Unbiased identification of novel transcription factors in striatal compartmentation and striosome maturation
Figures
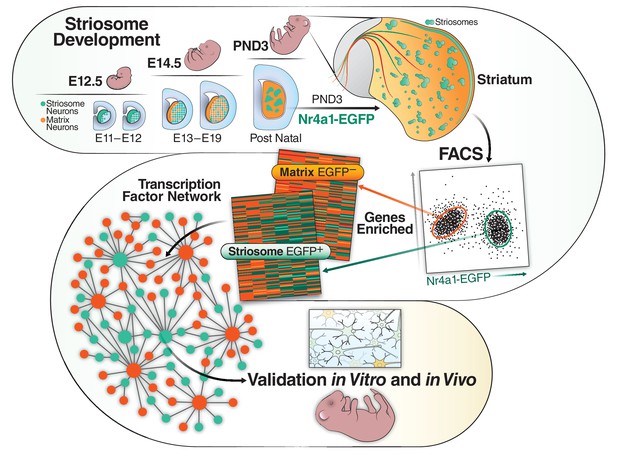
Schematic of the developmental cascade for the striosome and matrix compartments and our experimental approach.
Upper panel shows the cascade of the two compartments in the developing mouse. Lower panels show our experimental approach. We FACs sorted EGFP+ cells from Nr4a1 EGFP mice at PND3, performed RNAseq, identified top transcription factors in the striosome, validated them in vivo (mice) and in vitro (primary mouse cultures and human iPSCs).
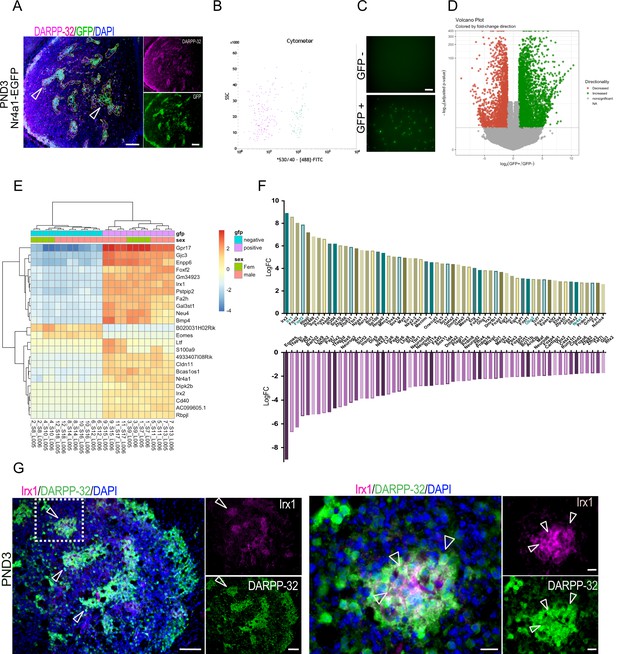
Identification of striatal striosome and matrix compartment-specific transcription factors using RNA-seq analysis in FACs EGFP+ and EGFP– cell populations from PND3 Nr4a1- EGFP mice.
(A) Coronal section of PND3 Nr4a1-EGFP mouse showing the striosomes (open arrowheads pointing to several examples) localization of spontaneous EGFP fluorescence, with DARPP-32 (magenta) colocalization. Scale bar = 100 µm. (B) Representative FACs cytometer panel indicating cell separation using the intensity of the 488-FITC channel with clear distribution of the EGFP+ population on the right side of the panel. (C) Detection of spontaneous EGFP fluorescence of EGFP+ and EGFP– cells seeded at 1000 cells/well 2 hr after FACs sorting, indicating the presence of EGFP exclusively in EGFP+ samples. Scale bar = 50 µm. (D) Volcano plot visualizing the differences in gene expression between the EGFP+ and EGFP– cell populations. Genes with an adjusted p-value below 0.01 with absolute log2 fold ratio greater than one are highlighted. Red indicates genes that were relatively decreased in expression in EGFP+ population (i.e. enriched in the EGFP– population), green indicates genes that were relatively enriched in expression in the EGFP+ population, and gray indicates those genes that were equally distributed in the two populations. (E) Heatmap of relative normalized counts across samples of genes with an absolute log2 fold-change > 7, and mean normalized counts > 40. (F) Representation of the 60 TFs most enriched in either population, according to their log2 fold change in the EGFP+ population (upper panel) and in the EGFP– population (lower panel), without regard to mean normalized counts. (G) Representative low- and high-power micrographs of Irx1 (magenta), DARPP-32 (green) and DAPI (blue) immunolabeling on 16-µm-thick coronal sections from wild-type PND3 showing the localization of Irx1 within a subset of the DARPP-32 immunopositive striosomes (arrowheads left panel), with double label of a subset of striosome neurons indicated by the arrowheads (right panel). Scale bars = 100 µm and 20 µm, respectively.
-
Figure 2—source data 1
RNA sequencing analysis of the transcripomes of striosome and matrix cells from the PND3 striatum striosome and matrix compartments expressed as a ratio of Nr4a1-EGFP+/Nr4a1-EGFP- following FACS sorting.
- https://cdn.elifesciences.org/articles/65979/elife-65979-fig2-data1-v1.xlsx
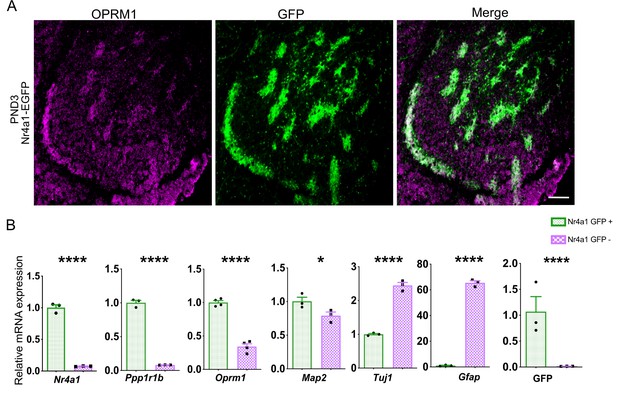
Validation of Nr4a1-EGFP striosome localization and characterization of EGFP+ and EGFP- cell population after FACS sorting.
