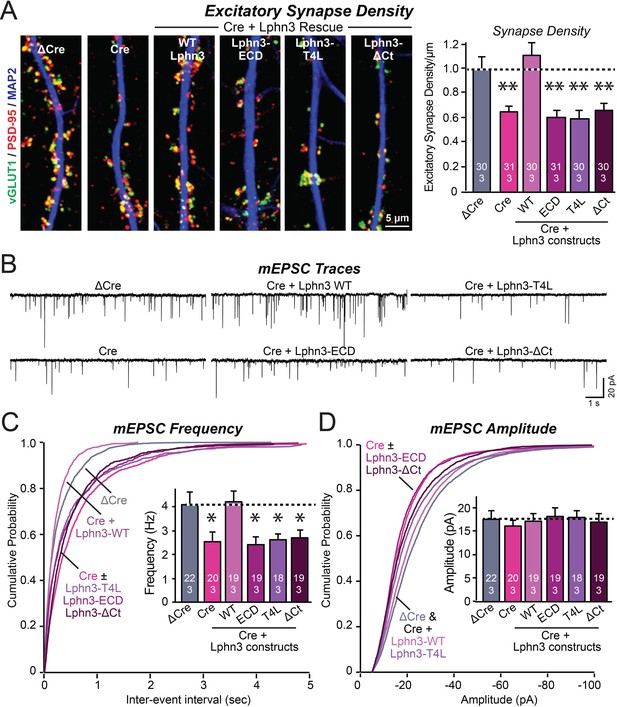
Mutations that disrupt Lphn3 signal transduction abolish the ability of Lphn3 to rescue the synapse formation in Lphn3-deficient cultured hippocampal neurons.
(A) Mutations of Lphn3 that delete its transmembrane regions (Lphn3-ECD), block GPCR signal transduction (Lphn3-T4L), or delete its cytoplasmic tail (Lphn3-ΔCt) abolish the ability of Lphn3 to rescue the decreased excitatory synapse density of Lphn3-deficient neurons despite engaging in surface-ligand interactions. Hippocampal neurons cultured from Lphn3 cKO mice were infected at DIV3 with lentiviruses encoding ΔCre (control) or Cre without or with co-expression of the indicated Lphn3 rescue proteins, and were analyzed at DIV14-16. (B–D) The same mutations as analyzed in A also abolish the ability of Lphn3 to rescue the decreased mEPSC frequency of Lphn3-deficient cultured hippocampal neurons. Data are means ± SEM (numbers of analyzed cells/experiments are indicated in bars). Statistical significance was assessed by one-way ANOVA with post hoc Tukey tests for multiple comparisons (For A, C and D) (** denotes p<0.01; * denotes p<0.05). See Figure 3—figure supplement 1 for additional expression controls and image data.
-
Figure 3—source data 1
Mutations that disrupt Lphn3 signal transduction abolish the ability of Lphn3 to rescue the synapse formation in Lphn3-deficient cultured hippocampal neurons.
