Impaired mRNA splicing and proteostasis in preadipocytes in obesity-related metabolic disease
Figures
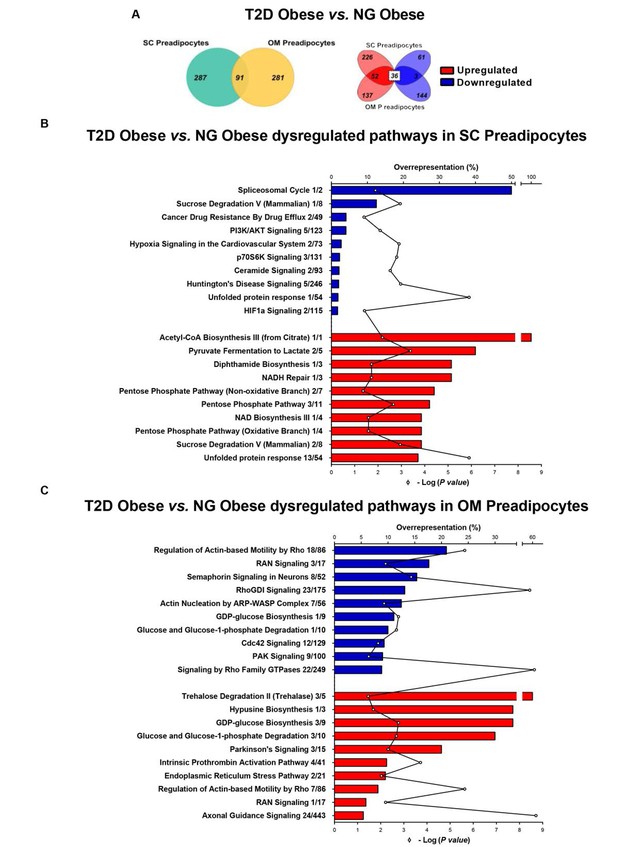
Comparative proteomic analysis of subcutaneous (SC) and omental (OM) preadipocytes from obese subjects with type 2 diabetes (T2D) vs. normoglycaemia (NG).
Data correspond to individuals from cohort 1. (A) Venn diagrams showing overlap of differentially regulated proteins between T2D vs. NG obese subjects in SC and OM preadipocytes (left panel). Up-regulated and down-regulated proteins are indicated in the right panel. (B) Top10 over-represented canonical pathways in T2D vs. NG obese subjects in SC preadipocytes and (C) in OM preadipocytes according to Ingenuity Pathway Analysis (IPA). Blue and red bars indicate down-regulated and up-regulated pathways, respectively, in T2D vs. NG obese subjects. Numbers indicate the number of identified proteins/total proteins annotated to the pathway. Black line indicates -Log2(p value). Samples from wo to three individuals per group and fat depot were pooled and used for two separate iTRAQ experiments ( n = 5–6 subjects/group/fat depot). Data normality was tested by Shapiro-Wilk test and Student´s t test was used, a ± 1.5 fold change with p < 0.05 was set as the threshold for categorizing up- and down-regulated proteins. Canonical pathway analysis was performed using IPA (see Materials and ethods section). The online version of this article includes the following figure supplements for Figure 1, Figure 1—figure supplement 1, Figure 1—figure supplement 2, and Figure 1—figure supplement 3; and the following source data for Figure 1—source data 1 and Figure 1—source data 2.
-
Figure 1—source data 1
Common pathways of normoglycemic (NG) and with type 2 diabetes (T2D) morbidly obese subjects (cohort 1) in the subcutaneous (SC) vs omental (OM) preadipocytes proteome according to Ingenuity Pathway Analysis (IPA).
- https://cdn.elifesciences.org/articles/65996/elife-65996-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Fat depot-specific canonical pathways in the type 2 diabetes (T2D) vs. normoglycemic (NG) morbidly obese subjects (cohort 1) proteome according to Ingenuity Pathway Analysis (IPA).
- https://cdn.elifesciences.org/articles/65996/elife-65996-fig1-data2-v2.xlsx
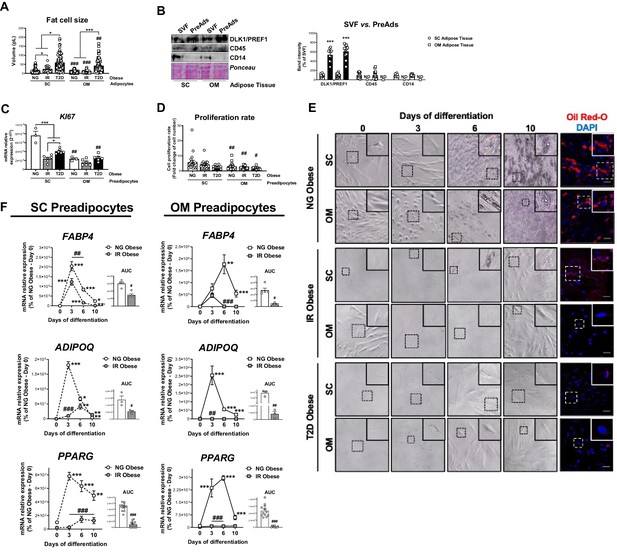
Characterization of SC and OM adipocytes and preadipocytes from NG, IR, and T2D morbidly obese individuals (cohort 1).
(A) Cell size of SC and OM mature adipocytes from NG, IR, and T2D morbidly obese subjects (cohort 1; = 6–8, 20 cells each). *p < 0.05, ***p < 0.001 vs. NG and/or IR obese subjects; ##p < 0.01, ###p < 0.001 vs SC preadipocytes from the same subjects. (B) Representative blot and protein level quantification of DLK1/PREF1, CD45, and CD14 in the stromal-vascular fraction (SVF) and preadipocytes (PreAds) from SC and OM adipose tissue from morbidly obese subjects (cohort 1; = 11, 1 technical replicate each). ***p < 0.001 vs. SVF. (C) mRNA levels of KI67 in SC and OM preadipocytes from NG, IR, and T2D morbidly obese subjects (cohort 1; n = 3–6, 1 technical replicate each). *p < 0.05, ***p < 0.001 vs NG and/or IR obese subjects; ##p < 0.01 vs. SC preadipocytes from the same subjects. (D) Proliferation rate of SC and OM preadipocytes from NG, IR, and T2D morbidly obese subjects (cohort 1; n = 20–22, 1 technical replicate each). #p < 0.05, ##p < 0.01 vs. SC preadipocytes from the same subjects. (E) Representative phase-contrast microscopy images (x100 magnification and zoom) during in vitro differentiation and representative confocal images at day 10 of differentiation of SC and OM preadipocytes from NG, IR and T2D morbidly obese subjects (cohort 1). Cells were stained with Oil Red-O (lipid droplets, red) and DAPI (nucleus, blue). Scale bar = 50 μm. (F) mRNA levels of FABP4, ADIPOQ, and PPARG during in vitro differentiation of SC and OM preadipocytes from NG and IR morbidly obese subjects (cohort 1); the values of the area under the curve (AUC) are also shown ( = 3–12, 3 technical replicates each). *p < 0.05, **p < 0.01, ***p < 0.001 vs. previous days; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. NG Obese. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively) were used for A, C, and D; unpaired t test or Mann Whitney test (for parametric or non-parametric data, respectively) were used for B and AUC values in F; and two-way ANOVA was used for time-course differentiation studies in F. Normality distribution was determined by Shapiro-Wilk normality test.
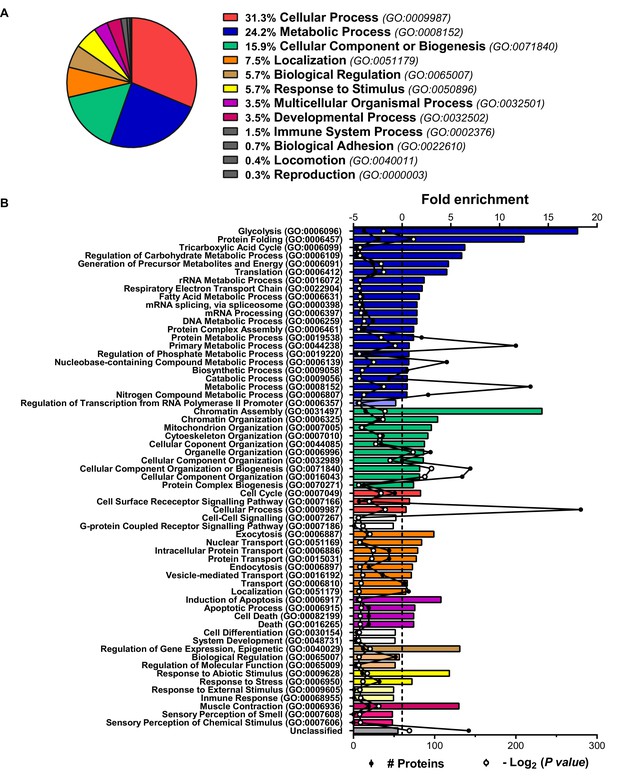
Characterization of the human preadipocyte proteome.
Analysis of the proteins identified by iTRAQ-LC-MS/MS in the proteome of preadipocytes isolated from morbidly obese subjects (cohort 1). Samples from two to three individuals per group and fat depot were pooled and used for two separate iTRAQ experiments ( = 5–6 subjects/group/fat depot). (A) Proteins were categorized according to biological processes using GO PANTHER classification system (http://www.pantherdb.org/). (B) The bar chart represents the GO classification and fold enrichment of significantly over- and under-represented Biological Processes (p ≤ 0.05 Bonferroni test) in preadipocyte proteome. The black line represents the number of identified proteins per each GO term and the red line represents -Log2(p value).