(A) Spontaneous EGFP fluorescence (green) co-localizes with immunolabeling of the striosome marker Oprm1 (magenta) in coronal sections of Nr4a1-EGFP PND3 mice. Scale bar = 100 µm. (B) RT- qPCR assay shows significant enrichment of striosome- and neural-specific markers in EGFP+ FACS sorted population, as well as EGFP. The difference in Oprm1 enrichment in RT-qPCR vs RNAseq may be due to the nature of the assays, in which only one sequence is being amplified in RT- qPCR. TuJ1 and GFAP expression is enriched in EGFP– population. n=three independent sorting sessions. *p<0.05 t-test. Error bars = standard deviation.
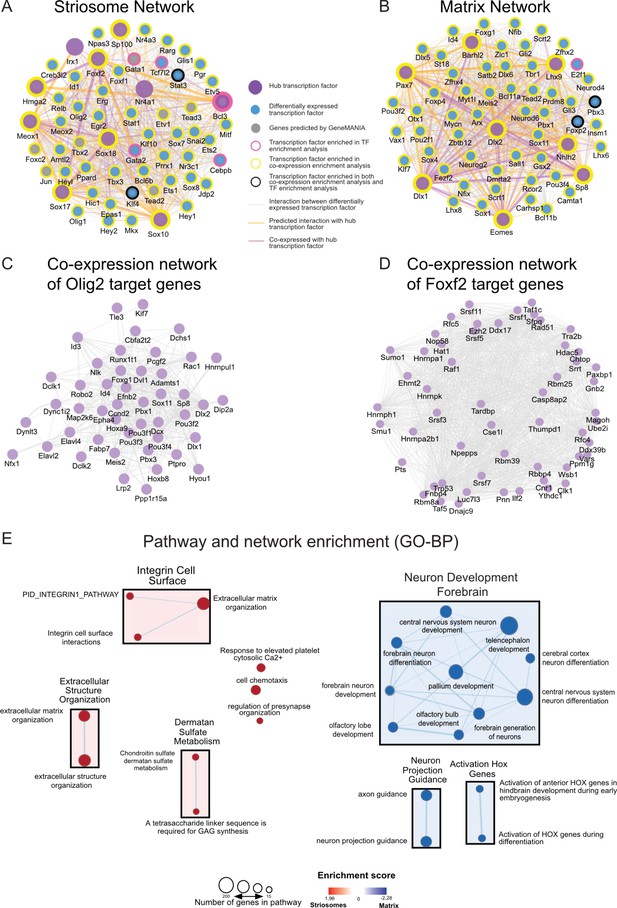
Transcription networks and enrichment maps of striosome and matrix.
(A,B) TF networks enriched in either striosome or matrix show interactions and co-expression among the regulatory genes. An arbitrary threshold of absolute log2 fold-change > five was set to designate hub genes. (C,D) Co-expression networks derived from striosome and matrix RNA-seq data show targets of Olig2 and Foxf2 are co-regulated in the striosome. Genes with higher connectivity to other target genes are designated as hub genes. (E) Pathway and network enrichment (GO-BP) for the striosome (left, red) and matrix (right, blue). The resulting enrichment map was annotated using the AutoAnnotate Cytoscape App.
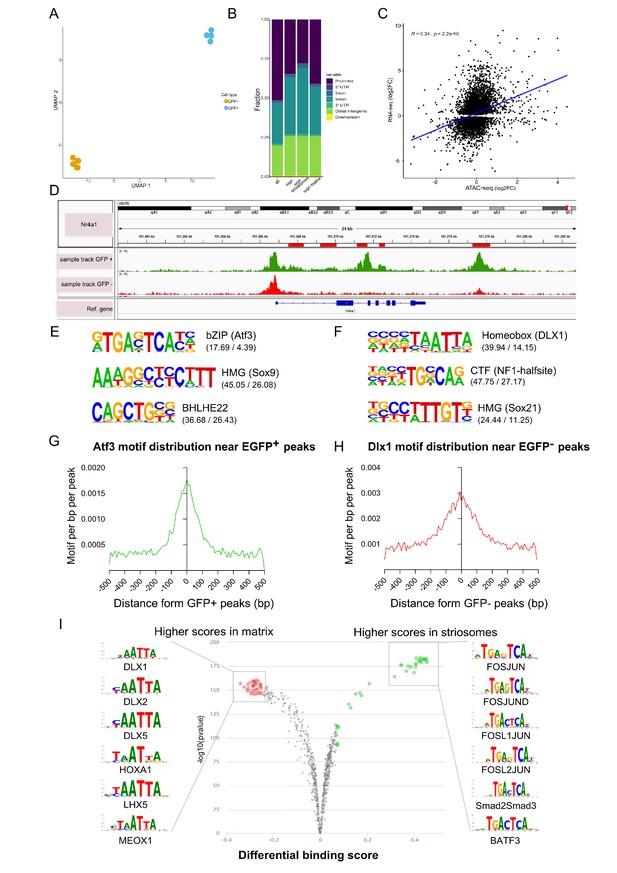
Open chromatin region (ATAC-seq) analysis identifies OCRs differentially located in striosome or matrix.
