Transient kinetic studies of the antiviral Drosophila Dicer-2 reveal roles of ATP in self–nonself discrimination
Figures
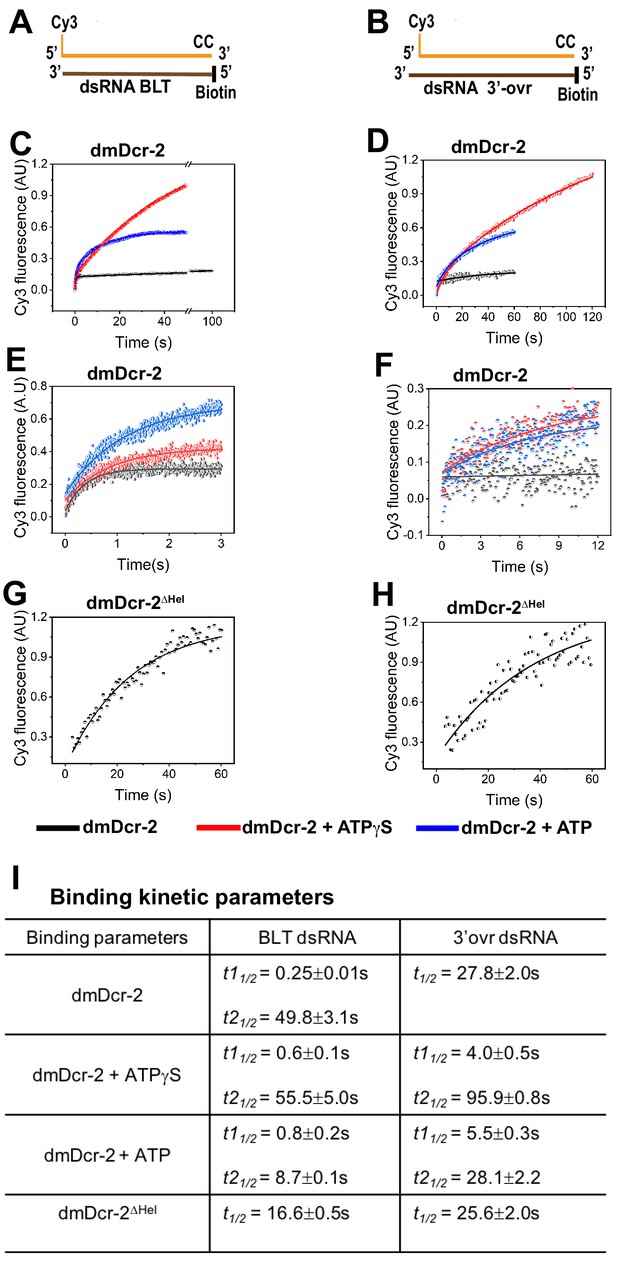
Association kinetics of BLT and 3’ovr dsRNA with dmDcr-2.
The time-dependent increase in the fluorescence signal of Cy3-end-labeled 52-dsRNA (0.2 μM) was monitored upon mixing with 10-fold excess of dmDcr-2 (2 μM), alone and bound with nucleotide (ATPγS or ATP), in stopped-flow syringes. Cartoons show Cy3-end-labeled 52-dsRNA with BLT (A) and 2 nt 3’ovr termini (B) with deoxynucleotides (CC) and biotin to prevent dmDcr-2 binding to one end. Representative kinetic traces for binding of Cy3-labeled BLT (C) and 3’ovr (D) 52-dsRNA to dmDcr-2 are shown, with independent experiments over shorter time courses for BLT (E) and 3’ovr dsRNA (F), as well as for binding of dmDcr-2ΔHel to BLT (G) and 3’ovr (H) dsRNA. At least four to ten traces were collected for each experimental condition, and averaged trace was analyzed with single or double exponential rate equations, yielding values for kinetic parameters (kobs = 0.693/t1/2) and associated standard error (I).
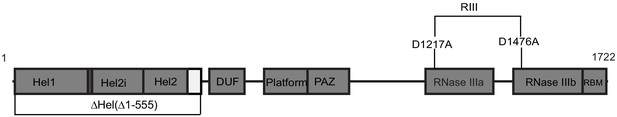
The domain organization of dmDcr-2.