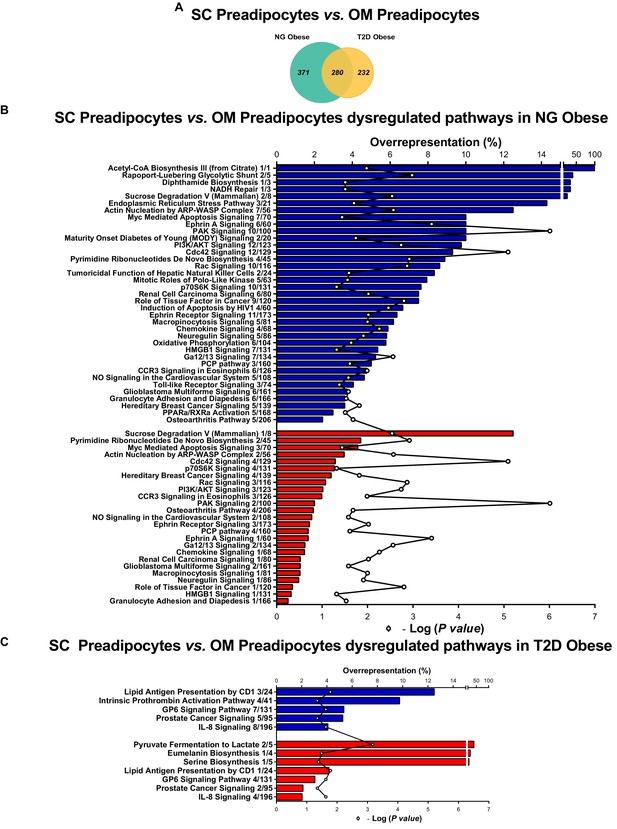
Comparative proteomic analysis of subcutaneous (SC) vs. omental (OM) preadipocytes from NG and T2D morbidly obese subjects (cohort 1).
(A) Venn diagrams showing overlap of differentially regulated proteins between SC and OM preadipocytes from NG and T2D morbidly obese subjects (cohort 1). Over-represented canonical pathways in SC vs. OM preadipocytes from NG (B), and T2D (C) obese subjects according to Ingenuity Pathway Analysis (IPA). Blue and red bars indicate down- and up-regulated pathways, respectively, in SC vs. OM preadipocytes. Numbers indicate the number of identified proteins/total proteins annotated to the pathway. Black line indicates –Log2(p value). Samples from two to three individuals per group and fat depot were pooled and used for two separate iTRAQ experiments ( n = 5–6 subjects/group/fat depot). Data normality was tested by Shapiro-Wilk test and Student´s t test was used, a ± 1.5-fold change with p < 0.05 was set as the threshold for categorizing up- and down-regulated proteins. Canonical pathway analysis was performed using IPA (see Materials and Methods section).
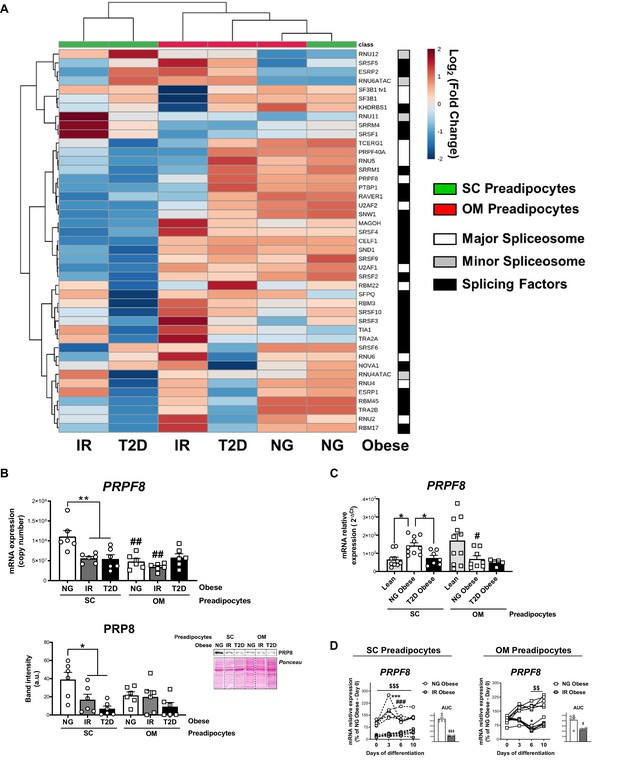
Downregulation of the splicing machinery in subcutaneous (SC) preadipocytes is associated with obesity-related insulin resistance (IR) and type 2 diabetes (T2D).
(A) Hierarchical clustering dendrogram heatmap analysis of splicing-related genes in SC (green) and OM (red) preadipocytes from normoglycemic (NG), IR, and T2D morbidly obese subjects (cohort 1) ( n = 6, 1 technical replicate each) measured by qPCR dynamic array. Rows stand for splicing-related genes (white, mayor spliceosome; grey, minor spliceosome; black, splicing factors), while columns stand for subject groups. The scale in the colour bar represents -Log2(Fold Change). (B) mRNA levels of PRPF8 measured by qPCR dynamic array (upper graph) and representative blot and protein level quantification of PRP8 (lower graph) in SC and OM preadipocytes from NG, IR, and T2D morbidly obese subjects (cohort 1; n = 6, 1 technical replicate each) *p < 0.05, **p < 0.01 vs. NG and/or IR subjects, ##p < 0.01 vs. SC preadipocytes from the same subjects. (C) mRNA levels of PRPF8 in SC and OM preadipocytes from lean, and NG and T2D subjects with simple obesity (cohort 2; n = 5–11, 1 technical replicate each). *p < 0.05 vs. lean and/or NG obese subjects, #p < 0.05 vs. SC preadipocytes from the same subjects. (D) PRPF8 mRNA levels and area under the curve (AUC) during in vitro differentiation of SC and OM preadipocytes from NG and IR morbidly obese subjects (cohort 1; n = 6, 1 technical replicate each). *p < 0.05, ***p < 0.001 vs. PRPF8 mRNA levels at Day 0; ###p < 0.001 vs. PRPF8 mRNA levels at Day 3; $< 0.05, $$p < 0.01, $$$p < 0.001 vs. NG. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively) were used for B and C; two-way ANOVA was used for D. Normality distribution was determined by Shapiro-Wilk normality test. The online version of this article includes the following figure supplements for Figure 2—figure supplement 1, Figure 2—figure supplement 2, and Figure 2—figure supplement 3.
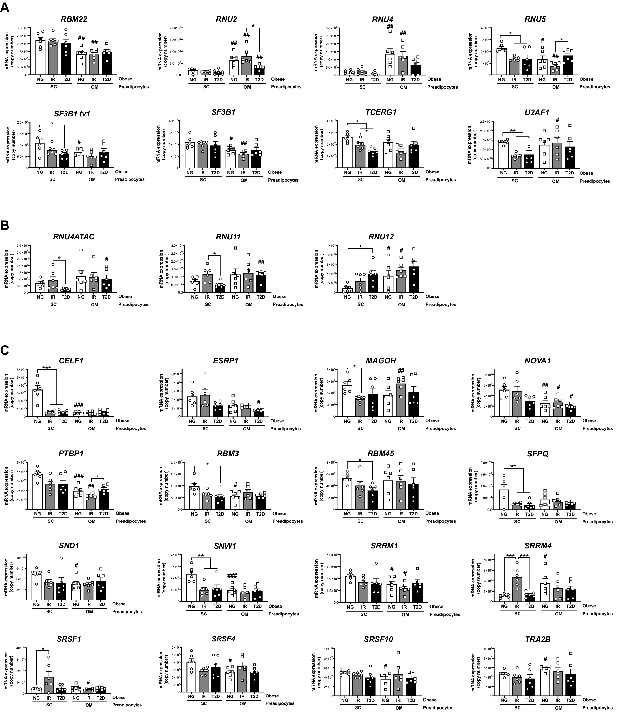
mRNA levels of components of the major spliceosome (A), minor spliceosome (B), and splicing factors (C) in SC and OM preadipocytes from NG, IR, and T2D morbidly obese subjects (cohort1; n = 6, 1 technical replicate each) measured by qPCR dynamic array.
Only the genes from the microarray that were significantly regulated are represented (27 out of 42; PRPF8 is shown in Figure 2B). *p < 0.05, **p < 0.01, ***p < 0.001 vs. NG and/or IR subjects; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. SC preadipocytes from the same subjects. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively). Normality distribution was determined by Shapiro-Wilk normality test.
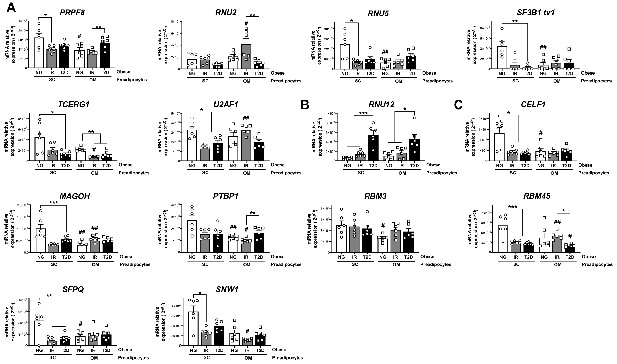
mRNA levels of components of the major spliceosome (A), minor spliceosome (B), and splicing factors (C) in SC and OM preadipocytes from NG, IR, and T2D morbidly obese subjects (cohort 1; n = 6, 1 technical replicate each) measured by RT-PCR.
*< 0.05, **p < 0.01, ***p < 0.001 vs. NG and/or IR subjects; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. SC preadipocytes from the same subjects. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively). Normality distribution was determined by Shapiro-Wilk normality test.
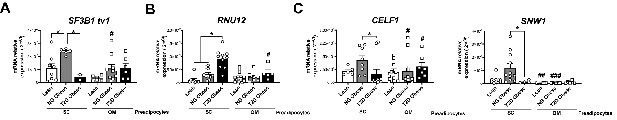
mRNA levels of representative components of the splicing machinery, including the major spliceosome (SF3B1 tv1) (A), minor spliceosome (RNU12) (B), and splicing factors (CELF1 and SNW1) (C) in SC and OM preadipocytes from lean individuals and NG and T2D subjects with simple obesity (cohort 2; n = 5–11, 1 technical replicate each).
*p < 0.05 vs. lean and/or NG obese subjects; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. SC preadipocytes from the same subjects. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively). Normality distribution was determined by Shapiro-Wilk normality test.

PRPF8 silencing impairs adipogenesis in SGBS cells.