(A) Clustering of the individual samples using UMAP. (B) Distribution of the location within the gene (see Materials and methods, Annotation of OCRs) of all and differentially distributed OCRs. (C) Correlation of log2 fold-changes in RNA-seq and ATAC-seq analyses. OCRs within TSS were considered for this analysis, and only genes with adjusted p-value <0.01 from RNA-seq analysis were included. (D) Nr4a1 ATAC-seq tracks in EGFP-positive and -negative populations showing the presence of multiple, possible striosome ‘specific’ Nr4a1 regulatory elements indicated by the red boxes in the gene annotation. (E,F) Motif analysis of OCRs in striosome (E) and matrix (F). Position weight matrix of enriched nucleotide sequence, the TF family that matches with the motif, and the predicted binding TF are shown. The values in parentheses indicate the percentage of the input sequences that contain the motif relative to the percentage of total background regions filtered by p-value of 0.01. (G, H) Frequency of predicted binding sites for Atf3 (left) and Dlx1 (right) as a function of distance from the center of striosome and matrix OCRs. (I) Pairwise comparison of TF activity between striosome and matrix. The volcano plots show the differential binding activity against the -log10(pvalue) of the investigated TF motifs; each dot represents one motif. For striosome-specific factors, the significant TFs are labeled in green, whereas matrix-specific factors are labeled in red.
-
Figure 4—source data 1
Uniquely mapped reads following ATAC seq of Nr4a1-EGFP+ and Nr4a1-EGFP- cells.
- https://cdn.elifesciences.org/articles/65979/elife-65979-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Comparison of striosome (Nr4a1-EGFP+) vs matrix (Nr4a1-EGFP-) peaks representing 44% of the total number of OCRs which remained statistically significant after multiple testing corrections.
- https://cdn.elifesciences.org/articles/65979/elife-65979-fig4-data2-v1.xlsx
-
Figure 4—source data 3
Transcription factors identified by HOMER which putatively bind to enriched binding motifs in Nr4a1-EGFP+ and Nr4a1-EGFP- cells.
- https://cdn.elifesciences.org/articles/65979/elife-65979-fig4-data3-v1.xlsx
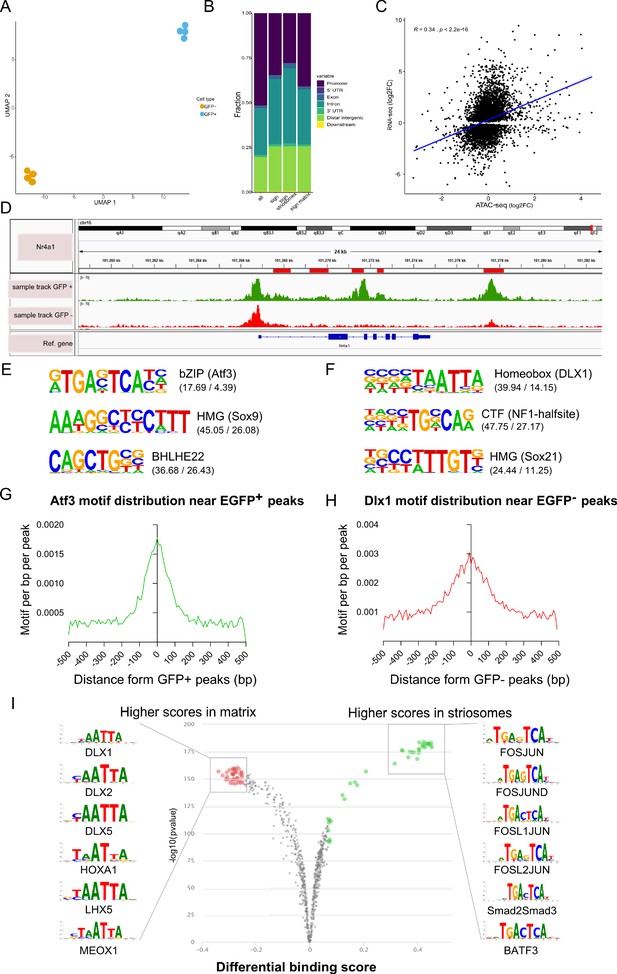
Analysis with ANANSE integrating RNAseq and ATACseq identifies key TFs required for striosome MSN differentiation.
(A) ANANSE analysis defines key transcription factors in striosomes. ANANSE is a network-based method that uses properties of enhancers and their gene regulatory networks to predict key TFs. The influence score vs. log 2Fc demonstrates Foxf2 is highly ranked. (B) The top 15 TFs ranked by influence score and the direct targets predicted for each TF. Notably, there is overlap with the TFs identified using only the RNA-seq data set.
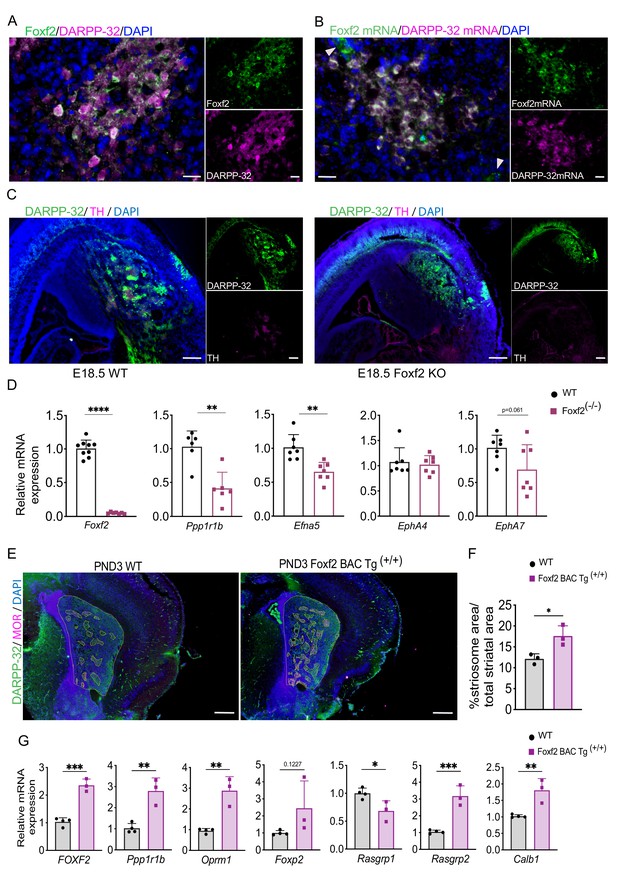
Foxf2 is required for striosome compartmentation.
