RNF43 inhibits WNT5A-driven signaling and suppresses melanoma invasion and resistance to the targeted therapy
Figures
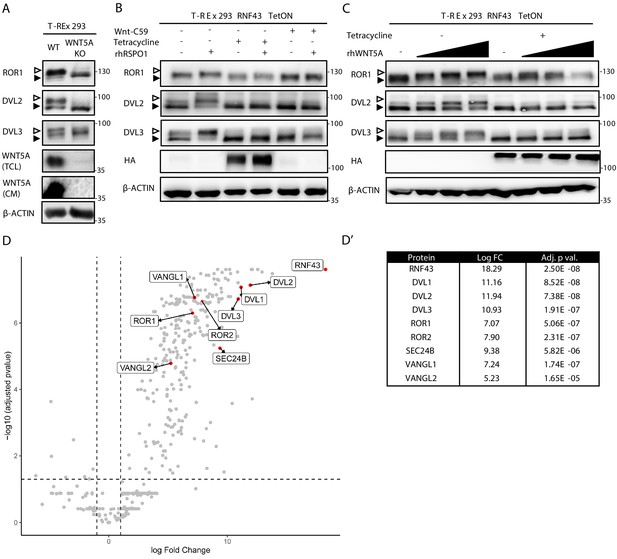
RNF43 interactome is enriched with the Wnt planar cell polarity pathway components.
(A) Western blot analysis of T-REx 293 wild type (WT) and WNT5A KO cells. Phosphorylation-dependent shifts of endogenous ROR1, DVL2, and DVL3 were suppressed upon WNT5A loss (TCL: total cell lysate; CM: conditioned medium). Signal of β-actin serves as a loading control. Empty arrowhead marks phosphorylation-dependent shift; black arrowhead indicates unphosphorylated protein. (B) Western blot showing activation of the noncanonical Wnt pathway components: ROR1, DVL2, and DVL3 upon rhRSPO1 overnight treatment (arrowheads as in A). Tetracycline-forced RNF43 overexpression (as visualized by HA tag-specific antibody) suppressed this effect. Inhibition of Wnt ligands secretion by the Porcupine inhibitor Wnt-C59 shows dependency of the rhRSPO1-mediated effect on endogenous Wnt ligands; representative blots from N = 3. (C) Western blot analysis of cellular responses to the increasing doses of rhWNT5A. ROR1 shift and phosphorylation of DVL2 and DVL3 (empty arrowhead) were inhibited upon tetracycline-induced RNF43-HA-BirA* overexpression. All samples were treated with Wnt-C59 Porcupine inhibitor to ascertain assay specificity to the exogenous rhWNT5A, N = 3. (D) Volcano plot of the RNF43 interactome identified by BioID and subsequent mass spectrometric detection (see Materials and methods for details). Significantly enriched proteins annotated as the components of the noncanonical Wnt signaling pathway are highlighted and their log fold change and adjusted p values are presented (D′). A full list of BioID-based identified interactors of RNF43 is presented in Figure 1—source data 1 and GO terms enrichment analysis in Figure 1—source data 2.
-
Figure 1—source data 1
BioID RNF43 interactors.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig1-data1-v1.xlsx
-
Figure 1—source data 2
gProfiler GO terms analysis.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig1-data2-v1.xlsx
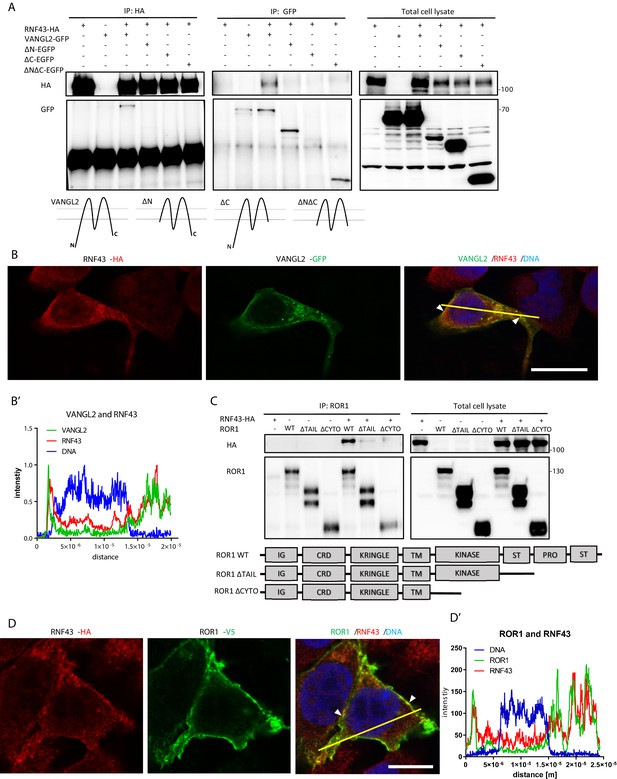
RNF43 interacts with Wnt/planar cell polarity (PCP) components.
(A) RNF43 interacts with VANGL2, but not with its mutants lacking N- or C-termini. VANGL2-EGFP and its variants (schematized) were overexpressed with RNF43-HA in Hek293 T-REx cells, immunoprecipitated by anti-HA and anti-GFP antibodies and analyzed by western blotting. Representative experiment from N = 3. Scheme illustrates secondary structure of the wild-type VANGL2 protein and its shortened variants used in this study. (B, B′) RNF43 (anti-HA, red) co-localized with transiently expressed VANGL2 (GFP, green). Co-localization was analyzed utilizing histograms of red, green, and blue channels signals along selection (yellow line) (B′). TO-PRO-3 Iodide was used to stain nuclei (blue). Scale bar: 25 μm. (C) RNF43 binds to the ROR1 and deletion of the intracellular part of ROR1 disrupts this interaction. RNF43-HA was detected in the ROR1 pull down prepared from lysates of Hek293 T-REx cells overexpressing RNF43-HA and ROR1-V5, N = 3. ROR1 wild-type and truncated mutants are represented in the scheme. (D, D′) RNF43 (anti-HA, red) co-localized with transiently expressed ROR1-V5 (anti-V5, green). Signals along selection (yellow line) were analyzed (D′). TO-PRO-3 was employed nuclei staining (blue). Scale bar: 25 μm. RNF43 interactions with VANGL1 and ROR2 are studied in Figure 2—figure supplement 1.
-
Figure 2—source data 1
RNF43 interacts with Wnt/planar cell polarity (PCP) components.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig2-data1-v1.xlsx
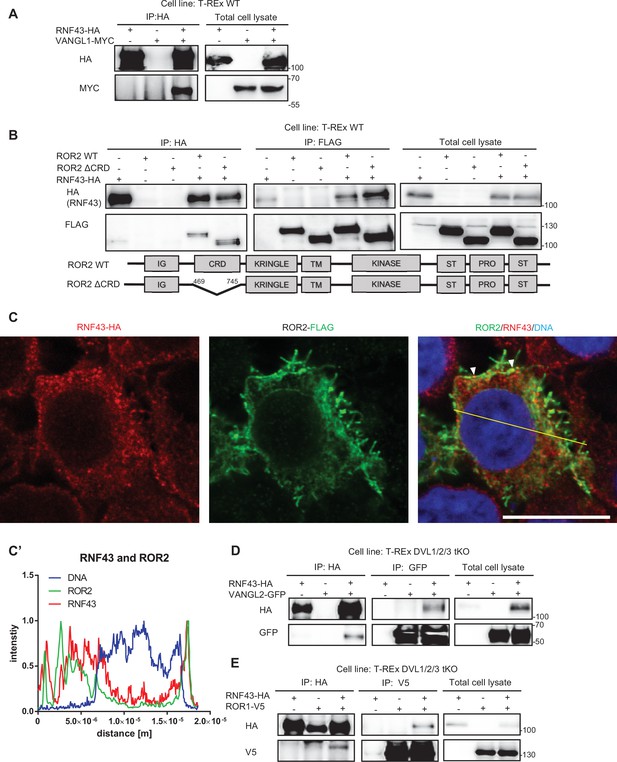
RNF43 interacts with Wnt/planar cell polarity (PCP) components.
A. RNF43 interacts with VANGL1. VANGL1-Myc was co-immunoprecipitated in the HA pull-down, prepared from lysate of Hek293 T-REx cells transiently overexpressing RNF43-HA and VANGL1-Myc, but not from the lysate containing only VANGL1-Myc overexpressed transgene, N=3. B. RNF43 interacts with the ROR2 in the CRD domain dispensable manner. Wild type ROR2 and ΔCRD-ROR2 mutant were detected in HA and FLAG pull downs, prepared from lysates of the Hek293 T-REx cells transiently overexpressing RNF43-HA and ROR2-FLAG or ΔCRD-ROR2, N=3. C., C’ Exogenous ROR2 (anti-FLAG, green) colocalizes with the RNF43-BirA*-HA (anti-HA, red) in the TetON Hek293 T-Rex cells. DNA was visualized by TO-PRO-3 Iodide. Scale bar represents 25 μm. Co-localization of ROR2 and RNF43 was analyzed utilizing histograms (C’) of red, green and blue channels along selection (yellow line). Data is present in the Figure 2—figure supplement 1—source data 1. D. RNF43 interacts with the VANGL2 in the absence of all three Disheveled isoforms. RNF43 binding to VANGL2 in the DVL1/2/3 deficient cells was confirmed in the two-directional co-IP assay, N=3. E. Interaction between ROR1-V5 and RNF43-HA is preserved in the DVL1-3 null cells. ROR1 was detected in the HA pull-down and RNF43 in the V5 immunoprecipitation. T-REx DVL1/2/3 tKO cells were transfected with highlighted plasmids, N=3.
-
Figure 2—figure supplement 1—source data 1
RNF43 interacts with Wnt/planar cell polarity (PCP) components.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig2-figsupp1-data1-v1.xlsx
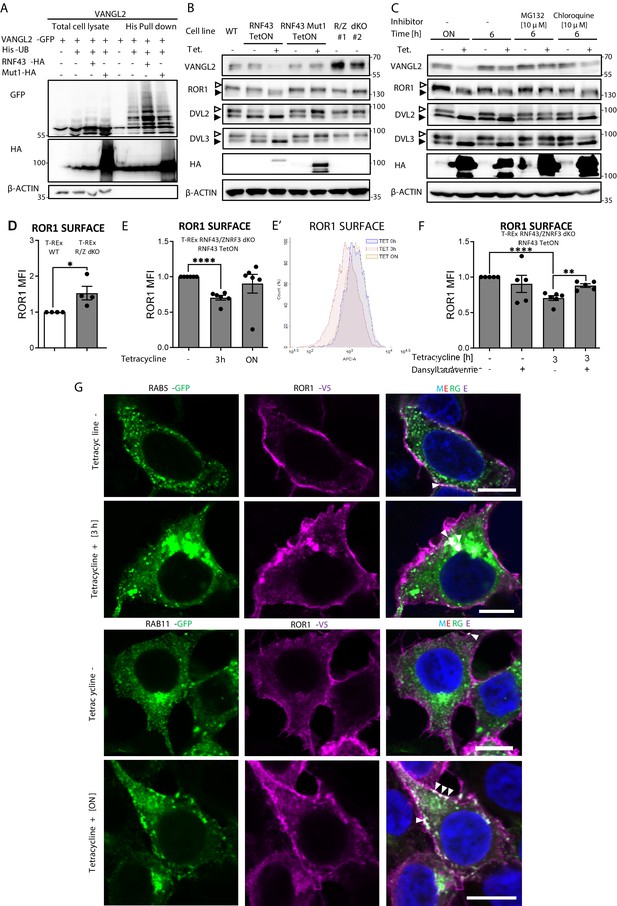
Mechanism of Wnt/planar cell polarity (PCP) inhibition by RNF43.