(A) PRPF8 mRNA levels in SGBS cells during adipogenesis. (B) Representative blot and protein quantification of PRP8 content in SGBS cells during adipogenesis. (4–6 replicate studies, 3 technical replicates each). (ND), non-detected. *p < 0.05, **p < 0.01, ***p < 0.001 vs. preceding differentiation days. (C) Representative confocal micrographs of SGBS adipocytes 3 days post-transfection (differentiation day 7) with control or PRPF8-siRNA stained with Oil Red-O [lipid droplets (LDs), red] and DAPI (nucleus, blue). Morphometric analysis of LDs was carried out using ImageJ software. Scale bar = 10 μm. (six replicate studies, 10 cells each). mRNA levels of splicing variants of the transcription factors, PPARG and SREBF1 (D), and representative blot and protein level quantification of PPARγ1 and PPARγ2 (E), and of the LD-related proteins, BSCL2, CIDEB, and CIDEC (F), in SGBS adipocytes 3 days post-transfection. (G) Representative blots and protein level quantification of ADRP, BiP, PSMB8, CHOP, DGAT2, FAS, PLIN1, pHSL(Ser563), HSL, Pro-caspase3, and Caspase-3 in SGBS adipocytes 3 days post-transfection. (3–6 replicate studies, 3 technical replicates each). *p < 0.05, **p < 0.01, ***p < 0.001 vs. control; ##p < 0.01 vs. PRPF8-siRNA; $p < 0.05 vs. PRPF8-pcDNA3.1. (H) Representative blots and protein level quantification of pAKT(Ser473) and AKT in SGBS adipocytes 3 days post-transfection treated with/without insulin (100 nmol/L, 15 min). (three replicate studies, 3 technical replicates each). ***p < 0.001 vs. Control-siRNA -Insulin; ###p < 0.001 vs. control-siRNA+ Insulin. (I) Representative confocal micrographs of SGBS adipocytes 5 days post-transfection (day 10 of differentiation) with control, PRPF8-siRNA or PRPF8-pcDNA3.1 alone, or in combination (PRPF8 Recovery) stained with Oil Red-O (LDs, red) and DAPI (nucleus, blue). Morphometric analysis of LDs was carried out using ImageJ software. Scale bar = 10 μm. (five replicate studies, 10 cells each). *p < 0.05, **p < 0.01, ***p < 0.001 vs. control; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. PRPF8-siRNA; $p < 0.05. $$p < 0.01 vs. PRPF8-pcDNA3.1. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively) were used for A and B; unpaired t test or Mann Whitney test (for parametric or non-parametric data, respectively) were used for C-G and I; and two-way ANOVA was used for H. Normality distribution was determined by Shapiro-Wilk normality test. The online version of this article includes the following figure supplements for Figure 3—figure supplement 1, Figure 3—figure supplement 2, and Figure 3—figure supplement 3, and the following source data for Figure 3—source data 1 and Figure 3—source data 2.
-
Figure 3—source data 1
Analysis of CLIP_Seq data (target binding sites and related pathways) from PRPF8-silenced HepG2 and K562 cells provided by The Encyclopedia of RNA Interactomes (ENCORI) database.
- https://cdn.elifesciences.org/articles/65996/elife-65996-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Adipose tissue-specific network of the functional interactions of PRPF8, SF3B1, and SFPQ according to HumanBase tool database, and binding sites of SFPQ according to SpliceAid-F.
- https://cdn.elifesciences.org/articles/65996/elife-65996-fig3-data2-v2.xlsx
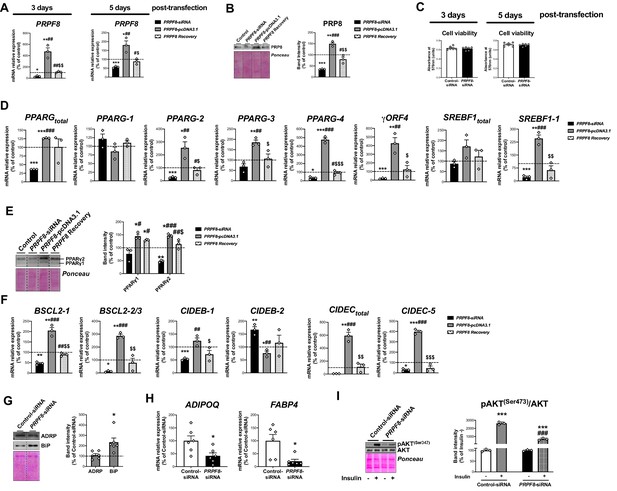
mRNA levels of PRPF8 in SGBS adipocytes at day 3 and at day 5 post-transfection (D7 and D10 of differentiation, respectively) (three replicate studies, 4 technical replicates each) (A), and representative blot and protein level quantification of PRP8 in SGBS adipocytes at day 5 post-transfection (three replicate studies, 3 technical replicates each) (B) with control, PRPF8-siRNA or PRPF8-pcDNA3.1, alone or in combination (PRPF8 Recovery).
Cell viability of SGBS adipocytes at day 3 and day 5 post-transfection (six replicate studies, 3 technical replicates each) (C). mRNA levels of splicing variants of the transcription factors, PPARG and SREBF1 (three replicate studies, 4 technical replicates each) (D), representative blot and protein level quantification of PPARγ1 and PPARγ2 (three replicate studies, 3 technical replicates each) (E), and mRNA levels of splicing variants of the lipid droplet-related proteins, BSCL2, CIDEB, and CIDEC (three replicate studies, 4 technical replicates each) (F) in SGBS adipocytes 5 days post-transfection with control, PRPF8-siRNA or PRPF8-pcDNA3.1, alone or in combination (PRPF8 Recovery). *p < 0.05, **p < 0.01, ***p < 0.001 vs. control; # < 0.05, ##p < 0.01, ###p < 0.001 vs. PRPF8-siRNA; $p < 0.05, $$p < 0.01, $$$p < 0.001 vs. PRPF8-pcDNA3.1. Representative blots and protein level quantification of ADRP and BiP (six replicate studies, 3 technical replicates each) (G), and mRNA levels of the adipokines, FABP4 and ADIPOQ (six replicate studies, 4 technical replicates each) (H), in SGBS adipocytes at day 5 post-transfection. *p < 0.05 vs. control. Representative blot and protein level quantification of pAKT(Ser473) and AKT in SGBS adipocytes 5 days post-transfection treated with/without insulin (100 nmol/L, 15 min) (three replicate studies, 3 technical replicates each) (I). ***p < 0.001 vs. without Insulin; ###p < 0.001 vs. control-siRNA+ Insulin. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively, determined by Shapiro-Wilk normality test) were used for A-F; unpaired t test or Mann Whitney test (for parametric or non-parametric data, respectively) were used for G and H; and two-way ANOVA was used for I.
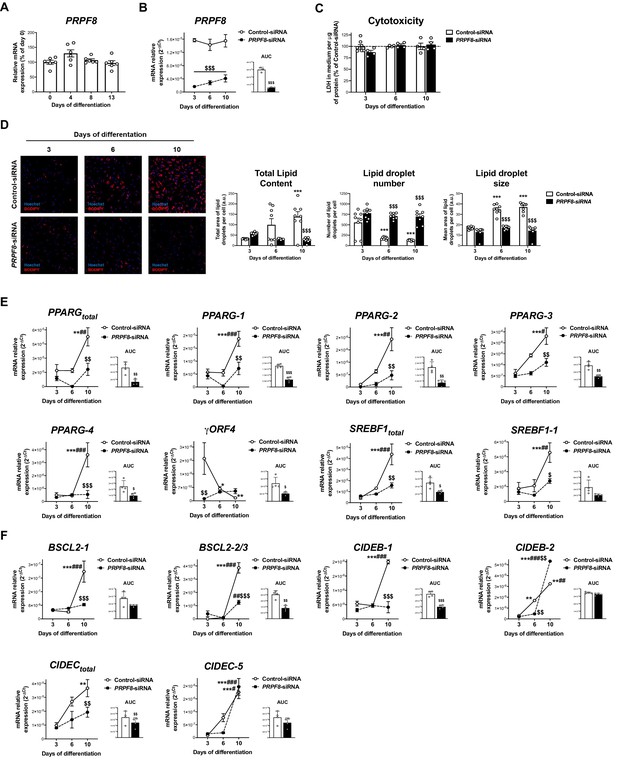
Effects of PRPF8 silencing in differentiating human adipose-derived stem cells (hADSCs).
(A) mRNA levels of PRPF8 in hADSCs cells during differentiation (days 0–13) (six replicate studies, 4 technical replicates each). (B) mRNA levels of PRPF8 in hADSCs cells transfected with PRPF8-siRNA or Control-siRNA at day 1 of differentiation and observed at days 3-6-10 of differentiation; the values of the area under the curve (AUC) are also shown (three replicate studies, 4 technical replicates each). (C) Cytotoxicity assay of hADSCs cells transfected with PRPF8-siRNA or Control-siRNA (4–8 replicate studies, 3 technical replicates each). (D) Representative micrographs of hADSCs cells transfected with PRPF8-siRNA or Control-siRNA after staining with BODIPY (LDs, red) and Hoechst (nucleus, blue) (eight replicate studies, 9 cell fields each). mRNA levels of splicing variants of the transcription factors, PPARG and SREBF1 (E), and of the LD-related proteins, BSCL2, CIDEB, and CIDEC (F) (four replicate studies, 3 technical replicates each) in hADSCs cells transfected with PRPF8-siRNA or Control-siRNA. *p < 0.05, **p < 0.01, ***p < 0.001 vs. day 3 of differentiation; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. day 6 of differentiation; $p < 0.05, $$p < 0.01, $$$p < 0.001 vs. Control-siRNA of the same day of differentiation. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively) were used for A; two-way ANOVA was used for B-F; and unpaired t test or Mann Whitney test (for parametric or non-parametric data, respectively) were used for AUC values in F. Normality distribution was determined by Shapiro-Wilk normality test.
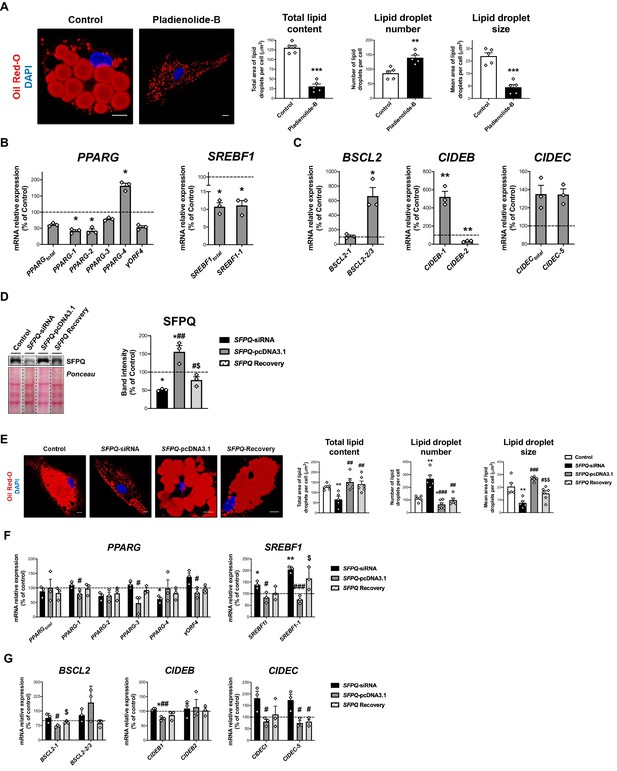
Contribution of the spliceosome components, SF3B1 and SFPQ, to SGBS adipocyte differentiation.