(A) Representative confocal micrographs of Foxf2 (green), DARPP-32 (magenta) and DAPI (blue) immunolabeling on 16-µm-thick coronal sections from wild-type PND3 showing the localization of Foxf2 within the DARPP-32-immunopositive striosomes. Scale bars correspond to 20 µm. (B) RNAscope analysis in 16-µm-thick coronal sections from wild-type PND3 mice shows Foxf2 mRNA (green) co-localization with Ppp1r1b/DARPP-32 mRNA (magenta). Arrow heads indicate vascular localization of Foxf2 mRNA, with absence of DARPP-32 mRNA. Scale bars = 20 µm. (C) DARPP-32 (green) and tyrosine hydroxylase (TH) (magenta) immunostaining in sagittal sections of E18.5 WT and Foxf2 KO mice show the absence of striosome assembly in Foxf2 KO mice. Scale bars = 100 µm (D) RT-qPCR from E18.5 Foxf2 WT and null littermates striatal RNA indicating that Foxf2 expression was abolished in the null mouse and that the levels of Ppp1r1b and Efna5 mRNA were significantly reduced by Foxf2 deletion. n=7 Foxf2-/- and n=9 WT. Unpaired t-test **p<0.001, ****p<0.0001. Error bars = standard deviation. (E,F) Comparison of area occupied by striosomes identified by merged DARPP-32 (green) and MOR (magenta) immunolabeling of 16-µm-thick coronal sections from PND3 WT and overexpressing Foxf2 (Foxf2 BAC Tg (+/+)) littermates showing an increase of striosome density (highlighted) mediated by Foxf2 overexpression. n=three individual mice per genotype. Unpaired t-test *p<0.05. Error bars = standard deviation. (G) RT- qPCR assay shows increase in the expression of both striosome (Ppp1r1b, Oprm1, Foxp2) and matrix (Rasgrp2) markers in the striatum of PND3 mice overexpressing Foxf2. n=three mice per genotype. Unpaired t-test *p<0.05, **p<0.01, ***p<0.001. Error bars represent standard deviation.
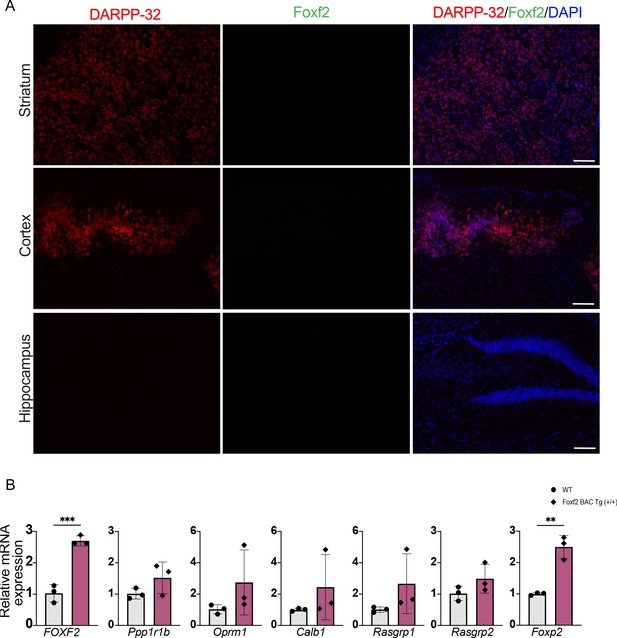
Foxf2 mRNA is undetectable in adult striatum but Foxf2 overexpression increased the level of FoxP2 mRNA in PND21 Foxf2 BAC Tg (+/+) striatum.
(A) Low-power magnification of RNAscope assay visualization of Foxf2 and Ppp1r1b/DARPP-32 mRNAs in adult mouse striatum, cortex, and hippocampus indicating that Foxf2 is not expressed in neurons in adult brain. Scale bar = 200 µm. (B) RT-qPCR from striatal RNA from PND21 Foxf2 BAC Tg (+/+) and WT littermates showing that Foxf2 mRNA level is increased almost threefold in in homozygote Foxf2-BAC-Tg(+/+) mice and that the striosome marker Foxp2 mRNA is increased whereas other MSN markers are unchanged. n=3/genotype. Unpaired t-test **p<0.001, ****p<0.0001. Error bars = standard deviation.
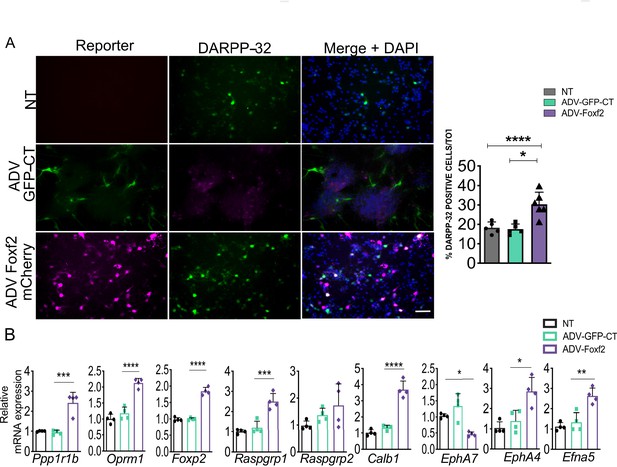
Foxf2 overexpression in medium spiny neurons promote their maturation in vitro.