(A) Hek293 T-REx cells were transfected with plasmid encoding His-tagged ubiquitin, VANGL2-GFP and HA-tagged wild-type or Mut1 RNF43 constructs. Ubiquitinated proteins were enriched by His pull down and analyzed by western blotting. VANGL2 is ubiquitinylated by the E3 ubiquitin ligase RNF43, but not by its enzymatically inactive variant (RNF43Mut1). Representative experiment from N = 3. RNF43-mediated ubiquitination of DVL1 and DVL2 together in Figure 3—figure supplement 1. (B) Tetracycline-induced overexpression of the wt RNF43 (HA), but not enzymatically inactive RNF43Mut1 (HA), decreased VANGL2 protein level and suppressed phosphorylation of ROR1 and DVL3 (empty arrowhead; full arrowhead indicates unphosphorylated protein). CRISPR/Cas9-derived RNF43/ZNRF3 (R/Z) dKO cell lines #1 and #2 display phenotype reversed to the RNF43 overexpression. Quantified in Figure 3—figure supplement 1B, N = 3. (C) Inhibition of the proteasomal degradation pathway by MG132 (but not by lysosomal inhibitor chloroquine) blocked the RNF43 effects on ROR1, DVL2, DVL3 (empty arrowhead: phosphorylated; full arrowhead indicates unphosphorylated) and VANGL2 as shown by the western blotting analysis, N = 3. (D) Flow cytometric analysis of surface ROR1 in wild-type (WT) and RNF43/ZNRF3 (R/Z) dKO cells; unpaired two-tailed t-test: p=0.0298, N = 4. ROR1 was stained using ROR1-APC conjugate on the nonpermeabilized cells. Validation of the a-ROR1-APC antibody is shown in Figure 3—figure supplement 1D. (E, E′’) Surface ROR1 levels upon 3 hr and overnight (ON) induction of RNF43 in RNF43 TetON RNF43/ZNRF3 dKO cells; unpaired t-test p<0.0001, N = 6. Representative histogram of ROR1-APC signal in the analyzed conditions is shown (E′). (F) Dansylcadaverine, inhibitor of clathrin-mediated endocytosis, blocked the effect of RNF43 overexpression on surface ROR1, performed as in (E); unpaired t-test p=0.0037 3 hr Tet. vs 3 hr Tet.+ dansylcadaverine; N = 5 and 6 (3 hr Tet. condition). (G) Immunofluorescence imaging showed enhanced ROR1 (anti-V5, magenta) co-localization with the marker of early endosomes RAB5 (GFP, green) after 3 hr tetracycline treatment in RNF43 TetON RNF43/ZNRF3 dKO cells. RAB11-positive (GFP, green) recycling endosomes were recruited to the ROR1 (anti-V5, magenta) at the plasma membrane after overnight tetracycline treatment. Cells were transfected, treated, fixed, and stained. DNA was visualized by Hoechst 33342 (blue). Other tetracycline time points are shown in Figure 3—figure supplement 3A. Similar results were obtained for T-REx RNF43 TetON cell line (Figure 3—figure supplement 1E). Raw data used in (D–F) are given in Figure 3—source data 1.
-
Figure 3—source data 1
Mechanism of Wnt/planar cell polarity (PCP) inhibition by RNF43.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig3-data1-v1.xlsx
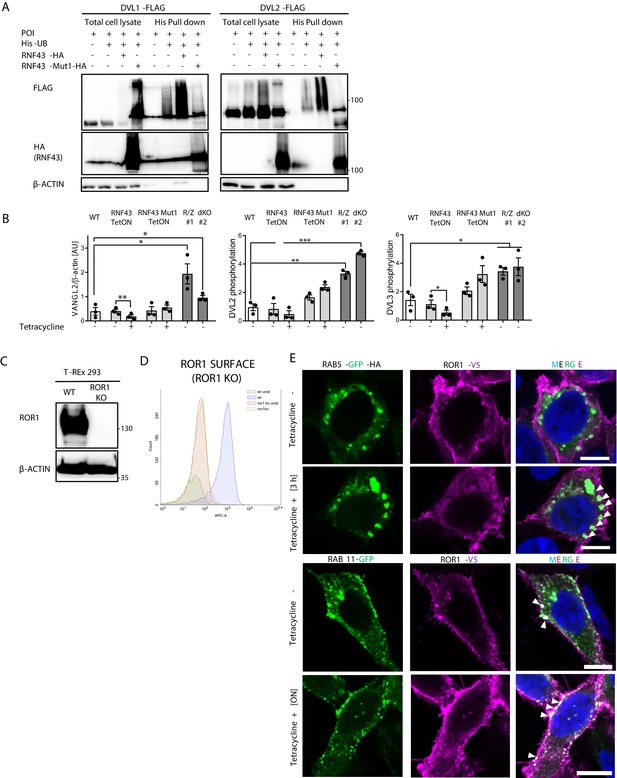
Mechanism of Wnt/planar cell polarity (PCP) inhibition by RNF43.
(A) DVL1 and DVL2 are ubiquitinylated by the E3 ubiquitin ligase RNF43, but not by its enzymatically inactive mutant (RNF43Mut1). Hek293 T-REx cells were transfected with plasmid encoding His-tagged ubiquitin, DVL1-FLAG or DVL2-FLAG, and wild-type or Mut1 RNF43 constructs and subjected to His-tag pull down and subsequent western blotting. N = 3. (B) Quantification of western blots from Figure 3B. Unpaired two-tailed t-test, *p<0.05, **p<0.01, ***p<0.001, N = 3. (C) Western blotting showing the lack of ROR1 protein in the T-REx 293 ROR1 KO cell lines. (D) T-REx ROR1 KO line was used for validation of the ROR1-APC antibody used for the flow cytometric determination of the ROR1 cell surface level. (E) Immunofluorescence imaging of enhanced ROR1 (anti-V5, magenta) co-localization with marker of early endosomes RAB5 (GFP, green) after 3 hr of tetracycline treatment in the T-REx RNF43 TetON cell line. RAB11 (GFP, green) was recruited to the ROR1 at plasma membrane upon overnight tetracycline exposition. DNA was visualized by Hoechst 33342 (blue). Scale bars represent 10 µm. Cells were treated 24 hr post-transfection for indicated time points. Images representing RAB5 control and ON tetracycline treatment together with RAB11 control and 3 hr tetracycline conditions are presented in Figure 3—figure supplement 3B. Data presented in (B) is presented in Figure 3—figure supplement 1—source data 1.
-
Figure 3—figure supplement 1—source data 1
Mechanism of Wnt/planar cell polarity (PCP) inhibition by RNF43.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig3-figsupp1-data1-v1.xlsx
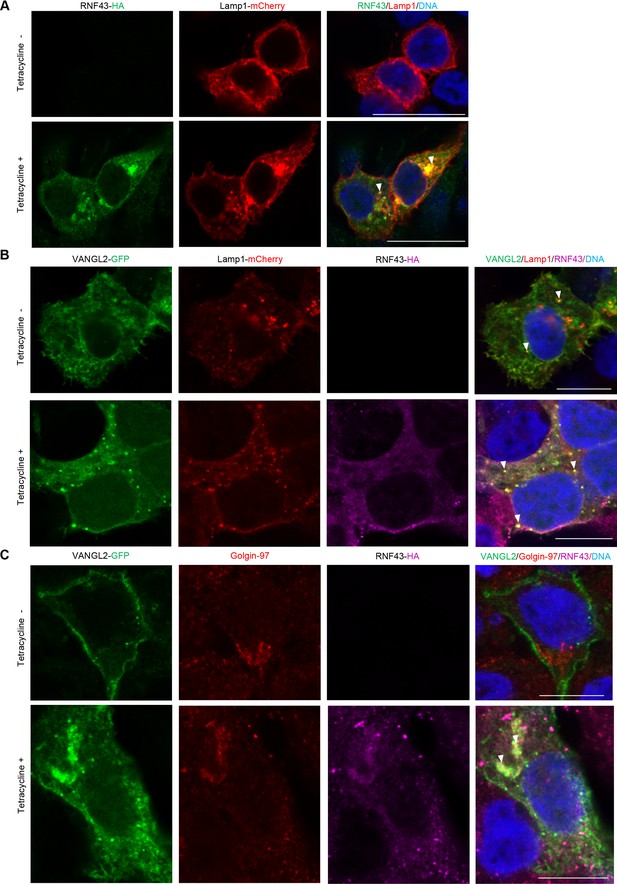
Mechanism of Wnt/planar cell polarity (PCP) inhibition by RNF43.
(A) Tetracycline-induced RNF43 (anti-HA, green) co-localized with transiently expressed lysosomal marker (Lamp1-mCherry, red). TO-PRO-3 Iodide was used to stain nuclei (blue). Scale bar: 25 μm. (B) Transiently overexpressed VANGL2 (GFP, green) and lysosomal marker (Lamp1-mCherry, red) in the presence and absence of tetracycline-induced RNF43 (anti-HA, magenta). Hoechst 33342 was used to stain nuclei (blue). Scale bar: 10 μm. (C) Tetracycline-forced expression of RNF43 (anti-HA, magenta) caused retention of VANGL2 (GFP, green) in Golgi complex (anti-endogenous Golgin-97, red). DAPI was used to stain nuclei (blue). Scale bar: 10 μm.
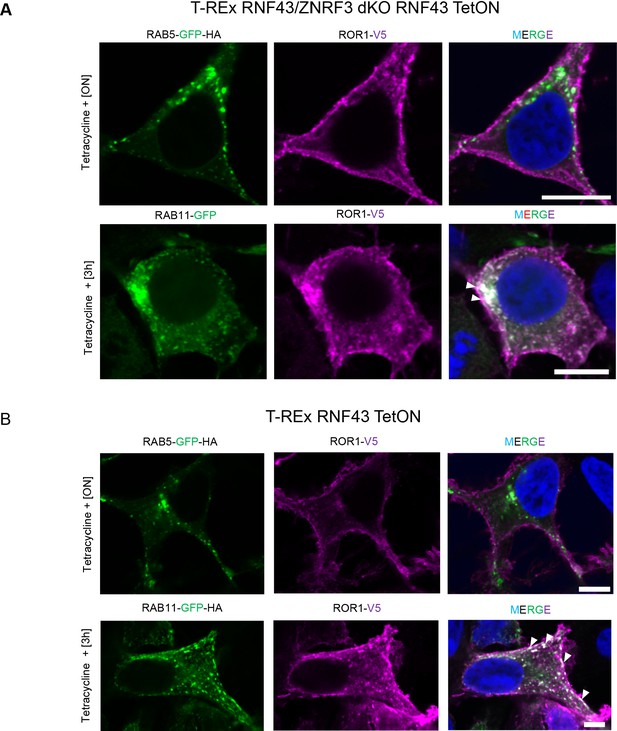
Mechanism of Wnt/planar cell polarity (PCP) inhibition by RNF43.
(A, B) Confocal imaging of the inducible T-REx RNF43/ZNRF3 dKO (A) and T-REx WT RNF43 TetON (B) and transfected with plasmids encoding ROR1-V5 (anti-V5, magenta) and RAB11-GFP-HA (GFP, green). 24 hr post-transfection, cells were treated with tetracycline for indicated time and then paraformaldehyde fixed, permeabilized, and stained for ROR1 (anti-V5) and nuclei detection (DAPI, blue). Scale bars represent 10 μm. Other tetracycline time points are presented in Figure 3 and Figure 3—figure supplement 1.