The boundaries of the Hel1, Hel2, and Hel2i subdomains of the helicase are indicated, as well as those for the Domain of Unknown Function (DUF), Platform, PAZ, tandem RNase III domains, and the C-terminal dsRNA binding motif (RBM). Variants used in this study are labeled.

The fluorescence decay curve of Cy3 attached to 5’-end of BLT (A) and 3’ovr (B) 52-dsRNA while bound to dmDcr-2RIII (C and D) or the dmDcr-2RIII•ATPγS complex (E and F).
dsRNA was incubated with dmDcr2RIII for 5 min to allow complete isomerization. The RNase III variant contained two-point mutations (D1217A, D1476A), each within one of the tandem active sites that each cleave one strand (Figure 1—figure supplement 1). Data were analyzed with single and double exponential rate equations yielding values for the fluorescence lifetime (𝜏) and amplitude (parenthesis) of Cy3 located in different micro-environments of dmDcr-2 (G). Also shown are the life-time values of unbound Cy3-BLT and Cy3-3’ovr dsRNA obtained under identical experimental conditions for comparison (G).
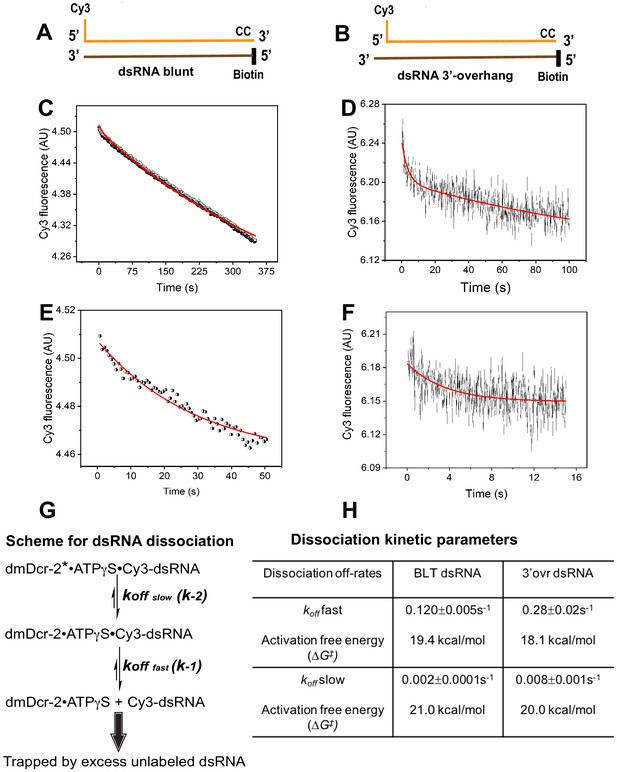
Dissociation kinetics of BLT and 3’ovr dsRNA bound to dmDcr-2•ATPγS.
Dissociation kinetics were measured by mixing enzyme-bound Cy3-end-labeled 52-dsRNA with 10-fold excess of unlabeled dsRNA in stopped-flow syringes, and monitoring decrease in Cy3 fluorescence over time. Cartoons show BLT (A) and 3’ovr (B) dsRNA with representative kinetic traces for dissociation of Cy3-labeled BLT (C) and 3’ovr (D) 52-dsRNA, and independent experiments over shorter time courses (BLT, E; 3’ovr F). (G) Reaction scheme for dissociation of enzyme-bound Cy3-dsRNA; in this scheme since k-1 >> k-2, the dmDcr-2•ATPγS•Cy3-dsRNA reaches steady-state level with rate constant k-1, and then decays slowly with rate constant k-2 (Fersht, 1999). (H) Kinetic traces were analyzed with single or double exponential rate equations, yielding values for dissociation off-rates. At least four to ten traces were collected for each condition, and averaged trace was analyzed with single or double exponential rate equations to obtain values for kinetic parameters (kobs = 0.693/t1/2) and associated standard error.