(A) Representative DARPP-32 immunostaining in DIV9 WT primary striatal neurons transduced at DIV5 with ADV-GFP (control) or ADV-FOXF2-mCherry demonstrating the increase in DARPP-32 immunopositive cells in the cultures overexpressing Foxf2, quantitated in the right panel. Scale bar = 50 µm. n=six images from three individual cultures. One-way ANOVA corrected for multiple comparisons (Bonferroni’s) *p<0.05, **p<0.01. (B) RT- qPCR assay shows increase in mRNA levels of both striosome (Ppp1r1b, Oprm1, Foxp2, Rasgrp1) and matrix (Calb1, Rasgrp2 and EphA4) markers in DIV- nine wild-type primary striatal neurons 96 hr after transduction with ADV-FOXF2mCherry.n=four individual cultures, one-way ANOVA corrected for multiple comparisons (Bonferroni’s) *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars = standard deviation.
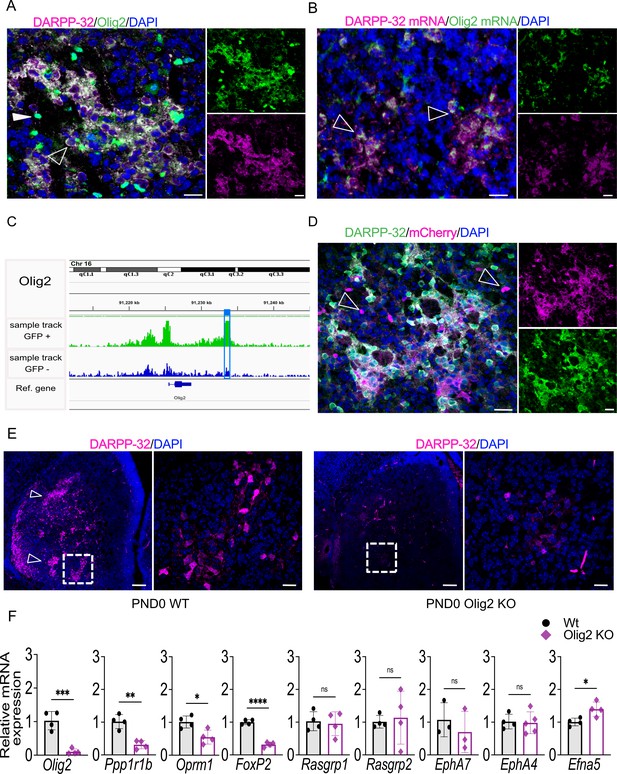
Olig2 is required for striosome compartmentation and a downstream intergenic Olig2 OCR drives transgenic reporter expression in striosomes.
(A) Confocal micrographs of Olig2 (green), DARPP-32 (magenta) and DAPI (blue) immunolabeling of WT PND3 16-µm-thick coronal sections showing localization of Olig2 within the DARPP-32-positive striosomes (unfilled arrowhead). An examples of a single Olig2-immunopositive and DARPP-32-negative cell is indicated by a filled overhead. Scale bars = 20 µm (B) RNAscope visualization of Olig2 (green) and Ppp1r1b/DARPP-32 (magenta) mRNAs showing colocalization in PND3 wild-type striosomes with individual double-labeled cells highlighted by arrowheads. (C) Olig2 ATAC-seq tracks in EGFP+ and EGFP– samples from PND3 Nr4a1-EGFP striatum showing the presence of an OCR 4.4 kb downstream of the Olig2 gene, restricted to EGFP+ samples (highlighted by the blue box). (D) Representative confocal micrographs of mCherry (magenta), DARPP-32 (green), and DAPI (blue) immunolabeling of 16-µm-thick coronal sections from PND3 Olig2 -OCR transgenic mice showing that the isolated OCR sequence drives expression of the mCherry reporter restricted in large part to DARPP-32-immunopositive neurons in striosomes. Individual magenta cells, examples indicated by arrowheads, likely represent cells of the oligodendroglia lineage. Scale bars = 20 µm. (E) DARPP-32 (magenta) and DAPI immunostaining in coronal sections of PND0 WT and Olig2 KO mice showing a dramatic reduction of DARPP-32-immunopositive neurons and striosome assembly in Olig2 KO mice. Scale bars = 100 µm and 20 µm, respectively. (F) RT-qPCR from striatal RNA from PND0 Olig2 KO and WT littermates indicating that striosome marker mRNAs are decreased in the KO brains. Olig2-/-, n=5; WT, n=4. Unpaired t-test *p<0.05, **p<0.001, ****p<0.0001. Error bars = standard deviation.
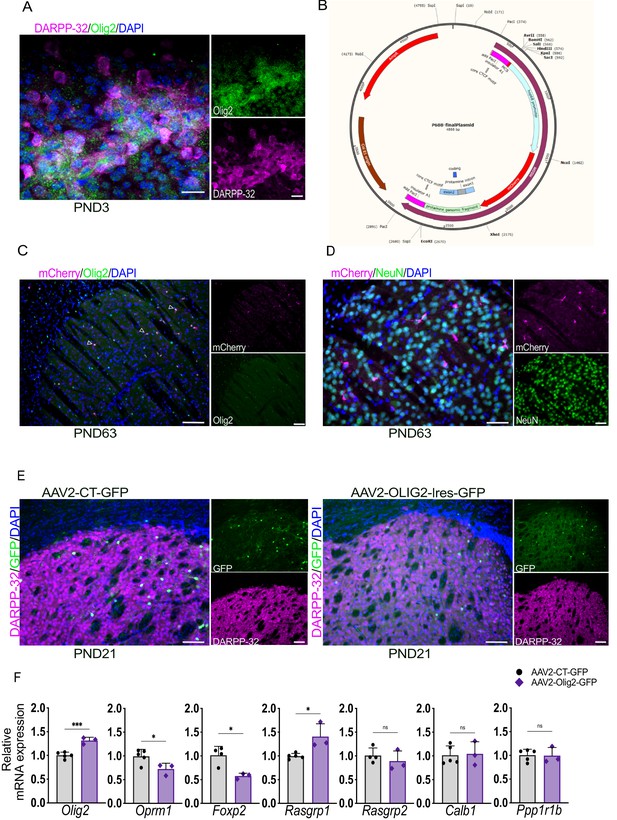
Impact of neonatal AAV2 driven Olig2 overexpression on the striatum of PND21 mice and analysis of Olig2 OCR-driven expression in adult founders.