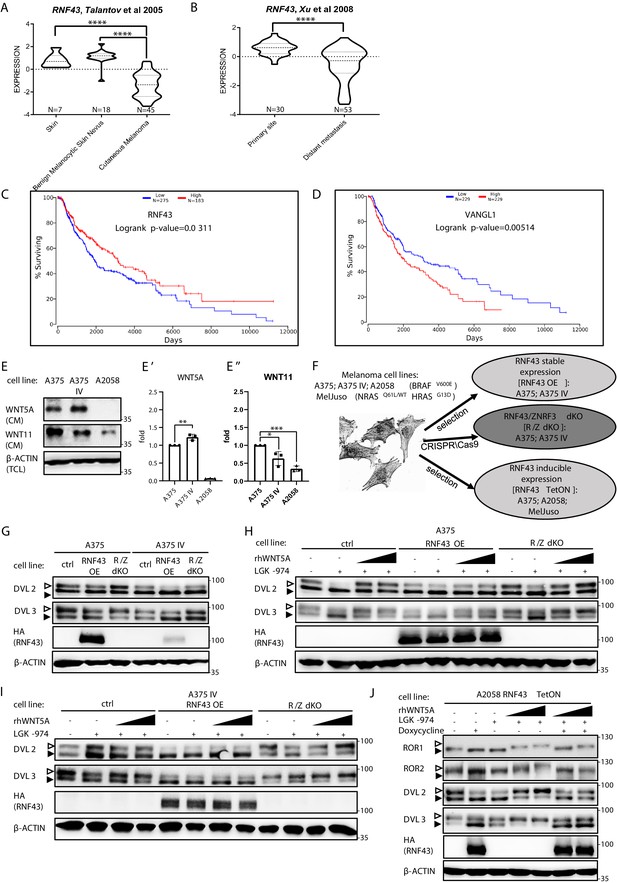
RNF43 in melanoma.
(A, B) RNF43 expression is lower in melanoma when compared with the skin and benign melanocytic skin nevus (A) and in the case of distant metastasis compared to the primary tumors (B), unpaired two-tailed t-test: ****p<0.0001. (C, D) RNF43 expression is a negative prognostic factor in melanoma. RNF43 low patients have shorter overall survival (logrank p-value=0.0311). Contrary, patients with low-expression VANGL1 (D) had longer survival (logrank test, p-value=0.00518). Expression of DVL3, VANGL1,and ZNRF3 is analyzed in Figure 4—figure supplement 1A–D. (E, E′, E′′) Culture media from melanoma A375, A375 IV, and A2058 cell lines were collected after 48 hr and analyzed by western blotting for the presence of WNT5A (E′) and WNT11 (E′′). Densitometric analysis has been done using the ImageJ software. Graphs represent ratio of corresponding WNT (medium) and β-actin (lysate) signals. Unpaired two-tailed t-test: p=0.0092 (E′); 0.0320 and 0.0001 (E′′); N = 3. (F) Schematic representation of the melanoma cell lines and their genetically modified variants used in this study. (G) Effects of the stable RNF43 overexpression and RNF43/ZNRF3 knockout in A375 and in its invasive derivate A375 IV. Exogenous RNF43 expression blocked DVL2 and DVL3 activation (arrowheads showing phosphorylation-dependent change of the electrophoretic mobility). Removal of endogenous RNF43 and ZNRF3 proteins had an opposite effect, N = 6. Quantification is given in Figure 4—figure supplement 1E,F. Expression of WNT5A, RNF43, ZNRF3, ROR1, DVL2, and DVL3 in tested cell lines was checked and shown in Figure 4—figure supplement 2. (H–J) Western blot showing inhibitory effect of RNF43 on A375 (H), A375 IV (I), and A2058 RNF43 TetON (J) cell lines in response to the 40 and 80 ng/ml 3 hr-long rhWNT5A treatments. RNF43/ZNRF3 dKO A375 (H), A375 IV (I) cell lines had stronger response to rhWNT5A than parental line. β-actin served as a loading control. Empty arrowhead: phosphorylated; full arrowhead: unphosphorylated protein. Porcupine inhibitor LGK-974 was used to block endogenous Wnt ligands secretion and RNF43 was probed by HA antibody, N = 3 and N = 4 (A2058). Quantification of DVL2 and DVL3 activation status and their protein levels in A2058 cells is given in Figure 4—figure supplement 1G and H. Figure 4—figure supplement 3 presents rhWNT5A treatment and consequences of RNF43-induced expression in RAS-mutant melanoma cell line MelJuso (A) and A375 (B) together with isogenic control (C). Figure 4—source data 1 contains raw data.
-
Figure 4—source data 1
RNF43 in melanoma.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig4-data1-v1.xlsx
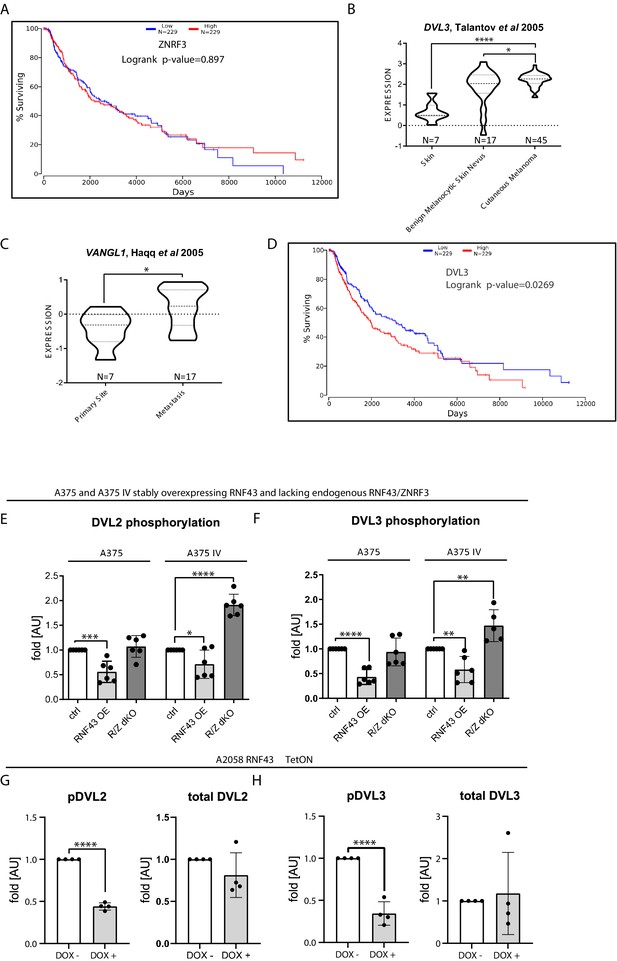
RNF43 in melanoma.
(A) ZNRF3 gene expression has no impact on melanoma patients’ survival (logrank p-value=0.897). (B) DVL3 expression level is elevated in human melanoma, unpaired two-tailed t-test, *p=0,0159, ****p<0.0001. (C) VANGL1 is more expressed in the metastasis than in primary melanoma, unpaired two-tailed t-test, p=0.0241. (D) High expression of DVL3 is a negative prognostic factor (50% lower and 50% upper percentiles). Logrank p-value=0.0269. (E, F) Quantification of western blots presented in Figure 4G. Exogenous RNF43 expression blocked in both tested cell lines DVL2 (E) DVL3 (F) phosphorylation-dependent shifts. CRISPR/Cas9-mediated knockout of RNF43/ZNRF3 resulted in the more activated DVL2 and DVL3 isoforms in case A375 IV cell line. Data were normalized to 1 for the parental cell lines values, unpaired two-tailed t-test: *p<0.05, **p<0.01, ***p<0.01, ****p<0.0001, N = 6 (F – A375 IV R/Z dKO N = 5). Total DVL2 and DVL3 protein levels accompanied by WNT5A, RNF43, ZNRF3, ROR1, DVL2, and DVL3 genes expression analysis are presented in Figure 4—figure supplement 2. (G, H) Phosphorylation and total protein levels of DVL2 (G) and DVL3 (H) in A2058 cells inducibly overexpressing RNF43, unpaired two-tailed t-test: ****p<0.0001, N = 4. Western blots are presented in Figure 4J. Figure 4—figure supplement 1—source data 1 contains raw results.
-
Figure 4—figure supplement 1—source data 1
RNF43 in melanoma.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig4-figsupp1-data1-v1.xlsx
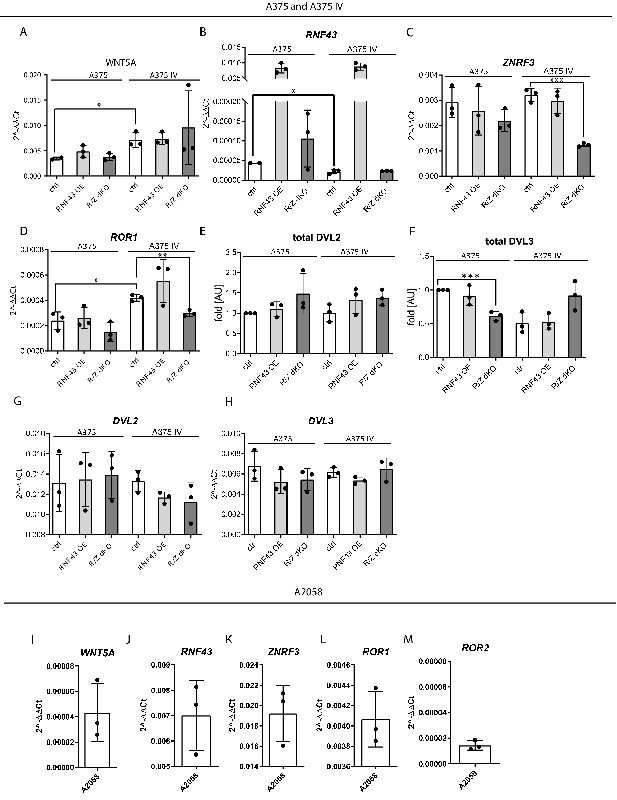
RNF43 in melanoma.
(A–D) RT-qPCR results – expression of the WNT5A (A), RNF43 (B), ZNRF3 (C), and ROR1 (D) genes was analyzed in the tested melanoma cells and presented as 2−ΔΔCt ± SD, two-tailed t-test: *p<0.05, **p<0.01, ***p<0.001; N = 3 (A375 N = 2 for A and B). Relative expression level was normalized to the B2M and GAPDH genes expression. (E, F) Western blot quantification results (Figure 4G) showing total levels of DVL2 (E) and DVL3 (F) not affected by RNF43 overexpression, nor by RNF43/ZNRF3 knockout. DVL3 protein level decreased only in the case of A375 R/Z dKO cells (F), unpaired two-tailed t-test: ***p<0.001, N = 3. Results were normalized to the A375 values. (G, H). RT-qPCR analysis of DVL2 (G) and DVL3 (H) genes expression in A375, A375 IV cell lines and their derivates. Relative expression level was normalized to the B2M and GAPDH genes expression values and presented as 2−ΔΔCt ± SD. Data used in (A–G) is presented in Figure 4—figure supplement 2—source data 1. (I–M) Expression analysis of WNT5A (I), RNF43 (J), ZNRF3 (K), ROR1 (L), and ROR2 (M) genes in A2058 cells. Relative expression levels are presented as 2−ΔΔCt ± SD and normalized to the levels of B2M and GAPDH genes.