- https://cdn.elifesciences.org/articles/65717/elife-65717-fig3-data1-v2.xlsx
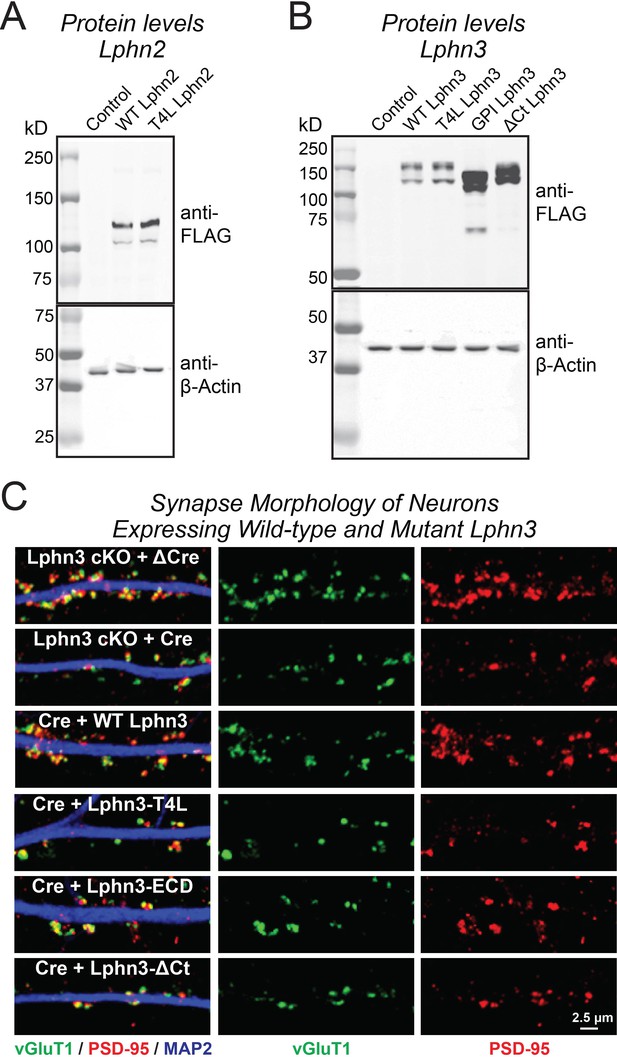
Immunoblotting analyses of neurons expressing Lphn2 (A) and Lphn3 constructs (B), and demonstration that expression of various Lphn3 constructs does not grossly alter synapse morphology (C).
(A and B) Immunoblotting for indicated FLAG-tagged Lphn2 (A) and Lphn3 proteins (B) in primary hippocampal cultures. β-Actin was used as a loading control. Wild-type and mutant Lphn2 and Lphn3 proteins used in rescue experiments are expressed at the expected molecular weight. (C) Representative high-magnification images of synapse morphology from Lphn3 rescue experiments shown in Figure 3.
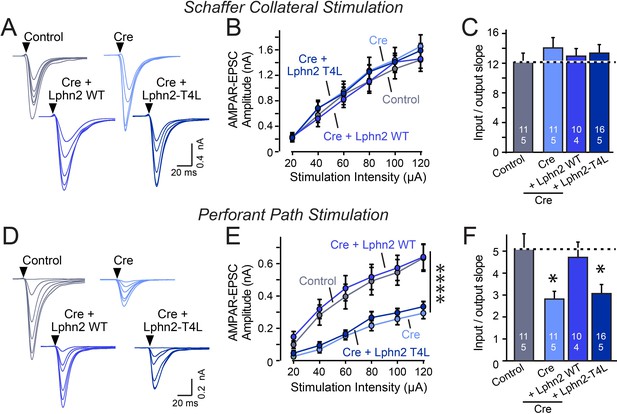
Abolishing GPCR signaling of Lphn2 by mutation of its third cytoplasmic loop blocks rescue of perforant-path synaptic connectivity in Lphn2-deficient CA1 region neurons in vivo.
Data are from patch-clamp whole-cell recordings in acute hippocampal slice from Lphn2 conditional KO mice at P21-25. The CA1 region of the mice was injected at P0 with low titers of lentiviruses expressing either Cre alone, or Cre together with the indicated rescue constructs to induce sparse infection of CA1 pyramidal neurons (Sando et al., 2019). Control recordings were from uninfected cells in slices in the opposite uninfected hemisphere. (A–C) Sparse conditional KO of Lphn2 in CA1-region neurons has no effect on the synaptic responses mediated by Schaffer-collateral inputs, and overexpression of wild-type or T4L-mutant Lphn2 does not alter these responses (A, sample traces; B, input-output curves; C, summary graph of the slopes of the input-output curves). (D–F) Sparse conditional KO of Lphn2 in CA1-region neurons impairs synaptic responses mediated by entorhinal cortex inputs in a manner that can be rescued by wild-type but not T4L-mutant Lphn2 (D, sample traces; E, input-output curves; F, summary graph of the slopes of the input-output curves). Data are means ± SEM (numbers of cells/mice are indicated in bars). Statistical significance was assessed by one-way (For C and F) or two-way ANOVA with post hoc Tukey tests for multiple comparisons (For B and E) (**** denotes p<0.0001; * denotes p<0.05). See Figure 4—figure supplement 1 for cellular capacitance and membrane resistance measurements.