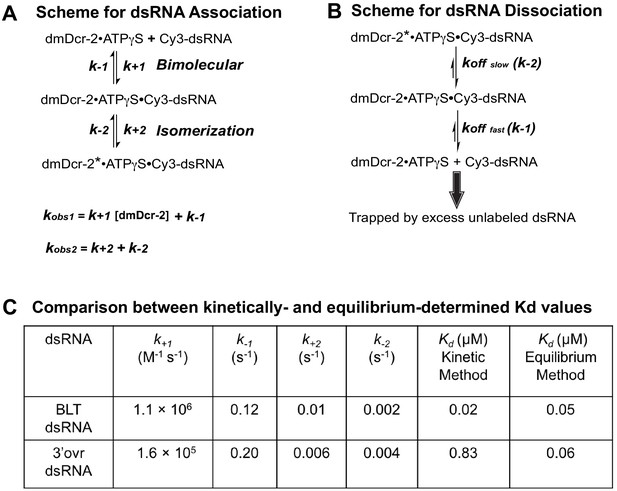
The schemes for dsRNA association and dissociation, and evaluation of kinetically determined Kd values for interaction of dsRNA with dmDcr-2•ATPγS.
Association (A) and dissociation (B) kinetics follow a two-step mechanism. Observed rate constants for bimolecular (kobs1) and isomerization (kobs2) steps are a function of microscopic rate constants. In association kinetics (A), kobs1 is linearly dependent on molar concentration of dmDcr-2 (1 µM after twofold dilution upon mixing, see Materials and methods), and therefore, accurate evaluation of the second-order rate constant (k+1, slope) requires kobs1 values to be measured over a fairly wide range of enzyme concentration. Since a non-ideal (aggregative behavior) of dmDcr-2 at higher micromolar concentration precludes determination of kobs1 as a function of enzyme concentration (see Materials and methods), we estimated the second-order rate constant from the kobs1 value measured at a single dmDcr-2 concentration. To enhance accuracy in values of estimated microscopic rate constants as reported in (C), we used the mean value of observed rate constants (BLT, kobs1 = 1.24 s−1, kobs2 = 0.013 s−1; 3’ovr, kobs1 = 0.36 s−1, kobs2 = 0.01 s−1) obtained from three independent experiments; values of kobs measured using different batches of protein and/or dsRNA, in general, varied by less than or equal to twofold. The Kd values for enzyme–substrate interaction were evaluated using all microscopic rate-constants; values of k-1 and k-2 reported in (C) represent an average of three independently determined values from dissociation off-rate measurements. The kinetically determined Kd values reported in (C), Kd = (k-1k-2)/(k+1k+2), are within experimental error of those previously determined with equilibrium binding studies (Donelick et al., 2020).
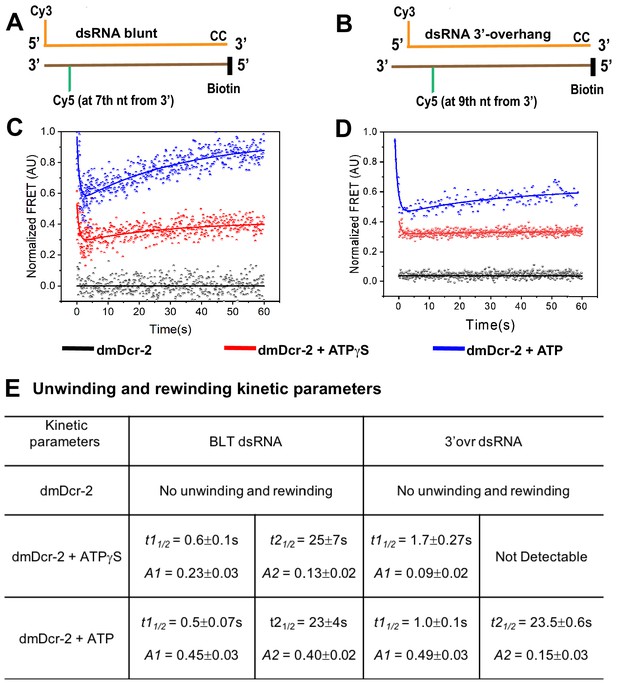
ATP-dependent transient unwinding and reannealing of dsRNA termini catalyzed by dmDcr-2.
The time-dependent change in FRET signal was monitored after mixing 0.2 μM dsRNA containing a FRET (donor–acceptor) pair with 2 μM dmDcr-2 in stopped-flow syringes. Cartoons showing BLT (A) and 3’ovr (B) 52-dsRNA indicating positions of Cy3 and Cy5, and modifications to block dmDcr-2 entry, as in Figure 1. Representative kinetic traces for unwinding and reannealing of FRET-labeled BLT (C) and 3’ovr (D) dsRNA by dmDcr-2 alone (black), with ATPγS (red), or ATP (blue). Kinetic traces were analyzed with a double exponential rate equation, and kinetic parameters are in (E). At least four to ten traces were collected for each experimental condition, and the averaged trace was analyzed with single or double exponential rate equations, yielding values for kinetic parameters (kobs = 0.693/t1/2), amplitude (A), and associated standard error.