-
Figure 4—figure supplement 2—source data 1
RNF43 in melanoma.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig4-figsupp2-data1-v1.xlsx
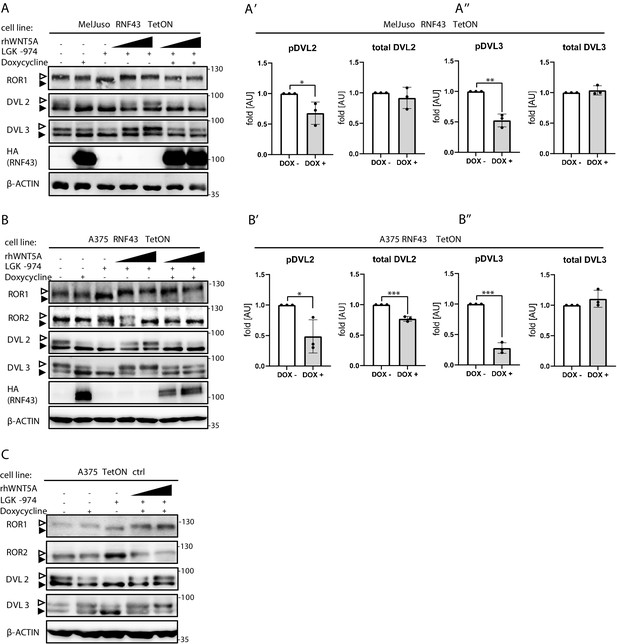
RNF43 in melanoma.
(A-A’’, B-B’’) Effects of the inducible RNF43 overexpression in RAS-mutant MelJuso (A) and BRAF V600E A375 (B) cells. Exogenous RNF43 expression blocked response to the 40 and 80 ng/ml 3 hr-long rhWNT5A treatments. Porcupine inhibitor LGK-974 was used to block endogenous Wnt ligands secretion and RNF43 was probed by HA antibody. β-actin is shown as a loading control. Phosphorylation status and total protein level of DVL2 (A’ B’) and DVL3 (A’’, B’’) were quantified, unpaired t-test *p<0.05, **p<0.01, ***p<0.001, N = 3. Results were normalized to 1, for the ‘DOX-’ condition. (C) Western blot analysis of control to A375 RNF43 TetON cell line. Cells were treated with doxycycline and Porcupine inhibitor LGK-974 for cells validation. Figure 4—figure supplement 3—source data 1 file presents raw data. Empty arrowhead: phosphorylated; full arrowhead: unphosphorylated protein.
-
Figure 4—figure supplement 3—source data 1
RNF43 in melanoma.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig4-figsupp3-data1-v1.xlsx
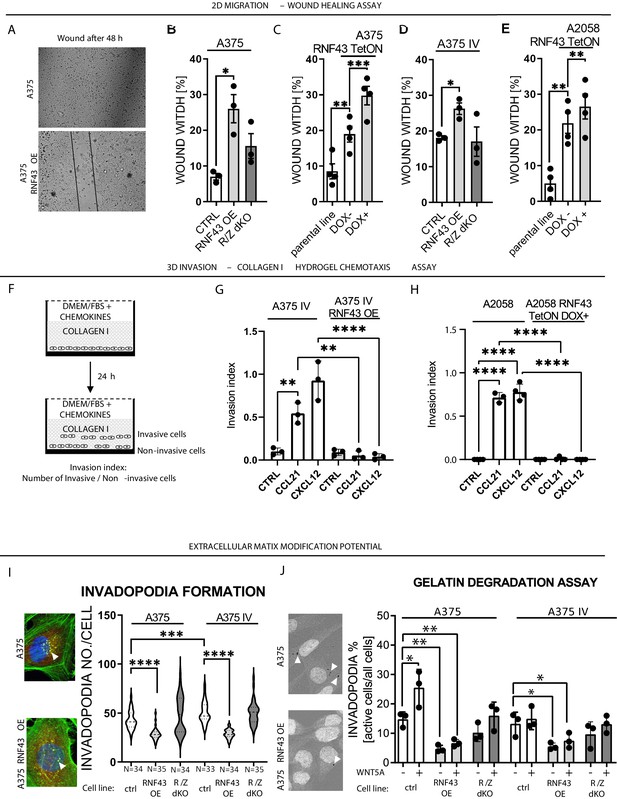
RNF43 inhibits WNT5A-dependent invasive properties of human melanoma.
(A–E) RNF43 reduced migration of A375 (B), A375 RNF43 TetON (C), A375 IV (D), and A2058 RNF43 TetON (E) in the wound healing assay. Wound was photographed 48 hr after scratch and presented as % of cell-free surface at the end of the experiment. Cells proliferation was suppressed by serum starvation, unpaired two-tailed t-test: *p<0.05, **p<0.01, ***p<0.001, N = 3 (B, D) or 4 (C, E). Representative photos at the end of the experiment are shown in (A) and in Figure 5—figure supplement 1A. (F–H) RNF43 blocked collagen I hydrogel 3D invasion in response to CCL21 (100 ng/ml) and CXCL12 (100 ng/ml) of A375 IV (G) and A2058 (H) cell lines. Cells were serum starved, collagen I (1.5 mg/ml) was overlaid and polymerized. Doxycycline was applied for RNF43 induction during starvation. After 24 hr, cells were fixed and stained for DNA (Hoechst 33342, blue) and F-actin (phalloidin, red) and imaged by confocal microscopy. Invasion index was calculated as the ratio of invaded cells at specified height to the number of noninvasive cells at the glass level, unpaired two-tailed t-test: **p<0.01, ****p<0.0001, N = 3 (G) or N = 4 (H). A375 cells did not invaded collagen I hydrogel (Figure 5—figure supplement 1B). Representative photos are presented in Figure 5—figure supplement 1C and C′. (I) RNF43 overexpression in A375 and A375 IV decreased number of invadopodia. Quantification of the invadopodia formed by melanoma cells, based on the analysis of confocal images. Number of cortactin/F-actin double-positive puncta in the individual cells was calculated in the ImageJ software, unpaired two-tailed t-test: ***p<0.001, ****p<0.0001. Examples of confocal imaging are shown: green, phalloidin; red, cortactin; blue, DNA. See Figure 5—figure supplement 2 for images from all experimental conditions. (J) Gelatin degradation assay; both A375 and A375 IV RNF43-overexpressing cell lines showed decreased capacity to locally degrade the extracellular matrix. rhWNT5A treatment induced gelatin degradation by A375 cells. Serum-starved cells were plated onto gelatin-Oregon Green-coated coverslips and incubated for 24 hr. Images obtained by Leica SP8 confocal microscope were analyzed for the presence of gelatin degradation by individual cells using ImageJ software, unpaired two-tailed t-test: *p<0.05, **p<0.01, N = 3. Example of gelatin degradation is shown; more pictures are presented in Figure 5—figure supplement 3 and Figure 5—figure supplement 4. Numerical data are given in Figure 5—source data 1 file.
-
Figure 5—source data 1
RNF43 inhibits WNT5A-dependent invasive properties of human melanoma.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig5-data1-v1.xlsx
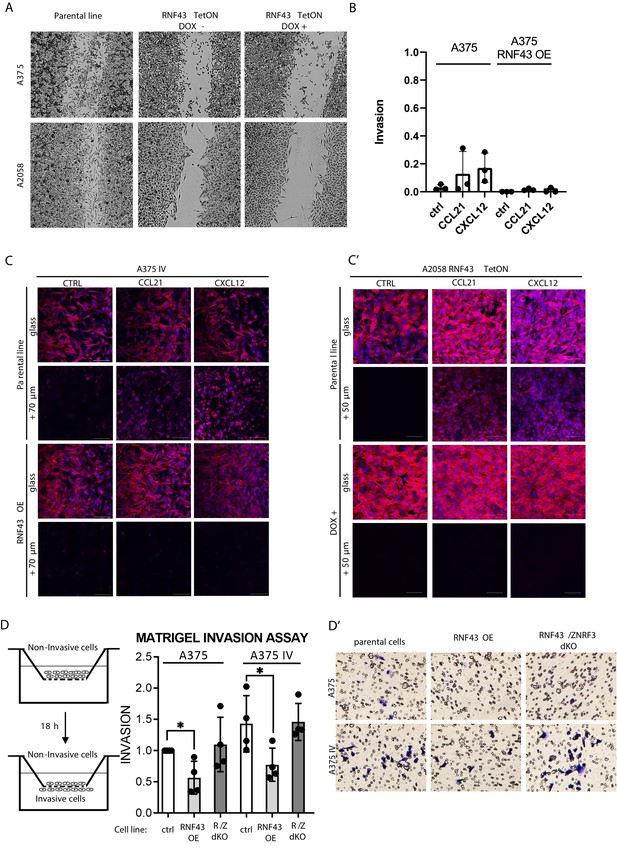
RNF43 inhibits Wnt5a-dependent invasive properties of human melanoma.
(A) Wound healing experiment – representative photos at the experimental end point (data Figure 5B–E). (B) Collagen I hydrogel chemotaxis assay A375 cell line results. After 24 hr, cells did not significantly respond to the treatments, unpaired two-tailed t-test: p>0.05, N = 3. (C, C′) Representative confocal photos of collagen I hydrogel chemotaxis assay for A375 IV (C) and A2058 (C′) cells (data presented in Figure 5G and H). (D, D′) Matrigel invasion assay – stable RNF43 overexpression-inhibited invasive properties of the A375 and A375 IV. Serum-starved cells were plated onto Matrigel-coated porous membrane. Medium containing 20% of serum was used as chemoattractant. After 18 hr of incubation, cells were fixed in methanol, noninvaded ones were removed from the upper part of transwell insert by cotton swab. Results were normalized to 1 for the number of invaded A375 cells, unpaired two-tailed t-test: *p<0.05, N = 4. (D′) Representative photos assay after crystal violet staining. Figure 5—figure supplement 1—source data 1 contains raw data.
-
Figure 5—figure supplement 1—source data 1
RNF43 inhibits Wnt5a-dependent invasive properties of human melanoma.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig5-figsupp1-data1-v1.xlsx
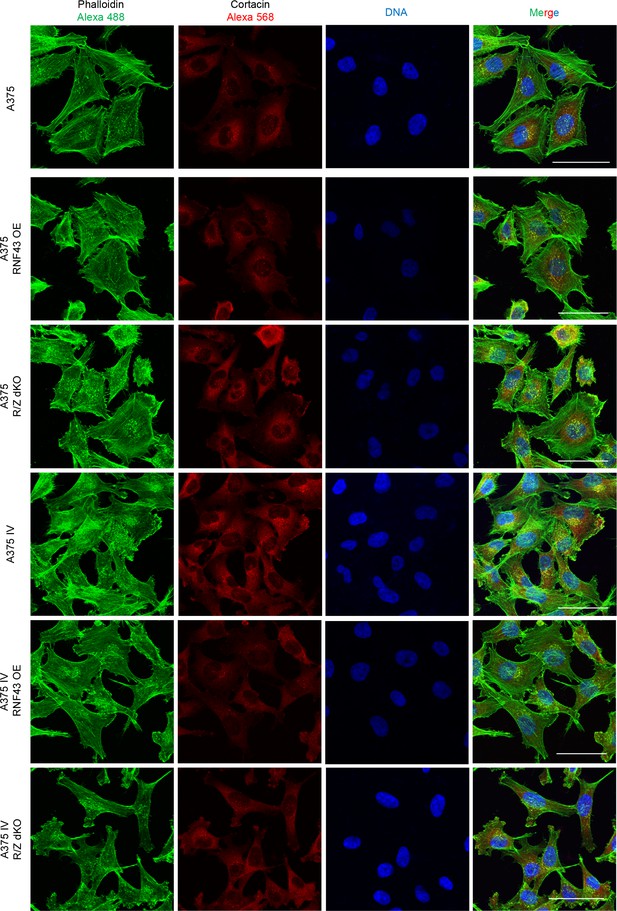
RNF43 inhibits Wnt5a-dependent invasive properties of human melanoma.