(A) Low- and high-power magnification of Olig2 (green), DARPP-32 (magenta) and DAPI (blue) immunolabeling of WT PND3 coronal sections (16 μm) using a second antibody against Olig2 (gift of Dr. Wichterle) (Wichterle et al., 2002) showing the localization of Olig2 within the DARPP-32-positive striosomes. Scale bars = 200 μm and 20 μm, respectively. (B) Map of the p688/mCherry plasmid used for the cloning of the Olig2 OCR shown in Figure 6C. (C,D) Representative low- and high-power micrographs of mCherry (magenta), Olig2 (green) (C) or NeuN (green) (D) and DAPI (blue) immunolabelling on coronal sections (30 µm) from adult Olig2 -OCR transgenic mice indicating co-localization with Olig2 but not with NeuN. Scale bar = 200 µm. (E) Representative images of DARPP-32 (magenta) and GFP (green) immunolabeling of 30 µm coronal sections from PND21 WT mice injected at PND0 with AAV2-CT-GFP or AAV2-Olig2-IRES-GFP virus, showing GFP expression in the striatum and co-localization with DARPP-32. Scale bars = 100 µm. (F) RT-qPCR from striatal RNA from PND21 WT mice injected at PND0 with AV2-CT-GFP or AAV2-Olig2-IRES-GFP virus showing that a low level of Olig2 overexpression reduces the mRNA levels of the striosomal markers Oprm1 andFoxP2 while increasing Raspgrp1 mRNA. n=3. Unpaired t-test **p<0.001, ****p<0.0001. Error bars = standard deviation.
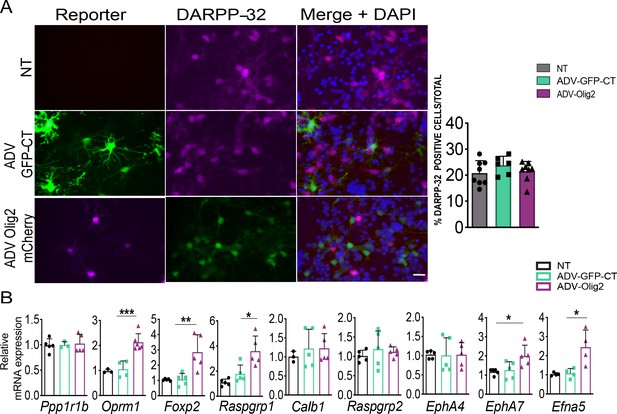
Olig2 overexpression in medium spiny neurons promote their maturation in vitro.
(A) Representative DARPP-32 immunostaining in DIV9 WT primary striatal neurons transduced at DIV5 with ADV-GFP (control) or ADV-Olig2-mCherry. Right panel shows quantification of the number of DARPP-32-immunopositve cells, which is equal between groups. Scale bar = 50 µm. (B) RT-qPCR assay shows increases of mRNA for striosome markers Oprm1, Foxp2 and Rasgrp1, EphA7 and its ligand Efna5 in DIV9 wild-type primary striatal neurons 96 hr after transduction with ADV-Olig2-mCherry. n=five individual cultures, one-way ANOVA corrected for multiple comparisons (Bonferroni’s) *p<0.05, **p<0.01, ***p<0.001. Error bars = standard deviation.
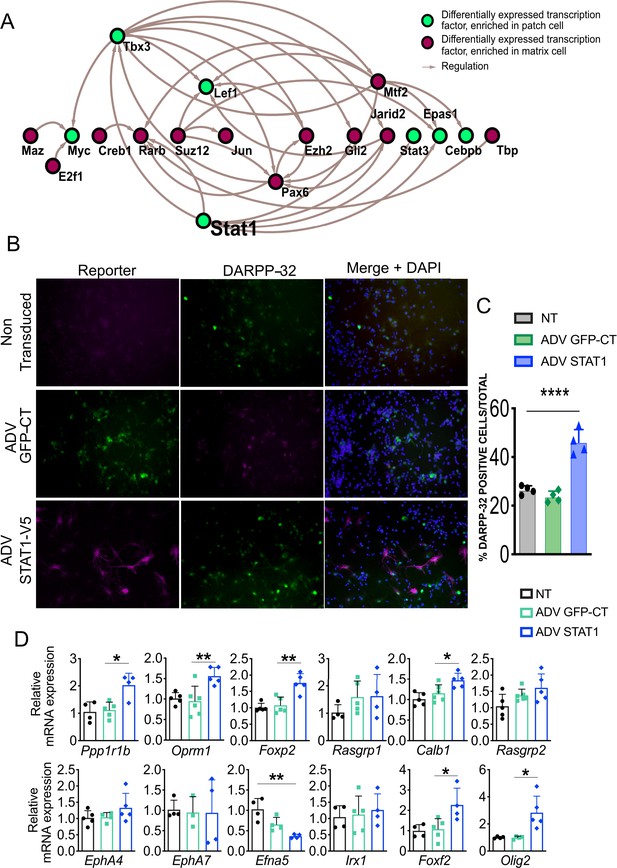
STAT1 overexpression in MSNs in vitro promotes maturation and increases levels of Foxf2 and Olig2 mRNAs.