-
Figure 4—source data 1
Abolishing GPCR signaling of Lphn2 by mutation of its third cytoplasmic loop blocks rescue of perforant-path synaptic connectivity in Lphn2-deficient CA1 region neurons in vivo.
- https://cdn.elifesciences.org/articles/65717/elife-65717-fig4-data1-v2.xlsx
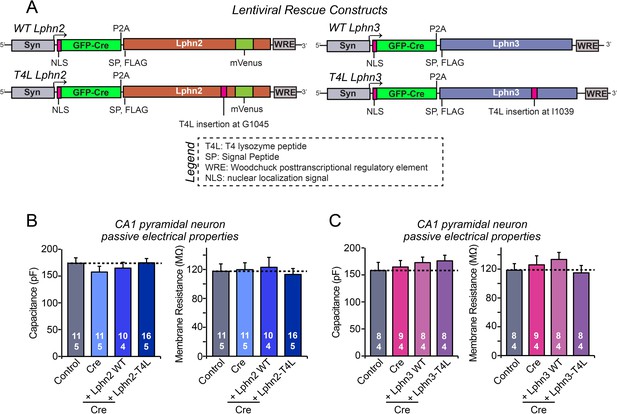
Intrinsic electrical properties of CA1 neurons are not significantly altered by the Lphn2 and Lphn3 deletions and by wild-type and T4L-mutant Lphn2 and Lphn3 rescues in the CA1 region of the hippocampus in vivo.
(A) Diagrams of the lentiviral shuttle plasmids used for in vivo rescue experiments. (B) Capacitance (left) and input resistance measurements (right) of neurons analyzed in acute slices for the Lphn2 in vivo rescue experiments shown in Figure 4. (C) Same as B, except that the data are from neurons analyzed in the Lphn3 in vivo rescue experiments shown in Figure 5. Data are means ± SEM (numbers of cells/mice are indicated in bars). Statistical significance was assessed by one-way ANOVA with post hoc Tukey tests for multiple comparisons.
-
Figure 4—figure supplement 1—source data 1
Intrinsic electrical properties of CA1 neurons in Lphn2 and Lphn3 rescue experiments.
- https://cdn.elifesciences.org/articles/65717/elife-65717-fig4-figsupp1-data1-v2.xlsx
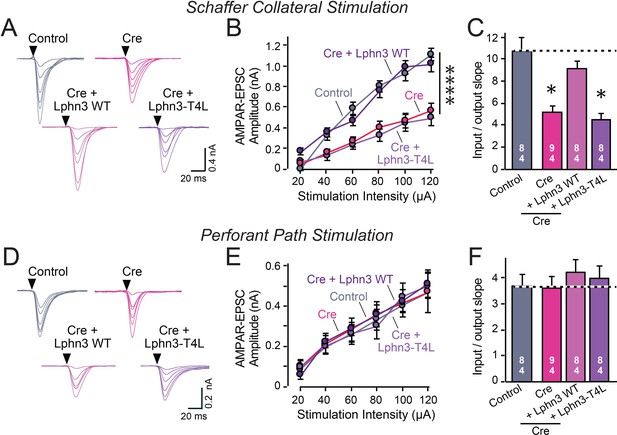
Abolishing Lphn3 GPCR activity by mutation of its third cytoplasmic loop blocks rescue of Schaffer collateral synaptic connectivity in CA1 region neurons in vivo.
Data are from patch-clamp whole-cell recordings in acute hippocampal slice from Lphn3 conditional KO mice at P21-25. The CA1 region of the mice was injected at P0 with low titers of lentiviruses expressing either Cre alone, or Cre together with the indicated rescue constructs to induce sparse infection of CA1 pyramidal neurons (Sando et al., 2019). Control recordings were from uninfected cells in slices in the opposite uninfected hemisphere. (A–C) Sparse conditional KO of Lphn3 in CA1-region neurons impairs synaptic responses mediated by Schaffer-collateral inputs in a manner that can be rescued by wild-type but not T4L-mutant Lphn3 (A, sample traces; B, input-output curves; C, summary graph of the slopes of the input-output curves). (D–F) Sparse conditional KO of Lphn3 in CA1-region neurons has no effect on the synaptic responses mediated by perforant pathway inputs, and overexpression of wild-type or T4L-mutant Lphn3 does not alter these responses (D, sample traces; E, input-output curves; F, summary graph of the slopes of the input-output curves). Data are means ± SEM (numbers of cells/mice are indicated in bars). Statistical significance was assessed by one-way (For C and F) or two-way ANOVA with post hoc Tukey tests for multiple comparisons (For B and E) (**** denotes p<0.0001; * denotes p<0.05). See Figure 4—figure supplement 1 for cellular capacitance and membrane resistance measurements.