Confocal imaging of A375, A375 IV, and RNF43 overexpressing and RNF43/ZNRF3 double knockout cell lines modifications. Cells were paraformaldehyde fixed, Triton X-100 permeabilized and stained for cortactin by antibody (secondary antibody Alexa 568, red) and F-actin using fluorescent phalloidin conjugate (Alexa 488, green) and TO-PRO-3 Iodide for DNA visualization (blue). Number of double-positive puncta in single cells was quantified using ImageJ software. Scale bars represent 100 μm. Results are presented in Figure 5I.
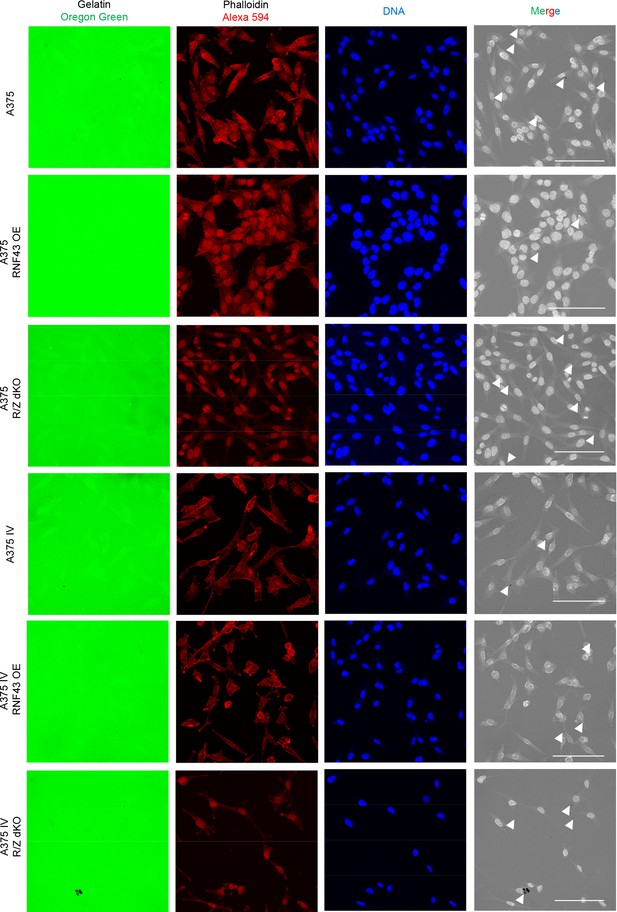
RNF43 inhibits Wnt5a-dependent invasive properties of human melanoma.
Confocal imaging of gelatin degradation assay without (Figure 5—figure supplement 3) and after (Figure 5—figure supplement 4) rhWNT5A treatment. Serum-starved cells were plated onto gelatin-Oregon Green (green)-coated coverslips and incubated for 24 hr. Fixed cells were stained with phalloidin-Alexa 594 for F-actin visualization (red) and TO-PRO-3 Iodide for nuclei (blue). Foci showing gelatin degradation are marked. Scale bars represent 50 μm. Experiment was repeated three times. Results are presented in Figure 5J.
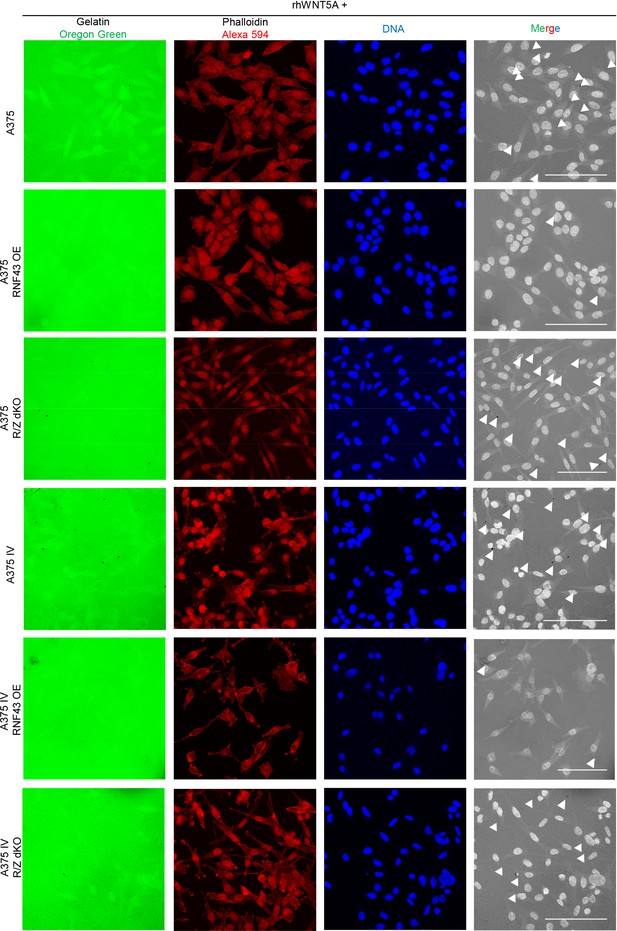
RNF43 inhibits Wnt5a-dependent invasive properties of human melanoma.
Confocal imaging of gelatin degradation assay after rhWNT5A treatment. Serum-starved cells were plated onto gelatin-Oregon Green (green)-coated coverslips and incubated for 24 hr. Fixed cells were stained with phalloidin-Alexa 594 for F-actin visualization (red) and TO-PRO-3 Iodide for nuclei (blue). Foci showing gelatin degradation are marked. Scale bars represent 50 μm. Experiment was repeated three times. Results are presented in Figure 5J.
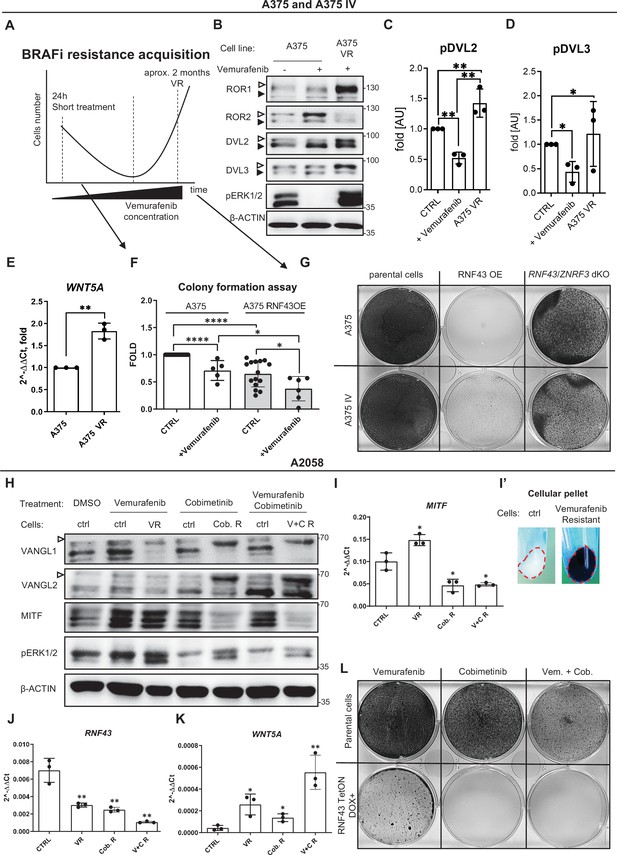
RNF43-overexpressing melanoma cells do not develop resistance to BRAF V600E targeted therapies.
(A) Scheme showing the experimental model used for the analysis of vemurafenib resistance (VR) acquisition. Melanoma cells are exposed to the increasing doses of the BRAF V600E inhibitor vemurafenib and following initial decrease in cell numbers recover and obtain capacity to grow in the presence of vemurafenib. (B–D) Western blot analysis of the cellular responses to the acute vemurafenib treatment (0.5 µM, 24 hr) in comparison to the signaling in VR cells growing in the presence of 2 µM vemurafenib. Transient treatment resulted in decreased DVL2 and DVL3 phosphorylation and increased ROR2 signal. In VR cells, ERK1/2 is constitutively phosphorylated in the vemurafenib presence. β-actin served as a loading control. A375 VR cells showed increased activation of DVL2 and DVL3 (empty arrowheads; quantifications in C and D) and higher expression of ROR1. Unpaired two-tailed t-test: *p<0.05, **p<0.01, N = 3. Figure 6—figure supplement 1A and B presents changes in ROR1 and ROR2 genes expression and quantification of ROR1 and ROR2 proteins signals. (E) Expression of WNT5A gene is elevated in the VR A375 cells in comparison to the parental line, unpaired two-tailed t-test: **p=0.0013, N = 3. Data presented are normalized to 1. Relative expression was normalized to the HSPCB and RPS13 genes (2−ΔΔCt). (F) Melanoma cell lines A375 and A375 IV overexpressing RNF43 showed decreased ability to grow and form colonies when seeded in the low density. After 7 days, colonies were fixed and stained with crystal violet. Paired (vemurafenib – vs+) and unpaired (A375 vs. IV) two-tailed t-tests: *p<0.05, ****p<0.0001, N ≥ 5. (G) RNF43-overexpressing A375 and A375 IV did not develop resistance to the BRAF V600E inhibition by vemurafenib treatment. Cells were cultured for approximately 2 months in the presence of increasing doses of the inhibitor. Photos show crystal violet-stained cultures at the end of the selection process. (H) A2058 cells (marked as ‘ctrl’) were exposed to the 24 hr-long treatments with vemurafenib (2.5 μM), cobimetinib (0.1 μM), or their combination. These short treatments were compared with cells cultured for approximately 2 months in the presence of vemurafenib (5 μM; VR), cobimetinib (0.5 μM; Cob. R) or their combination (V + C R). Experiment is schematized in Figure 6—figure supplement 1C’. Selected proteins were analyzed by western blot. Cells chronically exposed to inhibitors showed shifts of VANGL1 and VANGL2 bands (empty arrowhead) and changes in the MITF signal. Signal of pERK1/2 was used as treatments control. β-actin signal served as a loading control, N = 3. Figure 6—figure supplement 1C presents DVL2, DVL3 and total ERK1/2 signals. (I–K) Expression analysis of MITF (I), RNF43 (J), and WNT5A (K) genes. MITF is significantly upregulated in cells resistant to 5 μM Vemurafenib and downregulated in cells derived from the long-term culture in the presence of 0.5 μM cobimetinib alone and in combination with vemurafenib. (I′) VR A2058 show higher pigmentation. RNF43 expression decreased in the derived cells, while WNT5A increased. Relative expression levels are presented as 2−ΔΔCt ± SD and normalized to the levels of B2M and GAPDH genes, unpaired two-tailed t-tests: *p<0.05, **p<0.01, N = 3. Figure 6—figure supplement 1D–F shows expression analysis of ZNRF3, ROR1, and ROR2. (L) RNF43-overexpressing A2058 cells did not develop resistance to the cobimetinib and its combination with vemurafenib. Cells were cultured for approximately 2 months in the presence of increasing doses of the inhibitors (up to 5 μM for vemurafenib and 0.5 μM for cobimetinib), schematized in Figure 6—figure supplement 1C’. Figure 6—source data 1 presents raw data.