(A) Representative confocal micrographs of SGBS cells after exposition of vehicle (DMSO; control) or pladienolide-B 10–8 M for 24 hr stained with Oil Red-O (LDs, red) and DAPI (nucleus, blue). Morphometric analysis of LDs was carried out using ImageJ software. Scale bar = 10 μm (five replicate studies, 10 cells each). mRNA levels of splicing variants of the transcription factors, PPARG and SREBF1 (B), and the LD-related proteins, BSCL2, CIDEB, and CIDEC (C) in SGBS cells after exposure to vehicle (DMSO; control) or pladienolide-B 10–8 M for 24 h. (three replicate studies, 4 technical replicates each). *p < 0.05, **p < 0.01, ***p < 0.001 vs. control. (D) Representative blot and protein level quantification of SFPQ in SGBS cells after transfection with control, SFPQ-siRNA or SFPQ-pcDNA3.1, alone or in combination (SFPQ-Recovery (three replicate studies, 3 technical replicates each)). (E) Representative confocal micrographs of SGBS cells after transfection with control, SFPQ-siRNA or SFPQ-pcDNA3.1, alone or in combination (SFPQ-Recovery) stained with Oil Red-O (LDs, red) and DAPI (nucleus, blue). Morphometric analysis of LDs was carried out using ImageJ software. Scale bar = 10 μm (5–6 replicate studies, 10 cells each). mRNA levels of splicing variants of the transcription factors, PPARG and SREBF1 (F), and the LD-related proteins, BSCL2, CIDEB, and CIDEC (G) of SGBS cells after transfection with control, SFPQ-siRNA or SFPQ-pcDNA3.1, alone or in combination (SFPQ-Recovery) (three replicate studies, 4 technical replicates each). *p < 0.05, **p < 0.01 vs. control; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. SFPQ-siRNA; $p < 0.05. $$p < 0.01 vs. SFPQ-pcDNA3.1; . Data are presented as mean ± standard error of the mean (S.E.M.). Unpaired t test or Mann Whitney test (for parametric or non-parametric data, respectively) were used for A-C; One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively) were used for D-G. Normality distribution was determined by Shapiro-Wilk normality test.
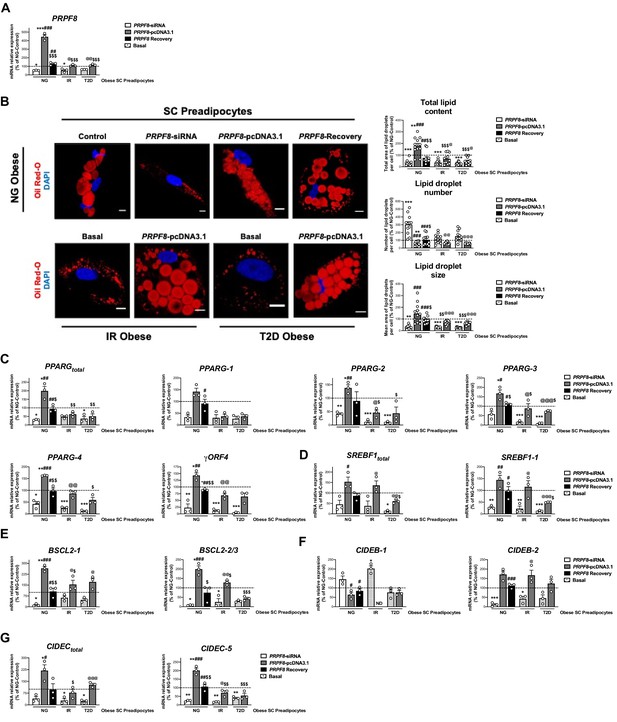
PRPF8 silencing alters differentiation of human primary SC preadipocytes.
Data corresponds to individuals from cohort 1. (A) Quantification of PRPF8 mRNA levels in subcutaneous (SC) preadipocytes from normoglycemic (NG), insulin resistant (IR), and with type 2 diabetes (T2D) morbidly obese individuals transfected with control, PRPF8-siRNA, or PRPF8-pcDNA3.1, alone or in combination (PRPF8 Recovery). (three replicate studies, 4 technical replicates each). (B) Representative confocal micrographs of SC preadipocytes from NG, IR, and T2D morbidly obese individuals after transfection with control, PRPF8-siRNA, or PRPF8-pcDNA3.1 alone or in combination (PRPF8 Recovery) stained with Oil Red-O [lipid droplets (LDs), red] and DAPI (nucleus, blue). Morphometric analysis of LDs was carried out using ImageJ software. Scale bar = 10 μm. (12 replicate studies, 10 cells each). mRNA levels of PPARG (C), and SREBF1 (D), and the LD-related proteins, BSCL2 (E), CIDEB (F), and CIDEC (G) and their splicing forms in SC preadipocytes from NG, IR, and T2D morbidly obese individuals after transfection with control, PRPF8-siRNA or PRPF8-pcDNA3.1, alone or in combination (PRPF8 Recovery). (three replicate studies, 4 technical replicates each). *p < 0.05, **p < 0.01, ***p < 0.001 vs. NG-Control; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. PRPF8-siRNA; $p < 0.05. $$p < 0.01, $$$p < 0.001 vs. PRPF8-pcDNA3.1; @p < 0.05, @@p < 0.01, @@@p < 0.001 vs. corresponding basal. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively) were used. Normality distribution was determined by Shapiro-Wilk normality test.

Unbalanced Unfolded Protein Response (UPR) in preadipocytes is associated with obesity-related insulin resistance (IR) and type 2 diabetes (T2D).
Data corresponds to individuals from cohort 1. Representative blots and protein level quantifications of BiP (A), ATF6αp90 and ATF6αp50 (B), pPERK(Thr980) and PERK (C), pEIF2α(Ser51) and EIF2α (D), CHOP €, pIRE1α(Ser724) and IRE1α (F), and PDI and GRP94 (H) in subcutaneous (SC) and omental (OM) preadipocytes from normoglycemic (NG), IR and T2D morbidly obese subjects ( = 6–12, 1 technical replicate each). (G) mRNA levels of XBP1s and XBP1u in SC and OM preadipocytes from NG, IR and T2D obese subjects ( = 12, 1 technical replicate each). *p < 0.05, **p < 0.01, ***p < 0.001 vs. NG and/or IR subjects; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. SC preadipocytes from the same subjects. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively, determined by Shapiro-Wilk normality test) were used.

Activation of Endoplasmic Reticulum-Associated Degradation (ERAD) in preadipocytes is associated with obesity-related insulin resistance (IR) and type 2 diabetes (T2D).
Data corresponds to individuals from cohort 1. mRNA levels of BIP (A), DERL1, SEC61A1, STT3A and STT3B (B), HRD1, RNF185 and SEL1L (C), and RAD23A and UBQLN1 (D), in subcutaneous (SC) and omental (OM) preadipocytes from normoglycemic (NG), IR and T2D morbidly obese subjects (n = 12, 1 technical replicate each). *p < 0.05, **p < 0.01, ***p < 0.001 vs. NG and/or IR subjects; #p < 0.05, #p < 0.01, ###p < 0.001 vs. SC preadipocytes from the same subjects. (E) Hierarchical clustering dendrogram heatmap analysis of ERAD-related genes in SC (green) and OM (red) preadipocytes from NG, IR, and T2D obese subjects. Rows stand for ERAD-related steps (white, recognition; light grey, retrotranslocation; dark grey, ubiquitination; black, targeting to proteasome), while columns stand for subject groups. The scale in the colour bar represents -Log2(Fold Change). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively, determined by Shapiro-Wilk normality test) were used. The online version of this article includes the following figure supplements for Figure 6—figure supplement 1 and Figure 6—figure supplement 2.
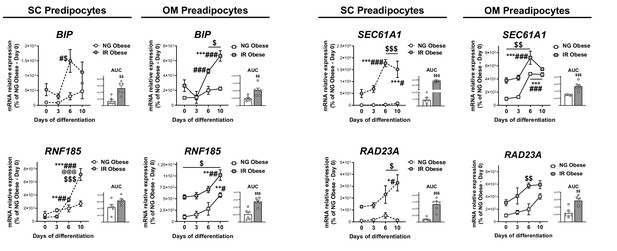
mRNA levels of ERAD-related genes during in vitro differentiation of SC and OM preadipocytes from NG and IR morbidly obese individuals (cohort 1; n = 6, 1 technical replicate each).
Area under the curve (AUC) is also shown. *p < 0.05, **p < 0.01, ***p < 0.001 vs. day 0 of differentiation; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. day 3 of differentiation; @@@p < 0.001 vs. day 6 of differentiation; $p < 0.05, $$p < 0.01, $$$p < 0.001 vs. NG Obese. Data are presented as mean ± standard error of the mean (S.E.M.). Two-way ANOVA was used; and unpaired t test or Mann Whitney test (for parametric or non-parametric data, respectively) was used for AUC values. Normality distribution was determined by Shapiro-Wilk normality test.
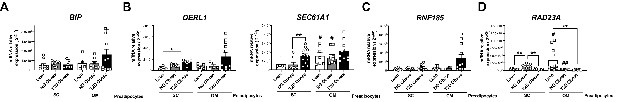
mRNA levels of representative components of the ERAD pathway, participating in the recognition (BIP) (A), retrotranslocation through the endoplasmic reticulum (ER) membrane (DERL1 and SEC61A1) (B), ubiquitination (RNF185) (C), and targeting of misfolded proteins to the proteasome (RAD23A) (D) in SC and OM preadipocytes from lean individuals and NG and T2D subjects with simple obesity (cohort 2; n = 5–11, 1 technical replicate each).
*p < 0.05, **p < 0.01 vs. lean and/or NG obese subjects; #p < 0.05, ##p < 0.01 vs. SC preadipocytes from the same subjects. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively). Normality distribution was determined by Shapiro-Wilk normality test.