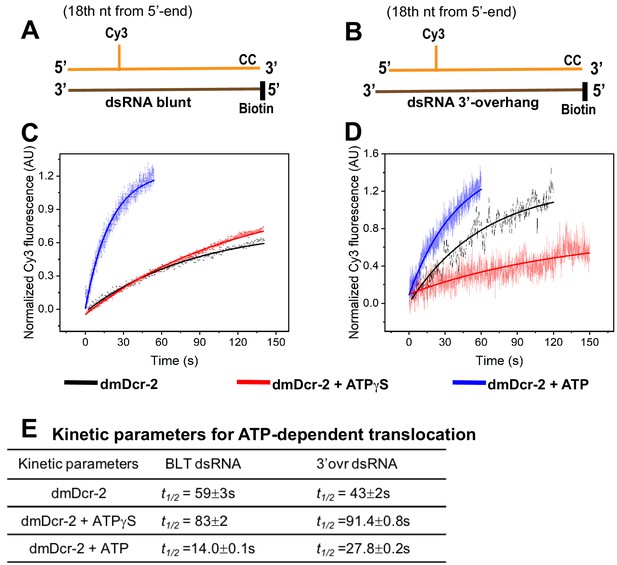
Real-time monitoring of ATP-dependent translocation/arrival of dmDcr-2 at the cleavage site.
Cartoons show BLT (A) and 3’ovr (B) 52-dsRNA with a covalently linked Cy3 at the 18th nucleotide of the top (sense) strand, and deoxynucleotide (CC) and biotin to prevent binding of dmDcr-2 to one end. Representative stopped-flow kinetic traces for translocation of dmDcr-2 alone (black trace), and while bound with ATPγS (red) and ATP (blue) on BLT (C) and 3’ovr (D) dsRNA. Kinetic traces were analyzed with a single exponential rate equation, yielding observed rate constants associated with translocation (E). At least four to ten traces were collected for each experimental condition, and averaged trace was analyzed with single or double exponential rate equations, yielding kinetic parameters (kobs = 0.693/t1/2) and associated standard error.
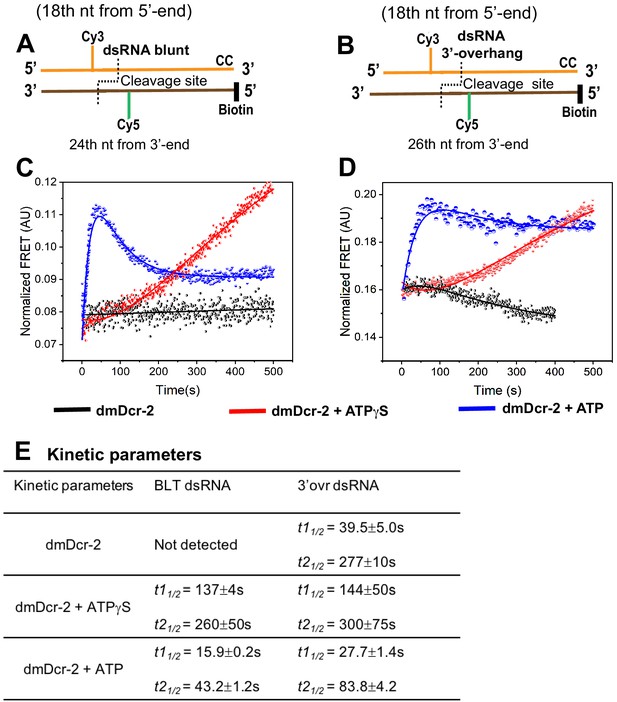
The arrival of dmDcr-2 at cleavage site poises the enzyme for ATP-dependent substrate cleavage and siRNA release.