-
Figure 5—source data 1
Abolishing Lphn3 GPCR activity by mutation of its third cytoplasmic loop blocks rescue of Schaffer collateral synaptic connectivity in CA1 region neurons in vivo.
- https://cdn.elifesciences.org/articles/65717/elife-65717-fig5-data1-v2.xlsx
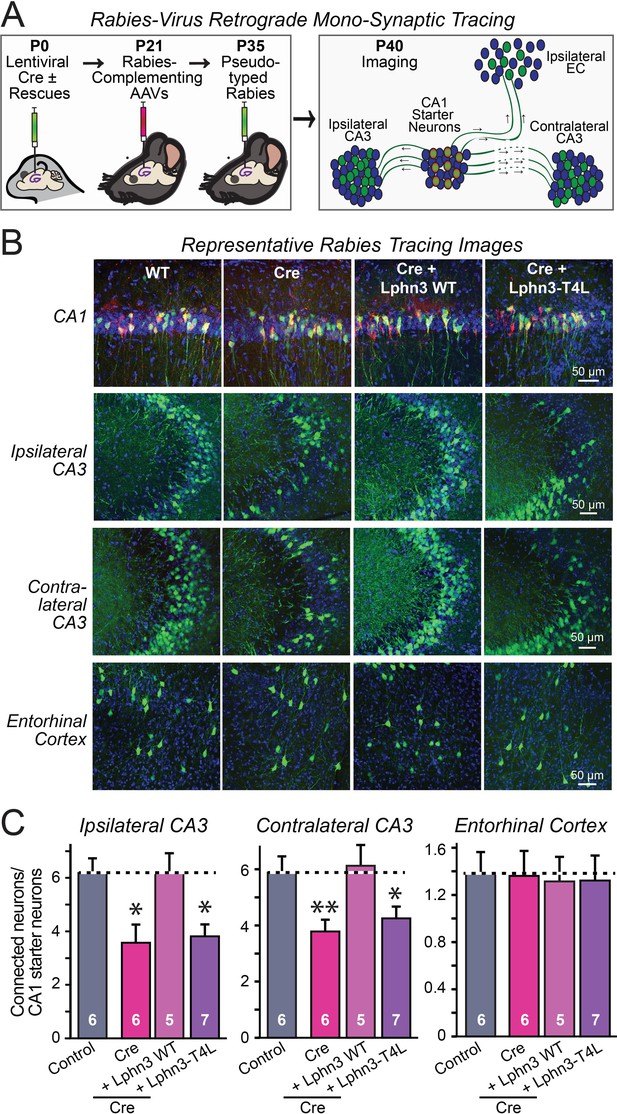
Retrograde mono-synaptic rabies virus tracing confirms that Lphn3 GPCR activity is essential for Schaffer-collateral synaptic connectivity in vivo.
(A) Schematic of the experimental approach. The CA1 region of the hippocampus of Lphn3 conditional KO or control mice was injected at P0 with lentiviruses encoding Cre without or with the indicated rescue constructs, at P21 with AAVs encoding Cre-dependent AAVs of rabies-complementing proteins, and at P35 with pseudotyped rabies virus. Mice were analyzed by imaging at P40. (B) Representative images of monosynaptic rabies-virus tracing experiments from the four conditions analyzed. (C) Synaptic connectivity quantifications for the indicated synaptic inputs to CA1 region pyramidal neurons starter cells in the hippocampal CA1 region. Rabies virus tracing demonstrates that the impairment of Schaffer-collateral synaptic connectivity induced by deletion of Lphn3 is rescued by wild-type Lphn3 but not by T4L-mutant Lphn3. Data are means ± SEM (numbers of mice are indicated in bars). Statistical significance was assessed by one-way ANOVA with post hoc Tukey tests for multiple comparisons (For C) (** denotes p<0.01; * denotes p<0.05).