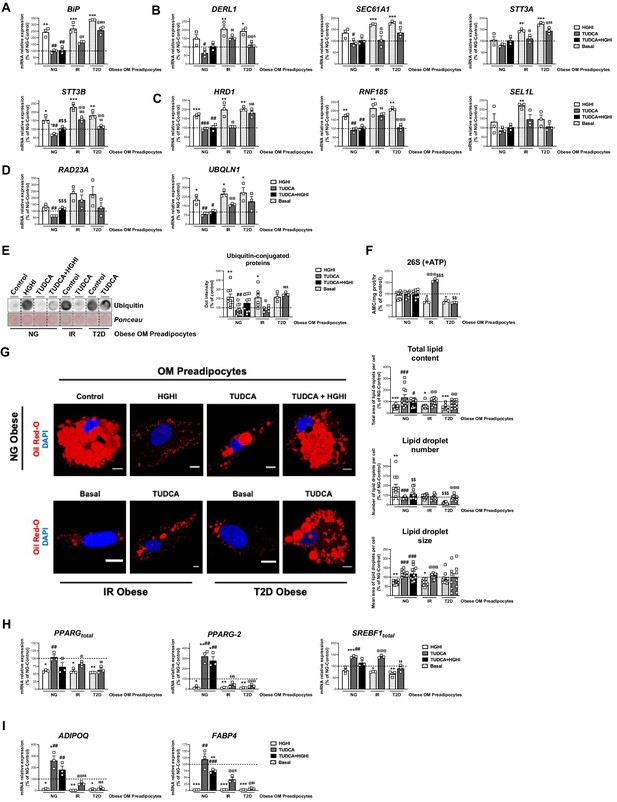
Activation of ERAD in human primary omental (OM) preadipocytes from normoglyceimc (NG) morbidly obese individuals by hyperglycemic / hyperinsulinemic (HGHI) conditions and reversal of ERAD activation in human primary OM preadipocytes from insulin resistant (IR) and with type 2 diabetes (T2D) morbidly obese individuals by TUDCA.
Data correspond to individuals from cohort 1. mRNA levels of BIP (A), DERL1, SEC61A1, STT3A, and STT3B (B), HRD1, RNF185 and SEL1L (C), and RAD23A and UBQLN1 (D). (three replicate studies, 4 technical replicates each). Representative dot-blot and protein quantification of ubiquitin-conjugated proteins (E), and chymotrypsin-like peptidase activity of the 26 S proteasome (+ ATP) (F) in OM preadipocytes from NG, IR, and T2D morbidly obese individuals exposed 14 hr to 0.5 mg/mL TUDCA, 24 hr to HGHI conditions, and/or a combination of both. (3–9 replicate studies, 3 technical replicates each). (G) Representative confocal micrographs of OM preadipocytes from NG, IR, and T2D morbidly obese individuals under the indicated experimental conditions stained with Oil Red-O [lipid droplets (LDs), red] and DAPI (nucleus, blue). Morphometric analysis of LDs was carried out using ImageJ software. Scale bar = 10 μm. (12 replicate studies, 10 cells each). (H–I) mRNA levels of adipogenesis-related genes in OM preadipocytes from NG, IR and T2D morbidly obese individuals under the indicated experimental conditions. (three replicate studies, 4 technical replicates each). *p < 0.05, **p < 0.01. ***p < 0.001 vs. control; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. HGHI; $ < 0.05, $$p < 0.01 vs. TUDCA; @p < 0.05, @@p < 0.01, @@@p < 0.001 vs. corresponding basal. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively) were used. Normality distribution was determined by Shapiro-Wilk normality test. The online version of this article includes the following figure supplements for Figure 7—figure supplement 1, Figure 7—figure supplement 2, and Figure 7—figure supplement 3.
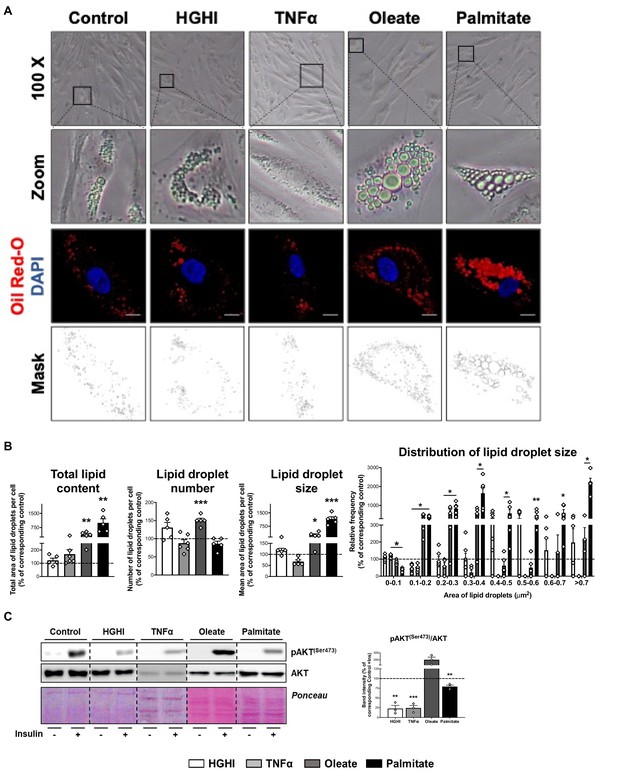
Representative phase-contrast microscopy (x100 magnification and zoom) and confocal micrographs of SGBS preadipocytes exposed during 30 hr to a combination of glucose (4.5 g/L) and insulin (100 nM) (HGHI; white bars), 5 nM TNFα (grey bars), 500 μM Oleate (dark grey bars), or 500 μM Palmitate (black bars) (A).
Cells were stained with Oil Red-O (LDs, red) and DAPI (nucleus, blue). Morphometric analysis of LDs was carried out in untreated and treated SGBS preadipocytes using ImageJ software. Scale bar = 10 μm (five replicate studies, 10 cells each) (B). *p < 0.05, **p < 0.01, ***p < 0.001 vs. corresponding control. Representative blots and protein level quantification of pAKT(Ser473) and AKT in SGBS preadipocytes after exposure to insulin (100 nM, 15 min) or medium alone (three replicate studies, 3 technical replicates each) (C). **p < 0.01, ***p < 0.001 vs. corresponding control with insulin. Data are presented as mean ± standard error of the mean (S.E.M.). Unpaired t test or Mann Whitney test (for parametric or non-parametric data, respectively, determined by Shapiro-Wilk normality test) were used. Normality distribution was determined by Shapiro-Wilk normality test.
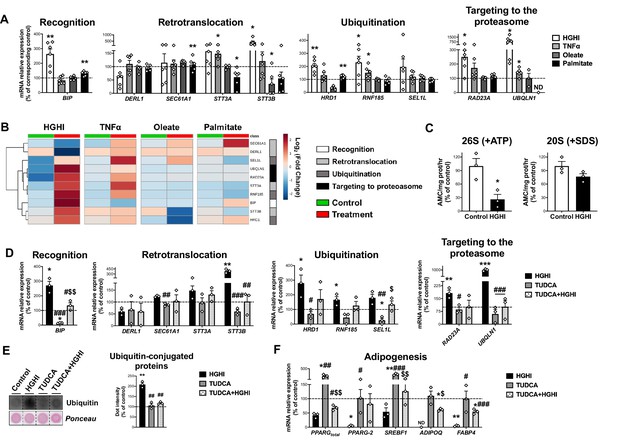
Regulation of Endoplasmic Reticulum-Associated Degradation (ERAD).
(A) mRNA levels of ERAD-related genes in SGBS preadipocytes exposed during 30 h to high glucose (4.5 g/L)/high insulin (100 nmol/L) (HGHI; white), 5 nM TNFα (light grey), 500 μM Oleate (dark grey) or 500 μM Palmitate (black) (six replicate studies, 4 technical replicates each). *p < 0.05, **p < 0.01 vs. corresponding control. (B) Hierarchical clustering dendrogram heatmap analysis of ERAD-related genes in SGBS preadipocytes in control (green) and after exposure to HGHI, TNFα, Oleate or Palmitate (red). Rows stand for ERAD-related steps (white, recognition; light grey, retrotranslocation; dark grey, ubiquitination; black, targeting to proteasome), while columns stand for subject groups. The scale in the colour bar represents -Log2(Fold Change). (C) Chymotrypsin-like peptidase activity of the 26 S proteasome (+ ATP) and 20 S proteasome (+ SDS) (three replicate studies, 3 technical replicates each). mRNA levels of ERAD (D) and adipogenesis-related (E) genes (three replicate studies, 4 technical replicates each), and representative dot-blot and protein quantification of ubiquitin-conjugated proteins (three replicate studies, 3 technical replicates each) (F) in SGBS preadipocytes exposed 14 hr to 0.5 mg/mL TUDCA (TUDCA, grey), 30 h to HGHI conditions (white), or a combination of both (TUDCA + HGHI, black). (ND), non-detected. *p < 0.05, **p < 0.01. ***p < 0.001 vs. control; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. HGHI; $p < 0.05, $$p < 0.01 vs. TUDCA. Data are presented as mean ± standard error of the mean (S.E.M.). Unpaired t test or Mann Whitney test (for parametric or non-parametric data, respectively, determined by Shapiro-Wilk normality test) were used for A and C; one-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively) were used for D-F. Normality distribution was determined by Shapiro-Wilk normality test.
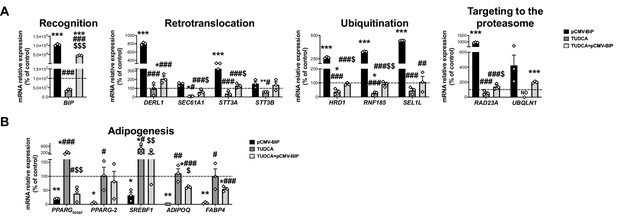
mRNA levels of ERAD (A) and adipogenesis-related genes (B) in SGBS preadipocytes overexpressing BiP (pCMV-BIP, black), exposed 14 h to 0.5 mg/mL TUDCA (TUDCA, grey), or a combination of both (TUDCA + pCMV BIP, white with black dots) (three replicate studies, 4 technical replicates each).
*p < 0.05, **p < 0.01. ***p < 0.001 vs. control; #p < 0.05, ##p < 0.01, ###p < 0.001 vs. pCMV-BIP; $p < 0.05, $$p < 0.01, $$$p < 0.001 vs. TUDCA. Data are presented as mean ± standard error of the mean (S.E.M.). One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively) were used. Normality distribution was determined by Shapiro-Wilk normality test.