Cartoons illustrate BLT (A) and 3’ovr (B) dsRNA containing the Cy3-Cy5 FRET pair at positions indicated, and deoxynucleotide (CC) and biotin to block dmDcr-2 binding at one end. Representative stopped-flow kinetic traces for cleavage of BLT (C) and 3’ovr (D) 52-dsRNA by dmDcr-2 alone (black), with ATPγS (red) or ATP (blue). Kinetic traces were analyzed with a double exponential rate equation, and kinetic parameters listed in (E). At least four to ten traces were collected for each condition, and averaged trace was analyzed with single or double exponential rate equations, yielding values for kinetic parameters (kobs = 0.693/t1/2) and associated standard error.

Stopped-flow kinetics using FRET substrates designed to monitor cleavage, but with cleavage-incompetent dmDcr-2RIII in the presence of ATP.
The RNase III mutant variant contains point mutations as indicated (Figure 1—figure supplement 1). Cartoons illustrate BLT (A) and 3’ovr (B) dsRNA as in Figure 5. Representative kinetic traces were analyzed with a single exponential rate equation yielding half-life values for BLT (C; 25 s) and 3’ovr (D; 50 s) dsRNA.
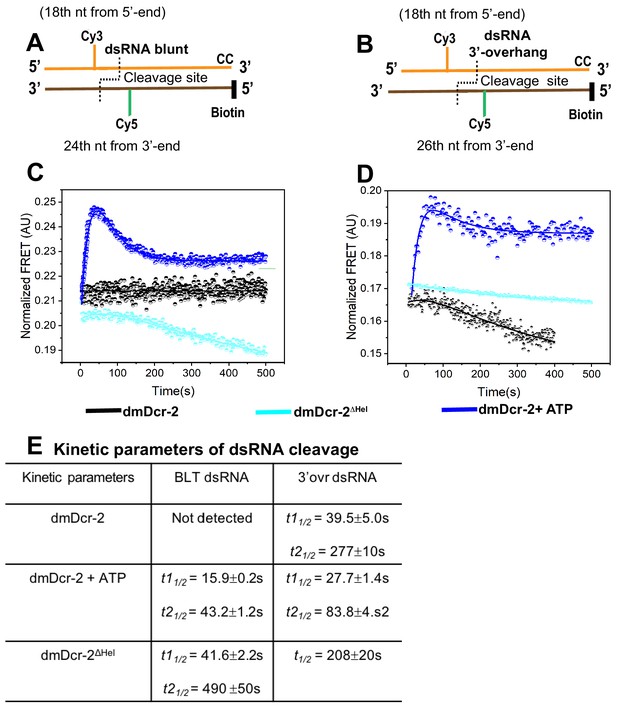
Stopped-flow kinetics monitoring cleavage of BLT and 3’ovr dsRNA with dmDcr2ΔHel, and for comparison, data for dmDcr-2 alone and with ATP from Figure 5 are shown.
Cartoons illustrating BLT (A) and 3’ovr (B) 52-dsRNA as in Figure 5. Representative stopped-flow kinetic traces for cleavage of BLT (C) and 3’ovr (D) dsRNA by dmDcr-2 alone (black trace), dmDcr2ΔHel (cyan trace), and dmDcr-2 with ATP (blue trace). Kinetic traces were analyzed with single or double exponential rate equations yielding half-life values (E).
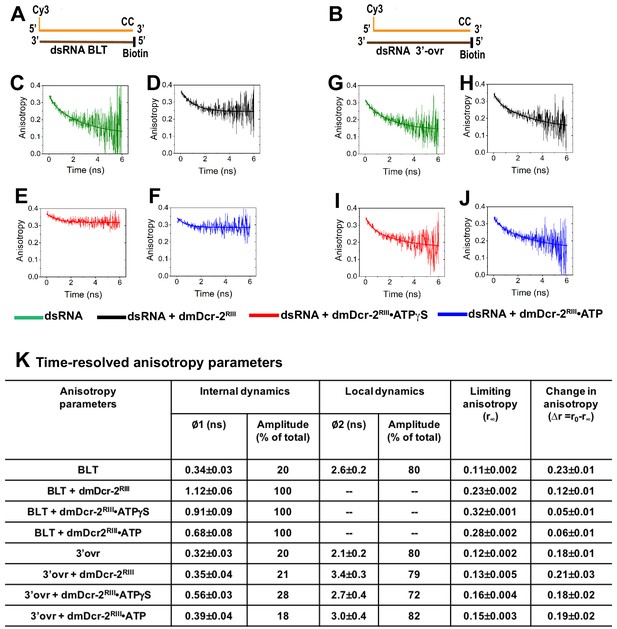
Time-resolved fluorescence anisotropy of Cy3-dsRNA alone and bound to the helicase and Platform–PAZ domains of dmDcr-2.