-
Figure 6—source data 1
Retrograde mono-synaptic rabies virus tracing confirms that Lphn3 GPCR activity is essential for Schaffer-collateral synaptic connectivity in vivo.
- https://cdn.elifesciences.org/articles/65717/elife-65717-fig6-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | Lphn2 cKO | PMID:28972101 | JAX ID 023401 | |
| Genetic reagent (Mus musculus) | HA-Lphn3 cKO | PMID:30792275 | JAX ID 026684 | |
| Genetic reagent (Mus musculus) | CD-1 | Charles River Laboratory | Pure CD-1 background mice used to generate primary hippocampal cultures for surface expression experiments. | |
| Cell line (Homo sapiens) | HEK293T | ATCC | CRL-11268 | Mycoplasma testing was not performed since cells were maintained for a maximum of 10 passages. |
| Cell line (Homo sapiens) | HEK293F | Thermo Fisher | R79007 | Mycoplasma testing was not performed since cells were maintained for a maximum of 10 passages. |
| Recombinant DNA reagent | Lentiviral Syn NLS-GFP-Δcre | PMID:21241895 | Also used in PMID:30792275 | |
| Recombinant DNA reagent | Lentiviral Syn NLS-GFP-CRE | PMID:21241895 | Also used in PMID:30792275 | |
| Recombinant DNA reagent | pCMV WT hLphn3 | PMID:30792275, 26235030 | N-terminal IgK signal peptide followed by FLAG | |
| Recombinant DNA reagent | pCMV hFLRT3 | PMID:30792275, 26235030 | N-terminal preprotrypsin signal peptide followed by MYC | |
| Recombinant DNA reagent | pcDNA hTen2 | PMID:30792275, 26235030 | C-terminal FLAG; extracellular HA prior to transmembrane region | |
| Recombinant DNA reagent | pCAG WT rLphn2 | PMID:28972101, 30792275 | N-terminal preprotrypsin signal peptide followed by FLAG; intracellular mVenus | |
| Recombinant DNA reagent | pCMV Lphn3-ECD | This paper | hLphn3 residues 20–923 with an N-terminal NCAM1 signal peptide and a C-terminal NCAM1 GPI-anchoring sequence | |
| Recombinant DNA reagent | pCMV Lphn3-ΔCt | This paper | hLphn3 with a stop codon at residue 1117, causing deletion of the C-terminal tail (amino acids 1117–1447) | |
| Recombinant DNA reagent | pCMV Lphn3-T4L | This paper | hLphn3 with T4 lysozyme peptide inserted at Ile1039 in intracellular loop 3 | |
| Recombinant DNA reagent | pCAG Lphn2-T4L | This paper | rLphn2 with T4 lysozyme peptide inserted at Gly1045 in intracellular loop 3 | |
| Recombinant DNA reagent | Lentiviral Syn WT Lphn3 | PMID:30792275 | Lentiviral shuttle plasmid containing FLAG-tagged hLphn3 | |
| Recombinant DNA reagent | Lentiviral Syn Lphn3-ECD | This paper | Lentiviral shuttle plasmid containing Lphn3-ECD for rescue experiments in primary cultures | |
| Recombinant DNA reagent | Lentiviral Syn Lphn3-ΔCt | This paper | Lentiviral shuttle plasmid containing Lphn3-ΔCt for rescue experiments in primary cultures | |
| Recombinant DNA reagent | Lentiviral Syn Lphn3-T4L | This paper | Lentiviral shuttle plasmid containing Lphn3-T4L for rescue experiments in primary cultures | |
| Recombinant DNA reagent | Lentiviral Syn NLS-GFP-CRE p2a WT Lphn3 | PMID:30792275 | Lentiviral shuttle plasmid co-expressing NLS-GFP-Cre and WT hLphn3 for in vivo rescue experiments | |