(A) Network analysis indicates that Stat1 may be a ‘master’ regulator in striosomes as it modulates the levels of the greatest number of TFs enriched in that compartment. (B,C) GFP and DARPP-32 immunolabeling in DIV9 WT primary striatal neurons either non-transduced (NT) or transduced for 96 hr with ADV-STAT1-V5 or ADV-GFP showing that STAT1 overexpression increases the number of DARPP-32-immunopositive cells. Scale bars = 50 µm. n=four images from four individual cultures, t-test ***p<0.001. (D) RT- qPCR assay shows increase of mRNA for striosome markers Ppp1r1b, Oprm1, Foxp2, Foxf2, and Olig2, matrix markers Calb1 and EphA4 and decrease in striosome enriched EphA7 ligand and Efna5 in DIV9 wild-type primary striatal neurons 96 hr after transduction with ADV-STAT1-V5. n=four individual cultures, one-way ANOVA corrected for multiple comparisons (Bonferroni’s) *p<0.05, **p<0.01. Error bars = standard deviation.
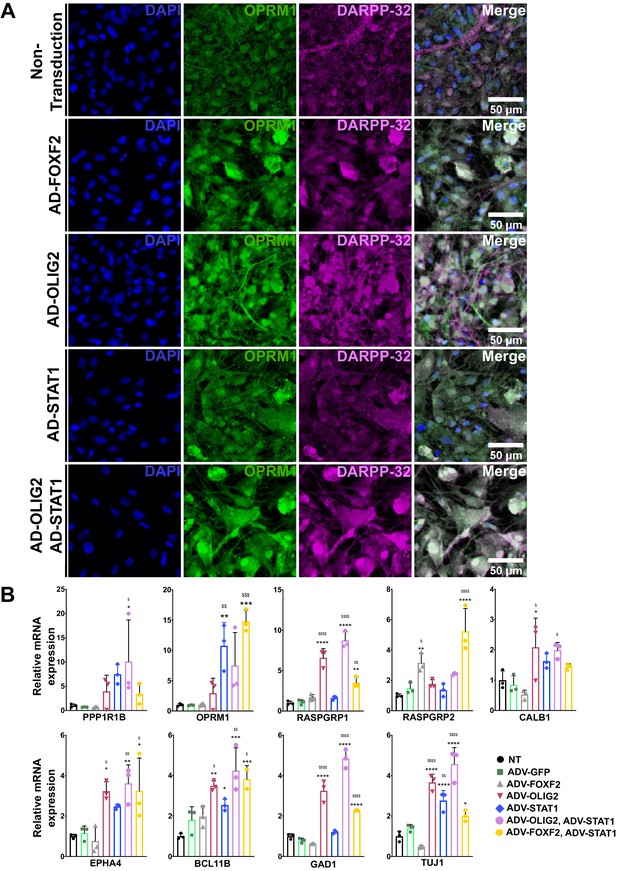
FOXF2, OLIG2, and STAT1, alone and in combination, promote MSN differentiation in NSCs from human HD induced pluripotent stem cells.
(A) HD72-NSCs transduced for 4 days with ADV-FOXF2, ADV-OLIG2 and ADV-STAT1 were immunostained with Oprm1 (green) and DARPP-32 (magenta). Non-transduced cells and AD-GFP were used as control. Scale bars = 100 µm. (B) RT-qPCR assay on HD72-NSCs transduced for 4 days with ADV-GFP, ADV-FOXF2, ADV-OLIG2 and ADV-STAT1. n=three individual cultures. One outlier (one of the three technical data points) was excluded in Bcl11b RT-qPCR when transduced with ADV- FOXF2 and ADV-STAT1. One-way ANOVA for multiple comparisons (Dunnett’s) *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. * for comparison to non-transduced and for comparison to ADV-GFP. Error bars are standard deviation.
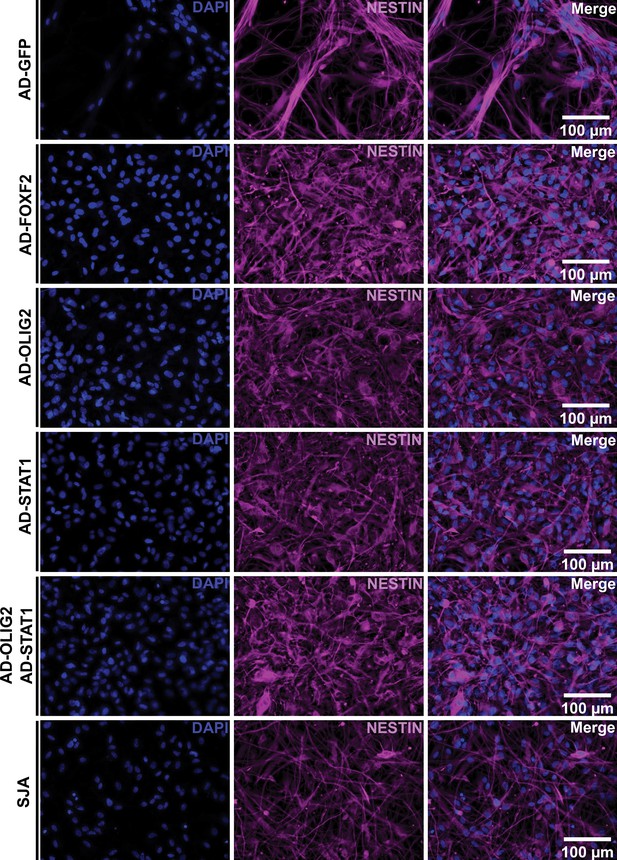
Expression of FOXF2, OLIG2 and STAT in HD NSCs resulted in an increase expression of pan neuronal marker Nestin in HD NSCs.
HD NSCs transduced for 4 days with ADV-FOXF2, ADV-OLIG2, and ADV-STAT1 were immunostained with Nestin. Non-transduced cells and AD-GFP, SynaptaJuice A were used as control. Scale bars: 100 µm.