-
Figure 6—source data 1
RNF43-overexpressing melanoma cells do not develop resistance to BRAF V600E targeted therapies.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig6-data1-v1.xlsx
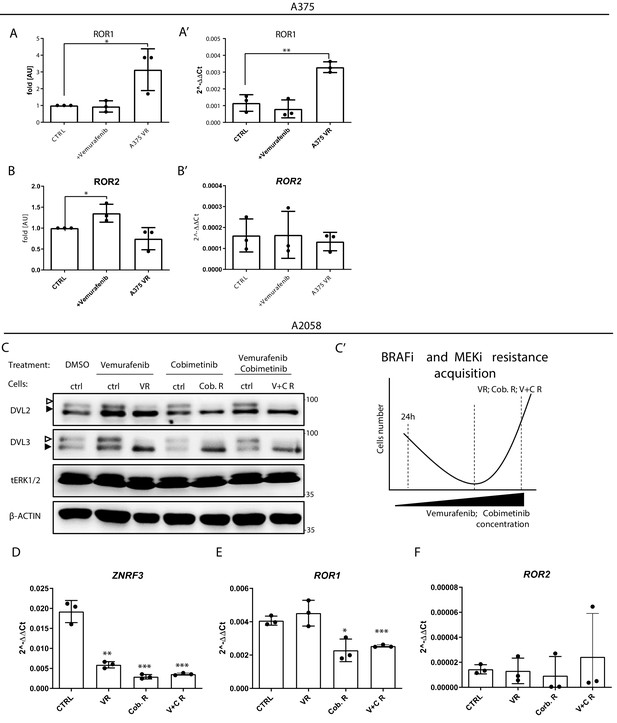
RNF43-overexpressing melanoma cells do not develop resistance to BRAF V600E targeted therapies.
(A, B) Protein levels of ROR1 (A) and ROR2 (B) (western blot shown in Figure 6B) were quantified using ImageJ software, two-tailed t-test: *p<0.05, **p<0.01; N = 3. Expression ROR1 (A′) and ROR2 (B′) genes in parental, cells transiently treated with BRAFi and vermurafenib-resistant A375 cells and presented as 2−ΔΔCt ± SD. Relative expression level was normalized to the HSPCB and RPS13. (C, C′) Western blot analysis of 24 hr and chronic response to vemurafenib, cobimetinib, and their combination in A2058 cells, continuation of Figure 6H. Empty arrowhead represents phosphorylation-dependent shift; full arrowhead indicates unphosphorylated status. Scheme of the experiment is presented in (C′). (D–F) Expression of ZNRF3 (D), ROR1 (E), and ROR2 (F) genes in A2058 control cells, resistant to vemurafenib (VR), cobimetinib (Cob. R), and double treatment (V + C R). ZNRF3 expression is decreased in the all-derived cells, while ROR1 in the cobimetinib and cobimetinib + vemurafenib-resistant lines. ROR2 did not change significantly. Relative expression levels are presented as 2−ΔΔCt ± SD and normalized to the levels of B2M and GAPDH genes, unpaired two-tailed t-tests: *p<0.05, **p<0.01, ***p<0.001, N = 3. Figure 6—figure supplement 1—source data 1 contains raw data.
-
Figure 6—figure supplement 1—source data 1
RNF43-overexpressing melanoma cells do not develop resistance to BRAF V600E targeted therapies.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig6-figsupp1-data1-v1.xlsx
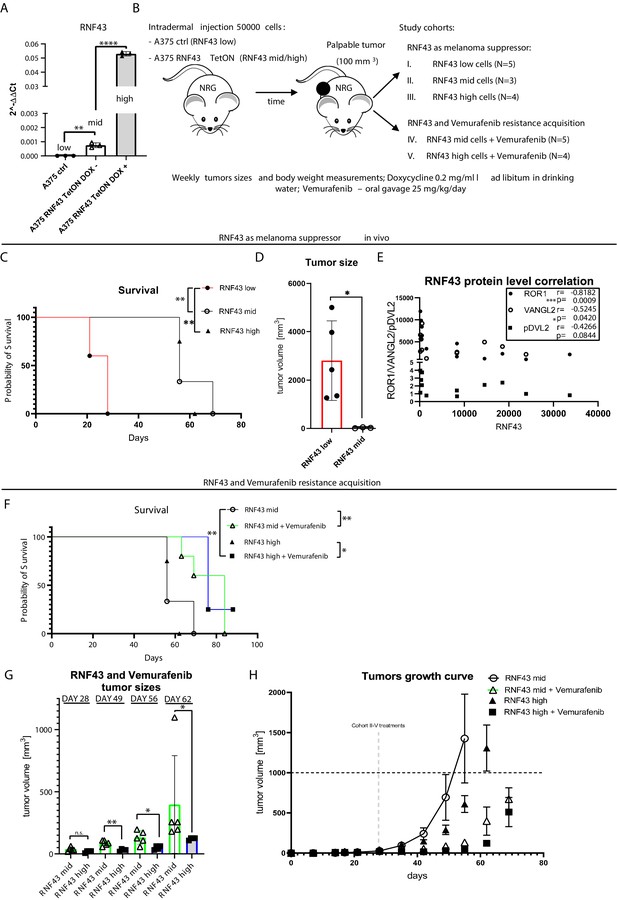
RNF43 inhibits melanoma proliferation and response to vemurafenib in vivo.
(A) RT-qPCR results – expression of the RNF43 gene in control (RNF43 low) and A375 RNF43 TetON cells in the absence (RNF43 mid) and presence of doxycycline (RNF43 high). Results are presented as 2−ΔΔCt ± SD, two-tailed t-test: **p<0.01, ****p<0.0001, N = 3. Relative expression level was normalized to the B2M and GAPDH genes expression. (B) Schematic representation of the in vivo experiment based in the immunodeficient NOD-Rag1null IL2rgnull mouse strain. A375 and its variants were injected intradermally (50,000 cells in PBS/mouse). Animals were divided into five cohorts once palpable tumors were formed. Ectopic RNF43 expression was induced by doxycycline presence in the drinking water. Vemurafenib was delivered daily by oral gavage as 25% Kolliphor in water formulation (see Figure 7—figure supplement 1A). Tumors sizes and animal weight were checked weekly. Mice were sacrificed when tumors reached approximately 1000 mm3. (C) Survival within cohorts I–III. Injection with cells having the lowest RNF43 expression led to the shortest mean survival (28 days). Experiment in cohorts II (56 days) and III (62 days) was significantly longer, Mantel–Cox test: *p<0.05, **p<0.01. (D) Cells expressing RNF43 higher than the endogenous level have impaired ability to proliferation in vivo. Tumor sizes at end point for the RNF43 low cohort (N = 5) with RNF43 mid (N = 3), two-tailed t-test: *p=0.0297. (E) RNF43 negatively regulates its targets also in vivo. Correlation analysis of RNF43 western blot signals with ROR1, VANGL2 protein levels, and phosphorylation status of DVL2 (shown in Figure 7—figure supplement 1D). Results for cohorts I–III were analyzed (N = 12), one-tailed Spearman correlation test, p and r values are shown in the graph. (F) Survival analysis showing vemurafenib treatment efficacy. Doxycycline treatment for RNF43 induction did not increase further vemurafenib effect. Mean survival: RNF43 mid: 56 days; RNF43 mid + vemurafenib: 84 days; RNF43 high: 62 days; RNF43 high + vemurafenib: 76 days, Mantel–Cox test: *p<0.05, **p<0.01. (G) RNF43 delays acquisition of vemurafenib resistance in vivo. Cohort V (N = 4) receiving doxycycline for RNF43 overexpression induction and vemurafenib (RNF43 high + vemurafenib) had significantly smaller tumors at days 49–62 than cohort IV (RNF43 mid + vemurafenib), tumors were not different at the treatments starting point (day 28) two-tailed t-test: *p<0.05, **p<0.01. (H) Tumor growth curve – caliper measurements of tumors sizes within experimental groups II–V. Treatment starting points are marked and presented in Figure 7—figure supplement 1B. All used data are presented in Figure 7—source data 1.
-
Figure 7—source data 1
RNF43 inhibits melanoma proliferation and response to vemurafenib in vivo.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig7-data1-v1.xlsx
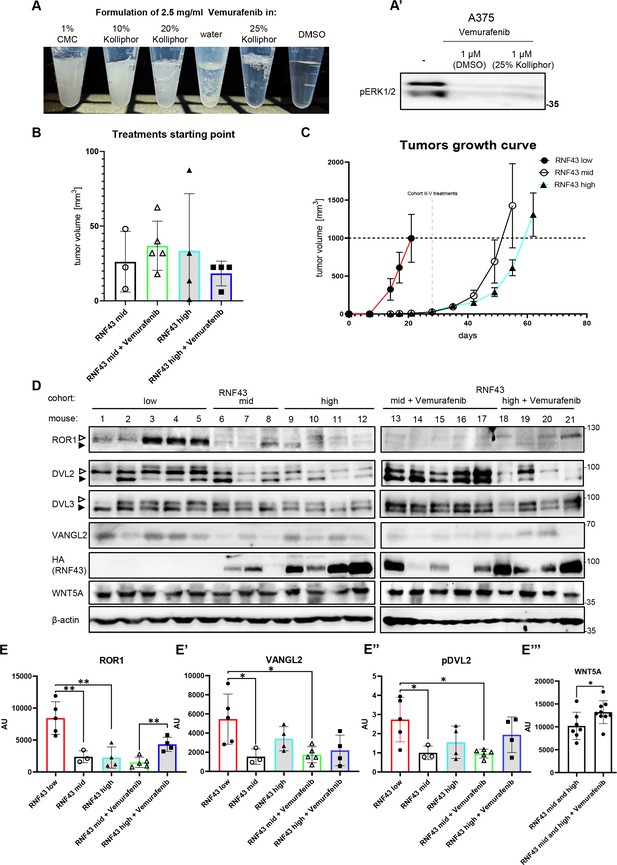
RNF43 inhibits melanoma proliferation and response to vemurafenib in vivo.
(A) Vemurafenib formulations test for the in vivo application. Vemurafenib 2.5 mg/ml was prepared in aqueous solutions of 1% carboxymethyl cellulose (CMC), 10, 20, and 25% Kolliphor. DMSO was used as reference. Vemurafenib was added to solutions from DMSO-based saturated stock. Formulation in 25% Kolliphor was selected for further tests. (A′) A375 cells were treated overnight with 1 μM vemurafenib in DMSO and in 25% Kolliphor formulation, lysed and phosphorylated ERK1/2 signal was used as readout for vemurafenib inhibitory properties. (B) Tumors sizes formed by A375 RNF43 TetON cells (RNF43 mid/high after following doxycycline supplementation) in cohorts II–V at the beginning of treatments. (C) Tumor growth curve – caliper measurements of tumors sizes within experimental groups I–III. (D) Western blot analysis of primary tumors at the experimental end points. Tissues were homogenized, lysed in the 1% SDS lysis buffer, sonicated and clarified by centrifugation, followed by addition of Laemmli buffer and thermal denaturation. Each sample contains 25 µg of total protein. RNF43 was probed by HA tag-specific antibody. Empty arrowhead represents phosphorylation-dependent shift; black arrowhead indicates unphosphorylated status. (E) Quantifications of tumor samples western blots. Tumors formed by RNF43 low cells had scientifically higher ROR1 (E) and VANGL2 levels (E′) and higher phosphorylation of DVL2 (E′′) than RNF43 mid-derived tumors. RNF43 high vemurafenib-resistant cells had higher ROR1 protein level than in the RNF43 mid cohort (E). Vemurafenib treatment led to the increased WNT5A protein level in tumors at the end of the experiment (E′′′). RNF43 low N = 5; RNF43 mid N = 3; RNF43 high N = 4; RNF43 mid + vemurafenib N = 5; RNF43 high + vemurafenib N = 4, unpaired two-tailed t-test *p<0.05, **p<0.01. All used data are presented in Figure 7—figure supplement 1—source data 1.