| Recombinant DNA reagent | Lentiviral Syn NLS-GFP-CRE p2a Lphn3-T4L | This paper | Lentiviral shuttle plasmid co-expressing NLS-GFP-Cre and hLphn3-T4L for in vivo rescue experiments | |
| Recombinant DNA reagent | Lentiviral Syn NLS-GFP-CRE p2a WT Lphn2 | PMID:30792275 | Lentiviral shuttle plasmid co-expressing NLS-GFP-Cre and WT rLphn2 for in vivo rescue experiments | |
| Recombinant DNA reagent | Lentiviral Syn NLS-GFP-CRE p2a Lphn2-T4L | This paper | Lentiviral shuttle plasmid co-expressing NLS-GFP-Cre and rLphn2-T4L for in vivo rescue experiments | |
| Recombinant DNA reagent | Lentiviral EF1a phospho-PKA tdTomato | This paper | A PKA phosphopeptide substrate (MRKVSKGEGGRRVSN) was fused to the N-terminus of tdTomato separated by a linker (GGSGGGSGG) and was detected with the PKA phosphopeptide substrate antibody (sc-56941) | |
| Recombinant DNA reagent | pink Flamindo cAMP reporter | PMID:28779099; Addgene | RRID:addgene_102356 | |
| Recombinant DNA reagent | AAV CAG FLEX TVA-mCherry | PMID:26232228; Addgene | RRID:addgene_48332 | Rabies complementing AAV shuttle plasmid |
| Recombinant DNA reagent | AAV CAG FLEX rG | PMID:26232228; Addgene | RRID:addgene_48333 | Rabies complementing AAV shuttle plasmid |
| Antibody | Anti-HA mouse monoclonal | Covance | #MMS101R | 1:500 IHC; 1:1,000 ICC; 1:2,000 IB |
| Antibody | Anti-HA rabbit monoclonal | Cell Signaling Technologies | #3724 | 1:2,000 ICC; 1:1,000 IHC |
| Antibody | Anti-FLAG mouse monclonal | Sigma | #F3165 | 1:2,000 IB |
| Antibody | Anti-beta Actin mouse monoclonal | Sigma | #A1978 | 1:10,000 IB |
| Antibody | Anti-MAP2 chicken polyclonal | Encor | #CPCA MAP2 | 1:2,000 ICC |
| Antibody | Anti-PSD95 mouse monoclonal | Synaptic Systems | #124011 | 1:2,000 ICC |
| Antibody | Anti-vGLUT1 guinea pig polyclonal | Millipore | #AB5905 | 1:2,000 ICC |
| Antibody | Anti-Homer1 rabbit polyclonal | Synaptic Systems | #160003 | 1:2,000 ICC |
| Antibody | Anti-PKA phosphopeptide substrate mouse monoclonal | Santa Cruz Biotechnology | #sc-56941 | 1:1,000 ICC |
| Chemical compound, drug | Papain | Worthington | #LS003127 | |
| Chemical compound, drug | Matrigel | Corning | #356235 | |
| Chemical compound, drug | B-27 supplement | Gibco | #17504044 | |
| Chemical compound, drug | Cytosine arabinofuranoside | Sigma | #C6645 | |
| Chemical compound, drug | DAPI | Roche | #10236276001 | |
| Chemical compound, drug | QX-314 | Tocris | #1014 | |
| Chemical compound, drug | Picrotoxin | Tocris | #1128 | |
| Chemical compound, drug | Tetrodotoxin | Tocris | #1069 | |
| Chemical compound, drug | Freestyle MAX reagent | Life Technologies | #16447100 | |
| Software, algorithm | SnapGene | GSL Biotech | previously existing | |
| Software, algorithm | pClamp10 | Molecular Devices | previously existing | |
| Software, algorithm | Clampfit10 | Molecular Devices | previously existing | |
| Software, algorithm | NIS-Elements AR | Nikon | previously existing | |
| Software, algorithm | ImageJ | National Institutes of Health | previously existing | |
| Software, algorithm | Adobe Photoshop | Adobe | previously existing | |
| Software, algorithm | Adobe Illustrator | Adobe | previously existing | |
| Software, algorithm | Graphpad Prism 8.0 | Graphpad software | previously existing |