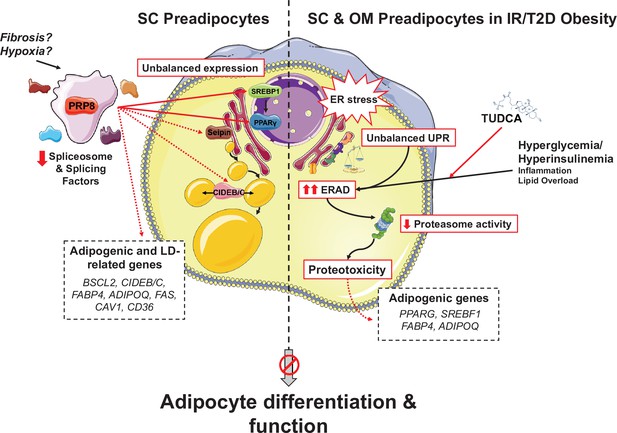
Schematic representation of the proposed mechanisms of action of PRP8/splicing and UPR/ERAD on adipocyte differentiation.
PRP8 mediates alternative splicing events in subcutaneous (SC) preadipocytes by regulating the expression and/or isoform balance of the master regulators of adipogenesis, SREBP1 and PPARγ (solid arrows), likely through a direct action on the corresponding pre-mRNAs. This, in turn, would result in changes in the expression of adipogenic genes down-stream SREBP1 and PPARγ (dashed arrows), yet a direct action of PRP8 on CIDEB splicing is also plausible. The observed down-regulation of PRP8 in SC preadipocytes from obese individuals with insulin resistance (IR) and type 2 diabetes (T2D) would contribute to the impaired adipogenic capacity of these cells. A second pathogenic mechanism is activated in SC preadipocytes from IR/T2D obese individuals that impairs adipogenesis, that is, dysregulated ER proteostasis leading to UPR/ERAD activation. This mechanism, which is also triggered in omental (OM) preadipocytes under conditions of IR/T2D obesity, alters the adipogenic program by modifying the expression of key adipogenic transcripction factors and down-stream adipocyte markers. Splicing dysregulation and UPR/ERAD activation could be differentially triggered by obesogenic insults. The model is based on published molecular mechanisms, in silico analysis of ENCORI and HumanBase databases, and the main findings shown in this article. This figure was created using graphical elements from Servier Medical Art repository (SMART; https://smart.servier.com/).

Representative dot-blot and protein quantification of ubiquitin-conjugated proteins in OM preadipocytes from NG obese individuals in control conditions (control, white), overexpressing BiP (pCMV-BIP, black), exposed 14 h to 0.
5 mg/mL TUDCA (TUDCA, grey), or to a combination of both (TUDCA+pCMV-BIP, white with black dots) (6-7 replicate studies, 1 technical replicate each). **P<0.01 vs. control; ###P<0.001 vs. pCMV-BIP; $P<0.05 vs. TUDCA. Data are presented as mean ± standard error of the mean (S.E.M.), n = 6. Kruskal-Wallis with Dunn's multiple comparisons test was used. Normality distribution was determined by Shapiro-Wilk normality test.

mRNA levels of ERAD-related genes in SGBS preadipocytes three post-transfection (differentiation day 7) with control (white) or PRPF8-siRNA (black) (6 replicate studies, 4 technical replicates each).
**P<0.01 vs. control-siRNA. Data are presented as mean ± standard error of the mean (S.E.M.). Unpaired t test or Mann Whitney test (for parametric or non-parametric data, respectively) were used. Normality distribution was determined by Shapiro-Wilk normality test.
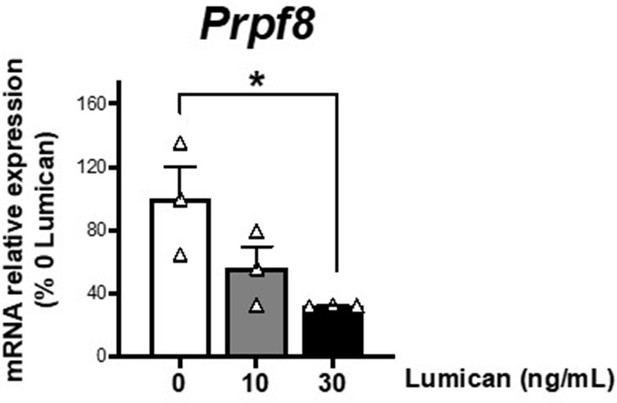
mRNA levels of Prpf8 in differentiated 3T3-L1 adipocytes (day 10 of differentiation) cultured in 3D matrices based on collagen I-enriched hydrogels containing or not increasing concentrations of the proteoglycan, lumican (0-30 ng/mL).
*P<0.05 vs. 0 ng/mL lumican. Data are presented as mean ± standard error of the mean (S.E.M.), n = 3. One-way ANOVA with Tukey’s multiple comparisons test was used. Normality distribution was determined by Shapiro-Wilk normality test.
Tables
Anthropometric and biochemical characteristics of study subjects from cohort 1.
| NG obese | IR obese | T2D obese | |
|---|---|---|---|
| N | 30 | 30 | 18 |
| Gender (female/male) | 15 / 15 | 15 / 15 | 11 / 7 |
| Post-menopause (n, %) | 2 (13) | 2 (13) | 3 (27) |
| Lipid-lowering therapy (n, %) | 0 (0) | 5 (17) | 4 (22) |
| Antidiabetic therapy (n, %) | 0 (0) | 3 (10) | 8 (44) |
| Antihypertensive therapy (n, %) | 1 (3) | 6 (20) | 5 (28) |
| Age (years) | 43 ± 2 | 44 ± 2 | 46 ± 2 |
| Weight (kg) | 140.4 ± 7.0 | 153.4 ± 10.0 | 145.4 ± 7.5 |
| Height (m) | 1.67 ± 0.03 | 1.69 ± 0.02 | 1.65 ± 0.03 |
| Body mass index (kg/m2) | 50.2 ± 2.1 | 52.8 ± 2.9 | 52.9 ± 1.9 |
| Fat mass (%) | 43.1 ± 1.9 | 41.2 ± 2.0 | 42.1 ± 1.7 |
| Lean mass (%) | 39.1 ± 3.2 | 36.1 ± 1.1 | 35.0 ± 1.0 |
| Water mass (%) | 23.0 ± 3.5 | 22.7 ± 1.6 | 22.9 ± 1.3 |
| Waist circumference (cm) | 144.2 ± 5.9 | 156.1 ± 8.1 | 149.9 ± 4.6 |
| Systolic pressure (mm/Hg) | 128.1 ± 2.3 | 127.0 ± 3.5 | 122.1 ± 2.6 |
| Diastolic pressure (mm/Hg) | 78.5 ± 4.4 | 75.5 ± 3.1 | 71.5 ± 2.6 |
| Fasting glucose (mg/dL) | 89.1 ± 1.7 | 105.4 ± 2.0 aaa | 157.3 ± 9.6 aaa, bbb |
| Fasting glucose (mmol/L) | 4.95 ± 0.10 | 5.85 ± 0.11 aaa | 8.73 ± 0.54 aaa, bbb |
| Fasting insulin (mU/L) | 15.4 ± 1.8 | 25.9 ± 2.9 aa | 17.9 ± 2.7 b |
| HbA1c (%) | 5.44 ± 0.06 | 6.22 ± 0.10 aaa | 8.43 ± 0.53 aaa, bb |
| HbA1c (mmol/mol) | 32.2 ± 0.6 | 40.9 ± 1.2 aaa | 64.8 ± 5.7 aaa, bb |
| HOMA-IR (units) | 3.42 ± 0.44 | 6.77 ± 0.74 aa | 6.74 ± 1.05 aa |
| Total cholesterol (mg/dL) | 167.4 ± 10.0 | 181.6 ± 8.3 | 198.1 ± 8.6 |
| LDL cholesterol (mg/dL) | 122.9 ± 11.1 | 112.9 ± 6.6 | 124.2 ± 8.5 |
| HDL cholesterol (mg/dL) | 39.5 ± 3.3 | 36.3 ± 2.1 | 36.7 ± 1.6 |
| Triglycerides (mg/dL) | 108.2 ± 7.0 | 132.7 ± 12.2 | 152.0 ± 12.9 a |
| Free fatty acids (mmol/L) | 66.7 ± 6.3 | 74.4 ± 7.2 | 83.5 ± 6.2 |
| C-reactive protein (mg/L) | 9.61 ± 1.68 | 11.4 ± 2.3 | 17.4 ± 5.3 |
| Uric acid (mg/dL) | 6.30 ± 0.32 | 7.03 ± 0.36 | 6.85 ± 0.68 |
-
NG, normoglycemic; IR, insulin-resistant; T2D, type 2 diabetes; LDL, low-density lipoprotein; HDL, high-density lipoprotein; HbA1c, glycated hemoglobin; HOMA-IR, homeostasis model assessment of insulin resistance. aP <0.05, aaP <0.01, aaaP <0.001 vs. NG Obese; bP <0.05, bbP <0.01, bbbP <0.001 vs. IR Obese. One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively) were used. Normality distribution was determined by Shapiro-Wilk normality test.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | Human preadipocytes from SC and OM adipose tissue | This paper | N/A | Primary cell line |
| Cell line (Homo sapiens) | Human adipose-derived stromal cells (hADSCs) | This paper and previous work from Dr. Mikael Rydén group Gao et al., 2014; Gao et al., 2017 | SC adipose tissue from a male donor (16 years old, BMI 24 kg/m2) | |
| Cell line (Homo sapiens) | Simpson-Golabi-Behmel syndrome (SGBS) cell line | Gift from Prof. Dr. José Manuel Fernández-Real (Institut d’Investigació Biomèdica; Girona, Spain) | N/A | SC adipose tissue from a 3 months male infant with SGBS |
| Biological sample (Homo sapiens) | Human SC and OM adipose tissue from NG, IR and T2D morbid obese individuals (cohort 1) | General and Digestive Surgery Unit and the Lipids and Atherosclerosis Unit of the Reina Sofía University Hospital (Córdoba, Spain) | Reina Sofía University Hospital Research Ethical Committee (439/2010) | Ethics Committee HURS, ref 3170 |
| Biological sample (Homo sapiens) | Human SC and OM adipose tissue from lean, NG and, T2D obese individuals (cohort 2) | Endocrinology ad Surgery Departments at the University Hospital Joan XXIII (Tarragona, Spain)Ejarque et al., 2017; Serena et al., 2016 | N/A | |
| Antibody | ADRP (B-6)(mouse monoclonal) | Santa Cruz Biotechnology | sc-377429, N/A | (1:1000) |
| Antibody | AKT(rabbit polyclonal) | Cell Signaling Technology | 9272, AB_329827 | (1:1000) |
| Antibody | ATF6α (H-280) (rabbit polyclonal) | Santa Cruz Biotechnology | sc-22799, AB_2242950 | (1:1000) |
| Antibody | CASPASE-3 (8G10) (rabbit monoclonal) | Cell Signaling Technology | 9665, AB_2069872 | (1:1000) |
| Antibody | CD14 (M-305) (rabbit polyclonal) | Santa Cruz Biotechnology | sc-9150, AB_2074171 | (1:1000) |
| Antibody | CD45 (H-230) (rabbit polyclonal) | Santa Cruz Biotechnology | sc-25590, AB_2174143 | (1:1000) |
| Antibody | CHOP (L63F7) (mouse monoclonal) | Cell Signaling Technology | 2895, AB_2089254 | (1:1000) |
| Antibody | DGAT2(goat polyclonal) | Novus | NB100-57851, AB_921135 | (1:1000) |