Cartoons show Cy3-end-labeled 52-dsRNA with BLT (A) and 2 nt 3’ovr termini (B) with deoxynucleotides (CC) and biotin to prevent dmDcr-2 binding to one end. Representative anisotropy decay curves for Cy3-end-labeled BLT (C–F) and 3’ovr dsRNA (G–J) alone (green) and bound to dmDcr-2RIII in the absence of nucleotide (black), or in the presence of ATPγS (red) or ATP (blue). All decay curves were analyzed with a minimum number of exponential terms (single and double exponential rate equations) to obtain the best fit (see Materials and methods), yielding values of anisotropy parameters (K). The precise values of anisotropy parameters associated with the fast correlation time (φ1) were determined from independent analyses of the fast phase of anisotropy decay curves using a single exponential equation (see Figure 6—figure supplement 1).
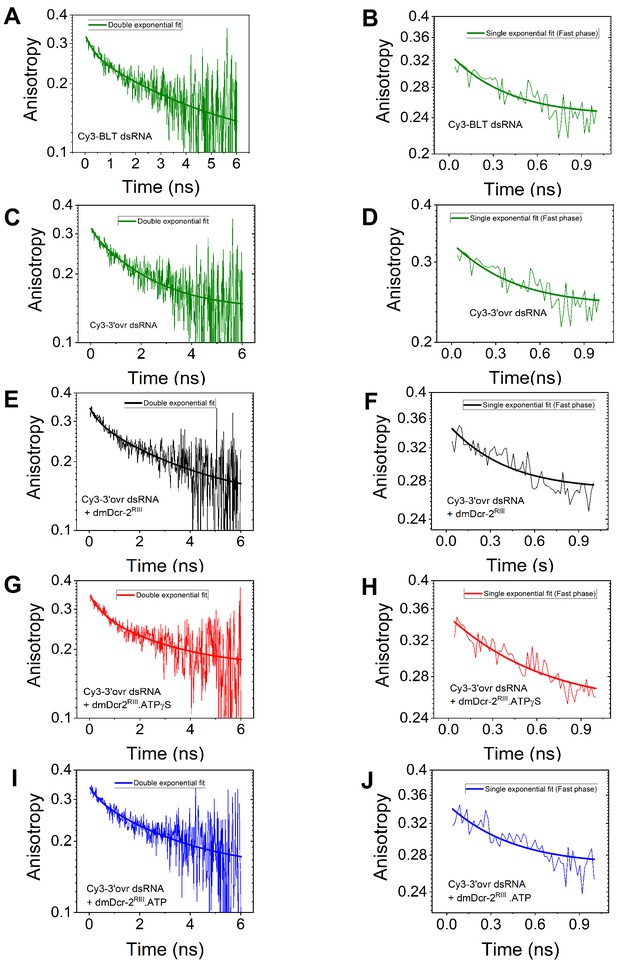
Fluorescence anisotropy decay curves of Cy3-end-labeled BLT (A and B) and 3’ovr (C–J) 52-dsRNA, −/+ enzyme, and nucleotide as indicated.
All biphasic anisotropy decay curves were analyzed using double-exponential equations, and to obtain precise values of anisotropy parameters associated with fast dynamics, the fast phase of decay curves was independently analyzed with a single-exponential equation, yielding anisotropy parameters listed in Figure 6.

The fluorescence decay curve of Cy3 attached to 5’-end of BLT (A) and 3’ovr (B) 52-dsRNA while bound to dmDcr2RIII•ATP (C and D).
Data were analyzed with single- and double-exponential rate equations yielding values for the fluorescence lifetime (𝜏) of Cy3 in different micro-environments of dmDcr-2 (E). Also shown are the life-time values of Cy3-BLT and Cy3-3’ovr dsRNA without protein (alone) obtained under identical experimental conditions to facilitate comparison (E). The life-time values listed here represent the averaged life-time.