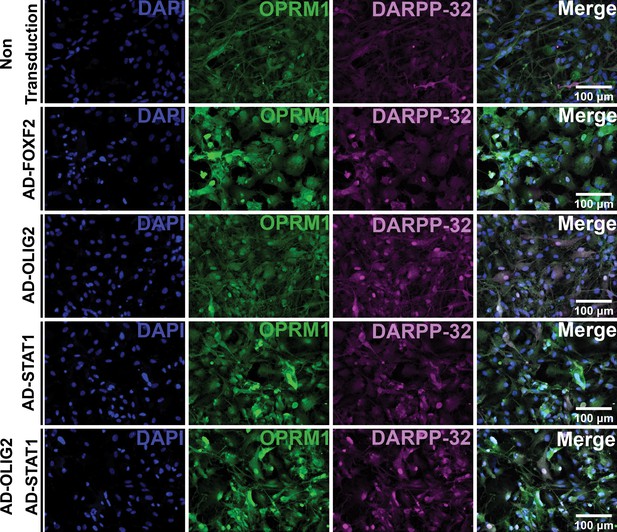
FOXF2, OLIG2, and STAT1, alone and in combination, promote MSN differentiation in NSCs from human isogenic control C116-induced pluripotent stem cells.
C116-NSCs transduced for 4 days with ADV-FOXF2, ADV-OLIG2, and ADV-STAT1 were immunostained with Oprm1 (green) and DARPP-32 (red). Non-transduced cells and ADV-GFP were used as control. Scale bars: 100 µm.
Tables
List of striosome and matrix markers.
| Gene symbol | Gene name | PND3 | Adult |
|---|---|---|---|
| Calb1 | Calbindin | Striosome | Matrix |
| EfnA5 | Ephrin A5 | Striosome | |
| EphA4 | Eph Receptor A4 | Matrix | Matrix |
| EphA7 | Eph Receptor A7 | Striosome | |
| FoxP1 | Forkhead Box P1 | Striosome | pan-MSN |
| FoxP2 | Forkhead Box P2 | Striosome | Striosome |
| Nr4a1 | Nuclear Receptor Subfamily 4 Group Member 1 | Striosome | Striosome |
| Oprm1 | Mu opioid receptor | Striosome | Striosome |
| Ppp1r1b | Protein phosphatase 1 regulatory subunit 1B | Striosome | pan-MSN |
| Rasgrp1 | CalDAG GEFII | Striosome | Striosome |
| Rasgrp2 | CalDAG GEFI | Matrix | Matrix |
RT-qPCR murine primer sequences.
| Gene | Primer forward 5'−3' | Primer reverse 5'−3' |
|---|---|---|
| GAPDH | AACGACCCCTTCATTGACCT | TGGAAGATGGTGATGGGCTT |
| Ppp1r1b | GAAGAAGAAGACAGCCAGGC | TAGTGTTGCTCTGCCTTCCA |
| Oprm1 | CCCTCTATTCTATCGTGTGTGT | AGAAGAGAGGATCCAGTTGCA |
| Foxp2 | AAGCAGCTTGCCTTTGCTAAG | GGATTGAATGTATGTGTGGCTGA |
| Rasgrp1 | GGACCTACCAAGAACTGGAAC | GATCCCAGTAAACCCGTCTG |
| Calb 1 | ACTCTCAAACTAGCCGCTGCA | TCAGCGTCGAAATGAAGCC |
| Rasgrp2 | CTTGGACCAGAACCAGGATG | GTGGCAGTTCACACCACAAG |
| Epha7 | TGCTCCGCTTTGCACACACAGG | TAAGTTCTCAATAATGGACCAGCAC |
| Epha4 | TCGTGGTCATTCTCATTG | TCTCTTCATCTGCTTCTTG |
| Ephna5 | CGATAGAACCAAGATAATACT | TAGAATCAGAGGACTCAG |
qRT-PCR human primer sequences.
| Gene | Primer forward 5'−3' | Primer reverse 5'−3' | Universal probe |
|---|---|---|---|
| PPP1R1b | CACACCACCTTCGCTGAAA | GAAGCTCCCCCAGCTCAT | 82 |
| OPRM1 | AGAAACAGCAGGAGCTGTGG | ACCGAGACTTTTCGGGTTC | 30 |
| BCL11b | CCCAGAGGGAGCTCATCAC | TTTGACACTGGCCACAGGT | 45 |
| CALB1 | CACAGCCTCACAGTTTTTCG | CCTTTCCTTCCAGGTAACCA | 36 |
| GAD1 | ATGGTGATGGGATATTTTCTCC | GCCATGCCCTTTGTCTTAAC | 46 |
| TUJ1 | GCAACTACGTGGGCGACT | CGAGGCACGTACTTGTGAGA | 78 |
| RASPGRP1 | GAGCCAAAGATCTGCTCCAT | GGTCCGATCCTTACTCTCCTC | 71 |
| RASPGR2 | TGAGCCACAGCTCCATCTC | CCGTCACTAGTTCCGTGAGAC | 75 |
| EPHA4 | AGCAGCCACTCAGGCAAC | ACGAAAATAGGGCGAAATAGAA | 51 |
| ACTB | CCAACCGCGAGAAGATGA | CCAGAGGCGTACAGGGATAG | 64 |
Additional files
-
Source data 1
Differential expression bioinformatics.
- https://cdn.elifesciences.org/articles/65979/elife-65979-data1-v1.xlsx
-
Source data 2
ATAC-seq samples.
- https://cdn.elifesciences.org/articles/65979/elife-65979-data2-v1.xlsx
-
Source data 3
ATAC-seq results.
- https://cdn.elifesciences.org/articles/65979/elife-65979-data3-v1.xlsx
-
Source data 4
Motif analysis.
- https://cdn.elifesciences.org/articles/65979/elife-65979-data4-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65979/elife-65979-transrepform-v1.docx