-
Figure 7—figure supplement 1—source data 1
RNF43 inhibits melanoma proliferation and response to vemurafenib in vivo.
- https://cdn.elifesciences.org/articles/65759/elife-65759-fig7-figsupp1-data1-v1.xlsx
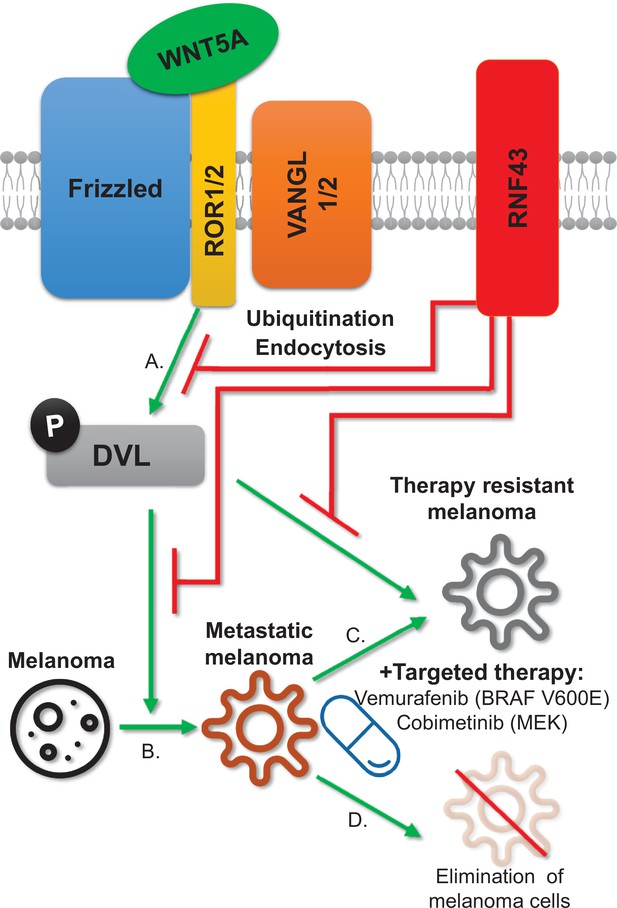
RNF43 inhibits WNT5A-driven signaling and suppresses melanoma invasion and resistance to the targeted therapy.
Graphical summary. RNF43 is an inhibitor of the noncanonical WNT5A-induced pathway. RNF43 interacts with receptor complexes of the Wnt/PCP signaling and its enzymatic activity results in the reduced cells sensitivity to WNT5A (A). In melanoma, WNT5A promotes invasion and metastasis (B) as well as resistance to targeted therapies, including treatments with vemurafenib – inhibitor of commonly mutated BRAF kinase and cobimetinib-targeting activity of the MEK enzyme (C). RNF43 blocks melanoma-invasive properties via interference with the nonconical Wnt pathway, leading to the increased sensitivity to treatment (D).
Tables
Cloning and mutagenesis primers.
| Primer | Sequence | Purpose |
|---|---|---|
| RNF43 BirA*F | ATGCAGTTAACATGAGTGGTGGCCACCAGCTG | RNF43 cDNA cloning into pcDNA3.1 MCS-BirA(R118G)-HA |
| RNF43 BirA*R | ATGCAGAATTCCACAGCCTGTTCACACAGCTCCT | |
| RNF43 InFusion F | GTTTAAACTTAAGCTTATGAGTGGTGGCCACCAG | RNF43-BirA(R118G)-HA into pcDNA4 |
| RNF43 InFusion R | AAACGGGCCCTCTAGACTATGCGTAATCCGGTACA | |
| RNF43-HA F | TTAAAGCTTATGAGTGGTGGCCACCAG | RNF43-HA cloning into pcDNA3 |
| RNF43-HA R | ATCGATATCTCAAGCGTAATCTGGAACATCGTATGGGTACACAGCCTGTTCACACAGCT | |
| pCW57-RNF43 InFusion F | ATTGGCTAGCGAATTATGAGTGGTGGCCACCAGC | pCW57-RNF43 generation |
| pCW57-RNF43 InFusion R | CGGTGTCGACGAATTTCAGGCGTAGTCGGGCACG |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | NOD-Rag1null IL2rgnull | Jackson Laboratory | RRID:BCBC_1261 | 9-week-old males |
| Cell line (Homo sapiens) | T-REx 293 | Thermo Fisher Scientific | R71007; RRID:CVCL_D585 | For TetON system using pcDNA4-TO backbone |
| Cell line (Homo sapiens) | T-REx 293 RNF43 TetON | This publication | Cells inducibly overexpressing RNF43 | |
| Cell line (Homo sapiens) | T-REx 293 RNF43 Mut1 TetON | This publication | Cells inducibly overexpressing inactive RNF43 | |
| Cell line (Homo sapiens) | T-REx 293 RNF43/ZNRF3 dKO | This publication | Cells lacking RNF43/ZNRF3; CRISPR/Cas9 | |
| Cell line (Homo sapiens) | T-REx 293 RNF43/ZNRF3 dKO RNF43 TetON | This publication | RNF43/ZNRF3 dKO inducibly overexpressing RNF43 | |
| Cell line (Homo sapiens) | T-REx 293 DVL1/2/3 tKO | Paclíková et al., 2017 | Cells lacking all DVL isoforms; CRISPR/Cas9 | |
| Cell line (Homo sapiens) | T-REx 293 WNT5A/B KO | This publication | Cells lacking WNT5A/B isoforms; CRISPR/Cas9 | |
| Cell line (Homo sapiens) | T-REx 293 ROR1 KO | This publication | Cells lacking ROR1; CRISPR/Cas9 | |
| Cell line (Homo sapiens) | A375 (amelanotic malignant melanoma) | Kucerova et al., 2014 | RRID:CVCL_0132 | BRAF V600E; GFP constitutive expression |
| Cell line (Homo sapiens) | A375 RNF43/ZNRF3 dKO | This publication | Cells lacking RNF43/ZNRF3; CRISPR/Cas9 | |
| Cell line (Homo sapiens) | A375+ RNF43 | This publication | Stable overexpression of RNF43 | |
| Cell line (Homo sapiens) | A375 RNF43 TetON | This publication | Cells inducibly overexpressing RNF43 | |
| Cell line (Homo sapiens) | A375 TetON ctrl | This publication | Control cells for A375 RNF43 TetON | |
| Cell line (Homo sapiens) | A375 IV (amelanotic malignant melanoma) | Kucerova et al., 2014 | BRAF V600E; GFP constitutive expression | |
| Cell line (Homo sapiens) | A375 IV RNF43/ZNRF3 dKO | This publication | Cells lacking RNF43/ZNRF3; CRISPR/Cas9 | |
| Cell line (Homo sapiens) | A375 IV + RNF43 | This publication | Stable overexpression of RNF43 | |
| Cell line (Homo sapiens) | A2058 (metastatic melanoma) | ECACC | 91100402; RRID:CVCL_1059 | BRAF V600E |
| Cell line (Homo sapiens) | A2058 RNF43 TetON | This publication | Cells inducibly overexpressing RNF43 | |
| Cell line (Homo sapiens) | MelJuso | Laboratory of Stjepan Uldrijan | RRID:CVCL_1403 | NRASQ61L/WT HRASG13D/G13D |
| Cell line (Homo sapiens) | MelJuso RNF43 TetON | This publication | Cells inducibly overexpressing RNF43 | |
| Peptide, recombinant protein | Recombinant human R-Sponidin-1 | PeproTech | 120-38 | Final concentration 50 ng/ml |
| Peptide, recombinant protein | Recombinant human WNT3A | R&D Systems | 5036-WN | Range 40–80 ng/ml |
| Peptide, recombinant protein | Recombinant human WNT5A | R&D Systems | 645-WN | Range 40–80 ng/ml |
| Chemical compound, drug | LGK-974, Porcupine inhibitor | PeproTech | 1241454 | 1 μM |
| Chemical compound, drug | C-59, Porcupine inhibitor | Abcam | ab142216 | 0.5 μM |
| Chemical compound, drug | Dansylcadaverine, inhibitor of clathrin-dependent endocytosis | Sigma-Aldrich | D4008 | 50 µM |
| Chemical compound, drug | Chloroquine, inhibitor of lysosomal hydrolases | Sigma-Aldrich | C662 | 10 μM |
| Chemical compound, drug | MG-132, proteasome inhibitor | Sigma-Aldrich | C2211 | 10 μM |
| Chemical compound, drug | Vemurafenib BRAF V600E inhibitor | MedChem Express | HY-12057 | Up to 5 μM |
| Chemical compound, drug | Cobimetinib, Mek1 inhibitor | MedChem Express | HY-13064 | Up to 0.5 μM |
| Antibody | β-actin (rabbit monoclonal) | Cell Signaling Technology | CS-4970; RRID:AB_2223172 | WB (1:3000) |
| Antibody | DVL-2 (rabbit polyclonal) | Cell Signaling Technology Mentink et al., 2018 | CS-3216; RRID:AB_2093338 | WB (1:1000) |
| Antibody | DVL-3 (rabbit polyclonal) | Cell Signaling Technology Mentink et al., 2018 | CS-3218; RRID:AB_10694060 | WB (1:1000) |
| Antibody | DVL-3 (rabbit monoclonal) | Santa Cruz Biotechnology Kaiser et al., 2019 | SC-8027; RRID:AB_627434 | WB (1:1000) |
| Antibody | Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) (rabbit polyclonal) | Cell Signaling Technology Radaszkiewicz et al., 2020 | CS-9101; RRID:AB_331646 | WB (1:1000) |
| Antibody | Total MAPK (Erk1/2) (rabbit monoclonal) | Cell Signaling Technology Radaszkiewicz and Bryja, 2020 | CS-4695; RRID:AB_390779 | WB (1:1000) |
| Antibody | ROR1 (rabbit polyclonal) | Kind gift from Ho et al., 2012 | WB (1:3000) | |
| Antibody | ROR2 (mouse monoclonal) | Santa Cruz Biotechnology Ozeki et al., 2016 | sc-374174; RRID:AB_10989358 | WB (1:1000) |
| Antibody | WNT5A (rat monoclonal) | R&D Systems Kaiser et al., 2019 | MAB645; RRID:AB_10571221 | WB (1:500) |
| Antibody | WNT11 (rabbit polyclonal) | LifeSpan BioSciences Kotrbová et al., 2020 | LS-C185754 | WB (1:500) |
| Antibody | VANGL2 2 G4 (rat monoclonal) | Merck Mentink et al., 2018 | MABN750; RRID:AB_2721170 | WB (1:500) |
| Antibody | MITF (rabbit monoclonal) | Cell Signaling Technology Lavelle et al., 2020 | CS-12590; RRID:AB_2616024 | WB (1:1000) |
| Antibody | HA-11 (mouse monoclonal) | Covance Paclíková et al., 2017 | MMS-101R; RRID:AB_291262 | WB (1:2000); IF (1:500); IP (1 µg) |
| Antibody | HA (rabbit polyclonal) | Abcam Paclíková et al., 2017 | ab9110; RRID:AB_307019 | WB (1:2000); IF (1:500); IP (1 µg); FC (1:1000) |
| Antibody | c-Myc (9E10) (mouse monoclonal) | Santa Cruz Biotechnology Hanáková et al., 2019 | sc-40; RRID:AB_2857941 | WB (1:500); IF (1:250); IP (1 µg) |
| Antibody | GFP 3 H9 (rat monoclonal) | Chromotek Harnoš et al., 2019 | 3 H9 | WB (1:2000); IP (1 µg) |
| Antibody | GFP (rabbit polyclonal) | Fitzgerald Hanáková et al., 2019 | 20R-GR-011; RRID:AB_1286217 | WB (1:2000); IP (1 µg) |
| Antibody | FLAG M2 (mouse monoclonal) | Sigma-Aldrich Paclíková et al., 2017 | F3165; RRID:AB_259529 | WB (1:2000), IF (1:500) |