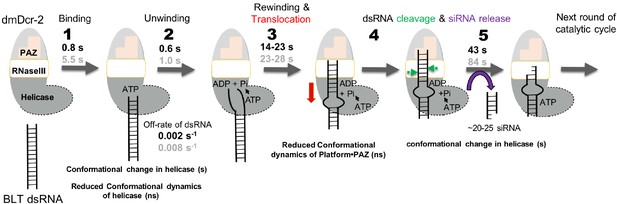
Real-time kinetic model for ATP-dependent dmDcr-2-catalyzed reaction with BLT and 3’ovr dsRNA.
Kinetic parameters for dmDcr-2 reacting with BLT (black font) and 3’ovr (gray font) dsRNA are reported in seconds (s), and conformational changes noted in seconds or nanoseconds (ns) for numbered steps. (1) ATP binding to the helicase domain promotes a closed conformational state that traps the dsRNA terminus, accompanied by reduced dynamics of the helicase domain; ATPγS allows measurement of off-rates. (2) ATP hydrolysis catalyzes dsRNA unwinding at the terminus. (3) Unwinding is followed by slow rewinding in concert with an ATP-dependent directional translocation of dmDcr-2 (red arrow) to the dsRNA cleavage site (~22 nt from dsRNA terminus). An ATP-dependent modulation in conformational dynamics of the Platform–PAZ domain poises the enzyme for catalysis. Platform–PAZ domains are shown in pink with a shape indicating optimal binding to 3’ovr termini compared to BLT termini. (4) The RNase III domain of dmDcr-2 cleaves (green arrowheads) dsRNA by measuring ~21–22 nts from the terminus bound to the Platform–PAZ domain. (5) The siRNA is released via an ATP-binding mediated conformational change in the helicase domain which shows long range communication with the Platform–PAZ domain bound to siRNA termini.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA agent | pFastBac (plasmid) | Thermo Fisher Scientific | Cat# 10360–014 | |
| Cell line (Spodoptera frugiperda) | Sf9 cells | Expression System | Cat# 94–001S RID:CVCL_0549 | |
| Strain (Escherichia coli) | DH10Bac Competent Cells | Thermo Fisher Scientific | Cat# 10361012 | Chemically competent cells |
| Protein purification reagent | Strep-Tactin | IBA Lifesciences | Cat# 2-1201-010 | |
| RNA labeling reagent | Sulfo-NHS-ester Cyanine3 | Lumiprobe Corporation | Cat# 21320 | |
| RNA labeling reagent | Sulfo-NHS-ester Cyanine5 | Lumiprobe Corporation | Cat# 23020 | |
| RNA synthesis reagent | Glen Research | http://www.glenresearch.com | Phosphoramidites | |
| Nucleotide | ATP | Thermo Fisher | Cat# R1441 | |
| Nucleotide-analog | ATPγS | Sigma | A-1388–25 MG | |
| Software, algorithm | Origin software package | Origin Lab | Origin Lab Corporation | |
| Fast-mixing device | Stopped-flow system | BioLogic Sciences Instruments | SFM 3000 | |
| Optical filter | Long pass- filter | NewPort | 10LWF-550B | |
| TCSPC fluorescence system | Time-correlated single photon counting (TCSPC) module | PicoQuant | PHR 800 and Picoharp 300 | |
| Sequence-based reagent | 52 nt sense strand for BLT and 3’ ovr dsRNA | This paper | ssRNA | 5’-GGAGGUAGUAGGUUGUAUAGUAGUAAGACCAGACCCUAGACCAAUUCAUGCC-3’ CC = deoxynucleotide |
| Sequence-based reagent | 52 nt antisense strand for BLT dsRNA | This paper | ssRNA | Biotin-5’- GGCAUGAAUUGGUCUAGGGUCUGGUCUUACUACUAUACAACCUACUACCUCC-3’ |
| Sequence-based reagent | 54 nt antisense strand for 3’ovr dsRNA | This paper | ssRNA | Biotin-5-GGCAUGAAUUGGUCUAGGGUCUGGUCUUACUACUAUACAACCUACUACCUCCCC-3’ |
| Sequence-based reagent | Cy3-end labeled 52 nt sense strand | IDT | ssRNA | 5’Cy3-GGAGGUAGUAGGUUGUAUAGUAGUAAGACCAGACCCUAGACCAAUUCAUGCC-3’ CC = deoxynucleotide |