| Antibody | DLK/PREF1 (H-118) (rabbit polyclonal) | Santa Cruz Biotechnology | sc-25437, AB_2292943 | (1:1000) |
| Antibody | eIF2α (3A7A8) (mouse monoclonal) | Santa Cruz Biotechnology | sc-517214, N/A | (1:1000) |
| Antibody | FAS (C-20)(goat polyclonal) | Santa Cruz Biotechnology | sc-16147, AB_2101097 | (1:1000) |
| Antibody | BiP/GRP78 (A-10) (mouse monoclonal) | Santa Cruz Biotechnology | sc-376768, N/A | (1:1000) |
| Antibody | GRP94(rabbit polyclonal) | Cell Signaling Technology | 2104, AB_823506 | (1:1000) |
| Antibody | HSL/LIPE(rabbit polyclonal) | Abcam | ab45422, AB_2135367 | (1:1000) |
| Antibody | IRE1α (B-12) (mouse monoclonal) | Santa Cruz Biotechnology | sc-390960, N/A | (1:1000) |
| Antibody | PDI (C81H6)(rabbit monoclonal) | Cell Signaling Technology | 3501, AB_2156433 | (1:1000) |
| Antibody | PERK (B-5)(mouse monoclonal) | Santa Cruz Biotechnology | sc-377400, AB_2762850 | (1:1000) |
| Antibody | Phospho-AKT (Ser473)(rabbit monoclonal) | Cell Signaling Technology | 4060, AB_2315049 | (1:750) |
| Antibody | Phospho-eIF2α (Ser51)(rabbit polyclonal) | Santa Cruz Biotechnology | sc-101670, AB_2096507 | (1:750) |
| Antibody | Phospho-HSL/LIPE (Ser563)(rabbit polyclonal) | Cell Signaling Technology | 4139, AB_2135495 | (1:750) |
| Antibody | Phospho-IRE1α (Ser724)(rabbit polyclonal) | Abcam | ab48187, AB_873899 | (1:750) |
| Antibody | Phospho-PERK (Thr981)(rabbit polyclonal) | Santa Cruz Biotechnology | sc-32577, AB_2293243 | (1:750) |
| Antibody | PLIN1 (guinea pig polyclonal) | Progen | GP29, AB_2892611 | (1:1000) |
| Antibody | PPARγ (D69)(rabbit polyclonal) | Cell Signaling Technology | 2430, AB_823599 | (1:1000) |
| Antibody | PRP8 (E-5)(mouse monoclonal) | Santa Cruz Biotechnology | sc-55533, AB_831685 | (1:1000) |
| Antibody | PSMB8/LMP7 (sheep polyclonal) | R&D Systems | AF7710, N/A | (1:1000) |
| Antibody | SFPQ(rabbit monoclonal) | Abcam | ab177149, N/A | (1:1000) |
| Antibody | UBIQUITIN(rabbit polyclonal) | Cell Signaling Technology | 3933, AB_2180538 | (1:1000) |
| Antibody | Goat/Sheep IgG (mouse monoclonal) | Sigma-Aldrich | A9452, AB_258449 | (1:2500) |
| Antibody | Guinea Pig IgG (goat polyclonal) | Sigma-Aldrich | A7289, AB_258337 | (1:2500) |
| Antibody | Mouse IgG(rabbit polyclonal) | Sigma-Aldrich | A9044, AB_258431 | (1:2500) |
| Antibody | Rabbit IgG(goat polyclonal) | Sigma-Aldrich | A8275, AB_258382 | (1:10000) |
| Recombinant DNA reagent | phrGFP-N1 | Agilent Technologies | 240,036 | |
| Recombinant DNA reagent | pcDNA3.1(+) | Invitrogen | V79020 | |
| Recombinant DNA reagent | PRPF8 (NM_006445) in pcDNA3.1(+) | GenScript | OHu19527C | |
| Recombinant DNA reagent | SFPQ(NM_005066.3) in pcDNA3.1(+) | GenScript | OHu23607C | |
| Recombinant DNA reagent | pCMV-Myc | Clontech | 635,689 | |
| Recombinant DNA reagent | pCMV-BiP-Myc-KDEL-wt | AddGene | 27,164Addgene_27164 | |
| Sequence-based reagent | Primers for RT-PCR, see Supplementary file 3 | This paper | N/A | |
| Sequence-based reagent | Primers for splicing-machinery components array | Gahete et al., 2018 | N/A | |
| Sequence-based reagent | PRPF8 Silencer Select Validated siRNA | Ambion | 4390824 | |
| Sequence-based reagent | SFPQ Silencer Select Validated siRNA | Ambion | s224606 | |
| Sequence-based reagent | Silencer Select Negative Control No. 1 siRNA | Ambion | 4390843 | |
| Peptide, recombinant protein | 3-Isobutyl-1-methylxanthine (IBMX) | Sigma-Aldrich | I5879 | |
| Peptide, recombinant protein | Antipain | Sigma-Aldrich | A6191 | |
| Peptide, recombinant protein | Biotin | Sigma-Aldrich | B4639 | |
| Peptide, recombinant protein | Bovine Serum Albumin (BSA) | Sigma-Aldrich | A8806, A7030 | |
| Peptide, recombinant protein | Chymostatin | Sigma-Aldrich | C7268 | |
| Peptide, recombinant protein | Collagenase Type V | Sigma-Aldrich | C9263 | |
| Peptide, recombinant protein | Dexamethasone | Sigma-Aldrich | D4902, D1756 | |
| Peptide, recombinant protein | Human 3,3’,5-Trihydrochloride sodium salt (T3) | Sigma-Aldrich | T5516, T6397 | |
| Peptide, recombinant protein | Human FGF2 | Sigma-Aldrich | F0291 | |
| Peptide, recombinant protein | Human Insulin | Sigma-Aldrich | I2643, I9278 | |
| Peptide, recombinant protein | Human Transferrin | Sigma-Aldrich | T8158 | |
| Peptide, recombinant protein | Human Tumor Necrosis Factor-α (TNFα) | Sigma-Aldrich | T6674 | |
| Peptide, recombinant protein | Hydrocortisone | Sigma-Aldrich | H0135 | |
| Peptide, recombinant protein | Leupeptin | Sigma-Aldrich | L2884 | |
| Peptide, recombinant protein | MG-132 | Calbiochem | 474,790 | |
| Peptide, recombinant protein | N-Succinyl-Leu-Leu-Val-Tyr-7-Amido-4-Methylcoumarin (Suc-LLVY-AFC) | Sigma-Aldrich | S6510 | |
| Peptide, recombinant protein | Pepstatin A | Sigma-Aldrich | P4265 | |
| Peptide, recombinant protein | Rosiglitazone | Sigma-Aldrich | R2408 | |
| Chemical compound, drug | (+)-Sodium L-ascorbate | Sigma-Aldrich | A4034 | |
| Chemical compound, drug | 1,4-dithiothreitol (DTT) | Thermo Scientific | R0862 | |
| Chemical compound, drug | Acetonitrile | Sigma-Aldrich | 271,004 | |
| Chemical compound, drug | Adenosine 5′-triphosphate (ATP) disodium salt hydrate | Sigma-Aldrich | A26209 | |
| Chemical compound, drug | BODIPY 500/510 C1, C12 (4,4-Difluoro-5-Methyl-4-Bora-3a,4a-Diaza-s-Indacene-3-Dodecanoic Acid) | Invitrogen | D3823 | |
| Chemical compound, drug | Calcium chloride (CaCl2) | Sigma-Aldrich | 449,709 | |
| Chemical compound, drug | CHAPS hydrate | Sigma-Aldrich | C3023 | |
| Chemical compound, drug | Chloroform | Sigma-Aldrich | C2432 | |
| Chemical compound, drug | Clarity Western ECL Substrate | Bio-Rad | 1705061 | |
| Chemical compound, drug | D-(+)-Glucose | Panreac Applichem / Siga-Aldrich | 1413411211, G8270 | |
| Chemical compound, drug | DAPI | Sigma-Aldrich | D9542 | |
| Chemical compound, drug | Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | D4540 | |
| Chemical compound, drug | D-Pantothenic acid hemicalcium salt | Sigma-Aldrich | P5155 | |
| Chemical compound, drug | Dulbecco’s Phosphate Buffered Saline (D-PBS) Solution | Sigma-Aldrich | D1408 | |
| Chemical compound, drug | Dulbecco’s Phosphate Buffered Saline (D-PBS) Solution (-) Ca2+, Mg2+ | HyClone | SH30028.02 | |
| Chemical compound, drug | Dulbecco’s Modified Eagle Medium (DMEM)/F-12 (1:1) | Gibco | 31330–038 | |
| Chemical compound, drug | EDTA disodium salt | Sigma-Aldrich | E5134 | |
| Chemical compound, drug | Exonuclease I Reaction Buffer | New England BioLabs | B0293S | |
| Chemical compound, drug | Fluorescence Mounting Medium | Dako | S3023 | |
| Chemical compound, drug | Formic acid | Scharlau | AC10760050 | |
| Chemical compound, drug | Glycerol | VWR | 97063–892 | |
| Chemical compound, drug | Ham’s F-12 Nutrient Mix | Gibco | 21765037 | |
| Chemical compound, drug | Hepes Buffer (1 M) | Gibco | 15630056 | |
| Chemical compound, drug | Hoechst 33342, Trihydrochloride, Trihydrate | Invitrogen | H3570 | |
| Chemical compound, drug | Immersol Immersion Oil | Carl Zeiss | 518 F | |
| Chemical compound, drug | Iodoacetamide (IAA) | Sigma-Aldrich | I1149 | |
| Chemical compound, drug | Isopropanol | Sigma-Aldrich | 33,539 | |
| Chemical compound, drug | iTRAQ Reagents | Applied Biosystems | PN4351918 | |
| Chemical compound, drug | Lipofectamine 2000 Transfection Reagent | Invitrogen | 11668019 | |
| Chemical compound, drug | Lipofectamine RNAiMAX Transfection Reagent | Invitrogen | 13778150 | |
| Chemical compound, drug | Magnesium Chloride (MgCl2) | Merck | 5,833 | |
| Chemical compound, drug | Magnesium Sulfate 7-hydrate (MgSO4) | Panreac Applichem | 141,404 | |
| Chemical compound, drug | Methanol (MeOH) | VWR Chemicals | 20864320 | |