| Antibody | FLAG (rabbit polyclonal) | Sigma Paclíková et al., 2017 | F7425; RRID:AB_439687 | WB (1:2000); IF (1:500) |
| Antibody | V5 (mouse monoclonal) | Thermo Fisher Scientific Kaiser et al., 2019 | R96025; RRID:AB_159313 | WB (1:1000), IF (1:1000); IP (1 µg) |
| Antibody | Cortactin (mouse monoclonal) | Santa Cruz Biotechnology Weeber et al., 2019 | sc-55579; RRID:AB_831187 | IF (1:250) |
| Antibody | Golgin-97 (mouse monoclonal) | Invitrogen | A-21270; RRID:AB_221447 | IF (1:500) |
| Antibody | a-mouse IgG HRP (goat polyclonal) | Sigma-Aldrich | A4416; RRID:AB_258167 | WB (1:4000) |
| Antibody | a-rabbit IgG HRP (goat polyclonal) | Sigma-Aldrich | A0545; RRID:AB_257896 | WB (1:4000) |
| Antibody | a-rat IgG HRP (goat polyclonal) | Sigma-Aldrich | A9037; RRID:AB_258429 | WB (1:4000) |
| Antibody | Streptavidin-HRP conjugate | Abcam | ab7403 | WB (1:4000) |
| Antibody | Ror1-APC (mouse monoclonal) | Miltenyi Biotec Kotašková et al., 2016 | 30-119-860 | FC (1:25) |
| Antibody | a-mouse Alexa Fluor 488 (goat polyclonal) and 568 (donkey polyclonal) | Thermo Fisher Scientific | A-11001 (RRID:AB_2534069) and A10037 (RRID:AB_2534013) | IF (1:600) |
| Antibody | a-rabbit Alexa Fluor 488 (donkey polyclonal) and 568 (goat polyclonal) | Thermo Fisher Scientific | A21206 (RRID:AB_2535792) and A11011 (RRID:AB_143157) | IF (1:600) |
| Antibody | Streptavidin, Alexa Fluor 488 conjugate | Thermo Fisher Scientific | S-32354 | IF (1:600) |
| Antibody | Phalloidine Alexa Fluor 594 | Thermo Fisher Scientific | A12381 | IF (1:600) |
| Antibody | Phalloidine 4 Alexa Fluor 488 | Thermo Fisher Scientific | A12379 | IF (1:600) |
| Recombinant DNA reagent | pcDNA4-TO-RNF43-2xHA-2xFLAG | Kindly gifted by Bon-Kyoung Koo (Koo et al., 2012) | Inducible expression of RNF43; backbone for cloning | |
| Recombinant DNA reagent | pcDNA4-TO-RNF43Mut1-2xHA-2xFLAG | Kindly gifted by Bon-Kyoung Koo (Koo et al., 2012) | Inducible expression of inactive RNF43-HA-FLAG | |
| Recombinant DNA reagent | pcDNA4-TO-RNF43-BirA*-HA | This publication | Inducible expression of RNF43 with BirA* and HA tags | |
| Recombinant DNA reagent | pcDNA3-RNF43-HA | This publication | Expression of RNF43-HA | |
| Recombinant DNA reagent | pCW57-RNF43 | This publication | Lentiviral plasmid allowing inducible expression of HA/FLAG tagged RNF43 | |
| Recombinant DNA reagent | myc-Vangl1, GFP-Vangl2, GFP-Vangl2ΔN, GFP-Vangl2ΔC, GFP-Vangl2ΔNΔC | Belotti et al., 2012 | Expression of VANGL1 and VANGL2 and their variants | |
| Recombinant DNA reagent | pLAMP1-mCherry | Addgene #45147 Van Engelenburg and Palmer, 2010 | RRID:Addgene_45147 | Expression of lysosomes marker |
| Recombinant DNA reagent | pEGFP-C1-Rab5a | Chen et al., 2009 | Expression of early endosomes marker | |
| Recombinant DNA reagent | GFP-rab11 WT | Addgene #12674 Choudhury et al., 2002 | RRID:Addgene_12674 | Expression of recycling endosomes marker |
| Recombinant DNA reagent | His-ubiquitin | Tauriello et al., 2010 | Tagged ubiquitin for His-Ub pulldown assay | |
| Recombinant DNA reagent | pcDNA3-Flag-mDvl1 | Tauriello et al., 2010 | Expression of Dvl1 with Flag tag | |
| Recombinant DNA reagent | pCMV5-3xFlag Dvl2 | Addgene #24802 Narimatsu et al., 2009 | RRID:Addgene_24802 | Expression of DVL2 with Flag tag |
| Recombinant DNA reagent | pCDNA3.1-Flag-hDvl3 | Angers et al., 2006 | Expression of DVL3 with Flag tag | |
| Recombinant DNA reagent | pcDNA3.1-hROR1-V5-His | gifted by Kateřina Tmějová | Expression of ROR1 with V5 tag | |
| Recombinant DNA reagent | pcDNA3-Ror2-Flag; pcDNA3-Ror2-dCRD-FLAG | Sammar et al., 2004 | Expression of ROR2 with FLAG tag and its mutant lacking CRD domain | |
| Recombinant DNA reagent | pRRL2_ROR1ΔCYTO and pRRL2_ROR1ΔTail | Gentile et al., 2011 | Expression of ROR1 and its truncated versions | |
| Recombinant DNA reagent | hCas9 | Addgene #41815 Mali et al., 2013 | RRID:Addgene_41815 | Humanized Cas9 |
| Recombinant DNA reagent | gRNA_GFP-T1 | Addgene #41819 Mali et al., 2013 | RRID:Addgene_41819 | gRNA expression plasmid |
| Recombinant DNA reagent | PiggyBack-Hygro; Transposase | Gifted by Bon-Kyoung Koo | RRID:Addgene_64324 | PiggyBack transposase system for stable cell lines generation |
| Recombinant DNA reagent | pU6-(BbsI)CBh-Cas9-T2A-mCherry | Addgene #64324 Chu et al., 2015 | RRID:Addgene_64324 | All-in-1 Cas9 plasmid |
| Recombinant DNA reagent | pSpCas9(BB)-2A-GFP (PX458) | Addgene #48138 Ran et al., 2013 | RRID:Addgene_48138 | All-in-1 Cas9 plasmid |
| Sequence-based reagent | RNF43 gRNA | This publication | gRNA | TGAGTTCCATCGTAACTGTGTGG |
| Sequence-based reagent | ZNRF3 gRNA | This publication | gRNA | AGACCCGCTCAAGAGGCCGGTGG |
| Sequence-based reagent | WNT5A gRNA | This publication | gRNA | AGTATCAATTCCGACATCGAAGG |
| Sequence-based reagent | ROR1 gRNA | This publication | gRNA | CCATCTATGGCTCTCGGCTGCGG |
| Sequence-based reagent | RNF43 gRNA | This publication | gRNA | AGTTACGATGGAACTCATGG |
| Sequence-based reagent | ZNRF3 gRNA | This publication | gRNA | CTCCAGACAGATGGCACAGTCGG |
| Sequence-based reagent | B2M_F | This publication | qPCR primer | CACCCCCACTGAAAAAGATG |
| Sequence-based reagent | B2M_R | This publication | qPCR primer | ATATTAAAAAGCAAGCAAGCAGAA |
| Sequence-based reagent | GAPDH_F | This publication | qPCR primer | GACAGTCAGCCGCATCTTCT |
| Sequence-based reagent | GAPDH_R | This publication | qPCR primer | TTAAAAGCAGCCCTGGTGAC |
| Sequence-based reagent | HSPCB_F | This publication | qPCR primer | TCTGGGTATCGGAAAGCAAGCC |
| Sequence-based reagent | HSPCB_R | This publication | qPCR primer | GTGCACTTCCTCAGGCATCTTG |
| Sequence-based reagent | RPS13_F | This publication | qPCR primer | CGAAAGCATCTTGAGAGGAACA |
| Sequence-based reagent | RPS13_R | This publication | qPCR primer | TCGAGCCAAACGGTGAATC |
| Sequence-based reagent | RNF43_F | This publication | qPCR primer | TTTCCTGCCTCCATGAGTTC |
| Sequence-based reagent | RNF43_R | This publication | qPCR primer | CAGGGACTGGGAAAATGAATC |
| Sequence-based reagent | ZNRF3_F | This publication | qPCR primer | GCTTTCTTCGTCGTGGTCTC |
| Sequence-based reagent | ZNRF3_R | This publication | qPCR primer | GCCTGTTCATGGAATTCTGAC |
| Sequence-based reagent | DVL2_F | This publication | qPCR primer | TCCTTCCACCCTAATGTGTCCA |
| Sequence-based reagent | DVL2_R | This publication | qPCR primer | CATGCTCACTGCTGTCTCTCCT |
| Sequence-based reagent | DVL3_F | This publication | qPCR primer | ACCTTGGCGGACTTTAAGGG |
| Sequence-based reagent | DVL3_R | This publication | qPCR primer | TCACCACTCCGAAATCGTCG |
| Sequence-based reagent | WNT5A_F | This publication | qPCR primer | GCAGCACTGTGGATAACACCTCTG |
| Sequence-based reagent | WNT5A_R | This publication | qPCR primer | AACTCCTTGGCAAAGCGGTAGCC |
| Sequence-based reagent | ROR1_F | This publication | qPCR primer | TCTCGGCTGCGGATTAGAAAC |
| Sequence-based reagent | ROR1_R | This publication | qPCR primer | TCCAGTGGAAGAAACCACCTC |
| Sequence-based reagent | ROR2_F | This publication | qPCR primer | GTGCGGTGGCTAAAGAATGAT |
| Sequence-based reagent | ROR2_F | This publication | qPCR primer | ATTCGCAGTCGTGAACCATATT |
| Sequence-based reagent | MITF_F | Su et al., 2020 | qPCR primer | TGCCCAGGCATGAACACAC |
| Sequence-based reagent | MITF_R | Su et al., 2020 | qPCR primer | GGGAAAAATACACGCTGTGAG |
-
WB: western blot; IF:immunofluorescence; IP: immunoprecipitation.
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65759/elife-65759-transrepform1-v1.docx
-
Supplementary file 1
Sequencing of the CRISPR/Cas9 derived cell lines.
- https://cdn.elifesciences.org/articles/65759/elife-65759-supp1-v1.pptx