| Chemical compound, drug | Newborn Calf Serum (NCS) | Gibco | 16010159 | |
| Chemical compound, drug | Nonfat dried milk powder | Panreac Applichem | Cat# A0830 | |
| Chemical compound, drug | Oil Red O | Sigma-Aldrich | O0625 | |
| Chemical compound, drug | Oleic acid | Sigma-Aldrich | O1383 | |
| Chemical compound, drug | Opti-MEM I Reduced Serum Medium | Gibco | 11058021 | |
| Chemical compound, drug | Paraformaldehyde (PFA) | Bosterbio | AR1068 | |
| Chemical compound, drug | Penicillin-Streptomycin (10,000 U/mL) | Sigma-Aldrich / Gibco | P4333, 15140122 | |
| Chemical compound, drug | Pladienolide-B | Santa Cruz Biotechnology | sc-391691 | |
| Chemical compound, drug | Potassium Chloride (KCl) | Merck | 104,936 | |
| Chemical compound, drug | Ponceau S | Sigma-Aldrich | P3504 | |
| Chemical compound, drug | RBC Lysis Buffer | Norgen Biotek Corp. | 21,201 | |
| Chemical compound, drug | RNase-Free DNase Set | Qiagen | 79,254 | |
| Chemical compound, drug | Sodium Chloride (NaCl) | Merk Millipore | 7647145 | |
| Chemical compound, drug | Sodium Dodecyl Sulfate (SDS) | Sigma-Aldrich | L3771 | |
| Chemical compound, drug | Sodium Hydroxide (NaOH) | Sigma-Aldrich | S4085 | |
| Chemical compound, drug | Sodium Palmitate | Sigma-Aldrich | P9767 | |
| Chemical compound, drug | Sodium Phosphate Monobasic Monohydrate (NaH2PO4) | Fisher scientific | BP330-1 | |
| Chemical compound, drug | Sodium Phosphate Monobasic Monohydrate Dihydrate (NaH2PO4 × 2H2 O) | Sigma-Aldrich | 71,505 | |
| Chemical compound, drug | TE Buffer | Invitrogen | 12090015 | |
| Chemical compound, drug | Tetraethylammonium Bromide (TEAB) | Sigma-Aldrich | 241,059 | |
| Chemical compound, drug | Thiazolyl Blue Tetrazolium Bromide (MTT) | Sigma-Aldrich | M5655 | |
| Chemical compound, drug | Trichloroacetic acid (TCA) | Sigma-Aldrich | T6399 | |
| Chemical compound, drug | Triton X-100 | Sigma-Aldrich | T8787 | |
| Chemical compound, drug | Trizma base (Tris base) | Sigma-Aldrich | T6066 | |
| Chemical compound, drug | Trizma hydrochloride (Tris-HCl) | Sigma-Aldrich | T3253 | |
| Chemical compound, drug | TRIzol Reagent | Invitrogen | 15596018 | |
| Chemical compound, drug | Trypan Blue solution | Sigma-Aldrich | T8154 | |
| Chemical compound, drug | Trypsin Gold, Mass Spectrometry Grade | Promega | V5280 | |
| Chemical compound, drug | Trypsin-EDTA solution | Sigma-Aldrich | T3924 | |
| Chemical compound, drug | Tween 20 | Panreac Applichem | A1389 | |
| Chemical compound, drug | Urea | Sigma-Aldrich | U1205 | |
| Commercial assay or kit | 96.96 DNA Binding Dye Sample / Loading Kit | Fluidigm | BMK-M10-96.96-EG | |
| Commercial assay or kit | AllPrep DNA/RNA/Protein Mini Kit | Qiagen | 80,004 | |
| Commercial assay or kit | Cytotoxicity Detection Kit Plus | Roche | 4744926001 | |
| Commercial assay or kit | GoTaq qPCR Master Mix | Promega | A6001 | |
| Commercial assay or kit | Mycoplasma gel detection kit | Biotools | 4,542 | |
| Commercial assay or kit | NEON Transfection System Kit | Invitrogen | MPK10096 | |
| Commercial assay or kit | Protein Assay Dye Reagent Concentrate | Bio-Rad | 5000006 | |
| Commercial assay or kit | RC DC Protein Assay | Bio-Rad | 5000122 | |
| Commercial assay or kit | RevertAid First Strand cDNA Synthesis Kit | Thermo Scientific | K1621 | |
| Commercial assay or kit | SsoFast EvaGreen Supermix | Bio-Rad | 1725200 | |
| Software, algorithm | Biomark & EP1 Software 4.5.2 | FluidigmSCR_015685 | https://www.fluidigm.com/software | |
| Software, algorithm | Basic Local Alignment Search Tool (BLAST) | NCBI Nowicki et al., 2018SCR_004870 | https://blast.ncbi.nlm.nih.gov/Blast.cgi | |
| Software, algorithm | ENCORI: The Encyclopedia of RNA Interactomes | Li et al., 2014N/A | http://starbase.sysu.edu.cn/index.php | |
| Software, algorithm | GeNorm 3.3 | Vandesompele et al., 2002SCR_006763 | https://genorm.cmgg.be/ | |
| Software, algorithm | GraphPad PRISM 7 | GraphPad Software, Inc Mitteer et al., 2018SCR_002798 | https://www.graphpad.com | |
| Software, algorithm | High-Content Screening (HCS) Studio Cell Analysis Software 2.0 | Thermo Fisher ScientificSCR_018706 | https://www.thermofisher.com/es/es/home/life-science/cell-analysis/cellular-imaging/high-content-screening/hcs-studio-2.html | |
| Software, algorithm | HumanBase | Greene et al., 2015SCR_016145 | https://hb.flatironinstitute.org/ | |
| Software, algorithm | Huygens Professional 2.4.4 | Scientific Volume ImagingSCR_014237 | https://svi.nl/Huygens-Professional | |
| Software, algorithm | ImageJ 1.50b | National Institute of Health (NIH) Schneider et al., 2012SCR_001935 | https://imagej.nih.gov/ij/ | |
| Software, algorithm | Ingenuity Pathways Analysis (IPA) 49309495 | Qiagen, provided by the Andalusian Bioinformatics Platform (PAB) center (University of Málaga, Spain;http://www.scbi.uma.es) Krämer et al., 2014SCR_008653 | http://www.ingenuity.com/ | |
| Software, algorithm | LightCycler 96 1.1.0.1320 | Roche Life ScienceSCR_012155 | https://lifescience.roche.com/en_es/products/lightcycler-381711.html | |
| Software, algorithm | Magellansoftware 7.2 SP1 | TecanSCR_008715 | https://lifesciences.tecan.com/software-magellan | |
| Software, algorithm | MetaboAnalyst 4.0 | Chong et al., 2018SCR_015539 | https://www.metaboanalyst.ca/docs/About.xhtml | |
| Software, algorithm | Primer3 Input 4.1.0 | Untergasser et al., 2012SCR_003139 | http://bioinfo.ut.ee/primer3/ | |
| Software, algorithm | Protein ANalysis THrough Evolutionary Relationships (PANTHER) classification system 14.1 | Mi et al., 2019SCR_004869 | http://www.pantherdb.org/ | |
| Software, algorithm | Real-Time PCR Analysis Software 3.0 | FluidigmSCR_015686 | https://www.fluidigm.com/ | |
| Software, algorithm | SoftMax Pro 2.2.1 | Molecular DevicesSCR_014240 | https://www.moleculardevices.com/ | |
| Software, algorithm | SpliceAid-F | Giulietti et al., 2013SCR_002082 | http://srv00.recas.ba.infn.it/SpliceAidF/ | |
| Software, algorithm | Statistic R module | Pearl softwareSCR_002394 | https://www.pearlsoftware.com/ | |
| Software, algorithm | StepOne Real-Time PCR System 2.3 | Thermo ScientificSCR_014281 | https://www.thermofisher.com/es/es/home.html | |
| Software, algorithm | Stratagene Mx3000p | Thermo ScientificSCR_020526 | https://www.thermofisher.com/es/es/home.html | |
| Software, algorithm | Venn Diagram Plotter | Pacific Northwest National LaboratorySCR_012842 | https://omics.pnl.gov/software/venn-diagram-plotter |
Additional files
-
Supplementary file 1
Lineal regression analyses of subcutaneous (SC) and omental (OM) preadipocytes proliferation rate and anthropometric and biochemical parameters, = 24–58.
r = Spearman’s correlation coefficient. *P < 0.05, **P < 0.01, ***P < 0.001. Normality distribution was determined by Shapiro-Wilk normality test.
- https://cdn.elifesciences.org/articles/65996/elife-65996-supp1-v2.docx
-
Supplementary file 2
Anthropometric and biochemical characteristics of study subjects from cohort 2.
NG, normoglycemic; T2D, type 2 diabetes; LDL, low-density lipoprotein; HDL, high-density lipoprotein; HOMA-IR, homeostasis model assessment of insulin resistance. aaP <0.01, aaaP <0.001 vs. Lean; bP <0.05 vs. NG Obese. One-way ANOVA with Tukey’s multiple comparisons test or Kruskal-Wallis with Dunn’s multiple comparisons test (for parametric or non-parametric data, respectively) were used. Normality distribution was determined by Shapiro-Wilk normality test.
- https://cdn.elifesciences.org/articles/65996/elife-65996-supp2-v2.docx
-
Supplementary file 3
Sequences and transcript sizes of primers used for RT-PCR studies.
bp, base pairs.
- https://cdn.elifesciences.org/articles/65996/elife-65996-supp3-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/65996/elife-65996-transrepform1-v2.pdf





