Chromatin topology defines estradiol-primed progesterone receptor and PAX2 binding in endometrial cancer cells
Figures
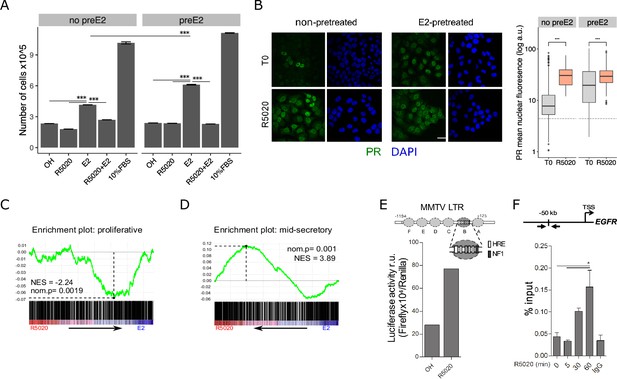
R5020 inhibits E2-induced Ishikawa cell proliferation through an active PR that is capable of transactivating an exogenous MMTV promoter sequence and an endogenous enhancer sequence located 50 kb upstream of EGFR gene.
(A) Proliferation of Ishikawa cells either pretreated with E2 10 nM for 12 hr (preE2) or not (no preE2) and later treated with vehicle (OH), E2 (E2), R5020 (R5020), E2 combined with R5020 (E2+R5020) and FBS (10%FBS), expressed as mean number of cells ± SE of three independent experiments. (***) p < 0.001. (B) Immunofluorescence of PR in untreated (T0; top left), 60 min R5020-treated (R5020; bottom left), 12 h E2-pretreated (top right) and 12 hr E2-pretreated 60 min R5020-treated (bottom right) Ishikawa cells. Scale bar is equivalent to 30 m. Mean nuclear signal of PR for every cell in all images was determined and shown to the right of the images as arbitrary units (log a.u.). Horizontal dashed lines in boxplots indicate background signal for secondary antibody. (***) p < 0.001. (C and D) Gene set enrichment analysis (GSEA) results using R5020- and E2-treated Ishikawa expression profiles as phenotypes for classification of normal endometrium (proliferative and secretory) samples. Enrichment profile (green) shows correlation of normal samples at the top or bottom of a ranked gene list (phenotypes). Normalized enrichment scores (NES) and nominal p values (nom.p) are shown in the graphs. (E) Ishikawa cells transfected with an MMTV-Luciferase reporter gene and treated with vehicle (OH) and R5020 (R5020) for 18 hr. Diagram at the top depicts MMTV LTR promoter features, including several hormone response elements (HRE) and a nuclear factor 1 (NF1) binding site within nucleosome B (dark gray circle and magnification). Numbers in the diagram indicate base pair position relative to transcription start site (TSS). Results are expressed as relative units (r.u.) of Luciferase activity. (F) Representation of EGFR TSS and the enhancer sequence located 50 kb upstream used to evaluate PR recruitment. Black arrows indicate position of qPCR primers employed on samples treated or not (0) with R5020 for 5, 30, and 60 min. Unspecific immunoprecipitation of chromatin was performed in parallel with normal rabbit IgG (IgG). Results are expressed as %input DNA and bars represent mean fold change in PR enrichment relative to time 0 (untreated cells) ± SE of two independent experiments. (*) p < 0.05.
-
Figure 1—source data 1
Proliferation assays in treated and untreated Ishikawa cells.
Measure represents number of cells x105 in the table.
- https://cdn.elifesciences.org/articles/66034/elife-66034-fig1-data1-v2.csv
-
Figure 1—source data 2
Ishikawa mean nuclear signal intensity.
- https://cdn.elifesciences.org/articles/66034/elife-66034-fig1-data2-v2.csv
-
Figure 1—source data 3
Ishikawa treated and untreated normalized expression dataset.
- https://cdn.elifesciences.org/articles/66034/elife-66034-fig1-data3-v2.txt
-
Figure 1—source data 4
PR binding Ct values in EGFR enhancer sequence.
- https://cdn.elifesciences.org/articles/66034/elife-66034-fig1-data4-v2.csv
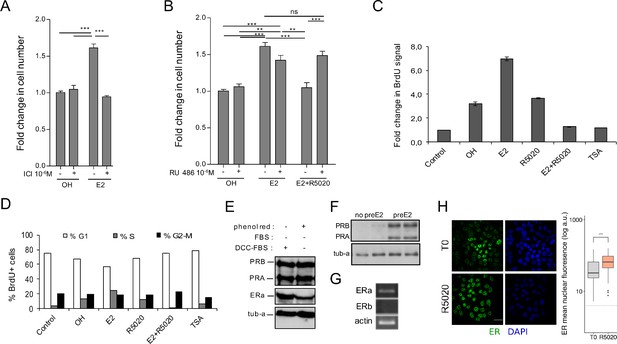
E2-dependent cell cycle progression and proliferation of Ishikawa cells is mediated by ERalpha.
R5020 antagonism on E2 is mediated by PR. (A) Cells were pretreated with ICI182780 (ICI ) and then treated with vehicle (OH) and E2 (E2) for 48 hr. Number of cells was determined and presented as in Figure 1A. (***) p < 0.001. (B) Cells were pretreated with RU486 (RU 486 ) and later treated with vehicle (OH), E2 10 nM (E2) and E2 and R5020 (E2+ R5020) for 48 hr. Number of cells was determined and expressed as in A. (**) p < 0.01; (***) p < 0.001; ns: not significant. (C) BdrU incorporation was measured in untreated cells (control) and treated with vehicle (OH), E2 10 nM (E2), R5020 10 nM (R5020), E2 and R5020 (E2+ R5020) and TSA 250 nM (TSA) for 15–18 hr. Mean fold change in BrdU-positive cells for each treatment over Control ± SE of three independent experiments is shown. (D) Percentage of BdrU-positive cells in each phase of the cell cycle for treatments shown in D: %G1 (white bars), %S (gray bars) and %G2-M (black bars). (E) PR (PRB and PRA) and ERalpha (ERa) protein levels of serum-deprived Ishikawa cells were assayed by western blot (Ishikawa) in two different conditions: phenol red-positive medium (phenol red +) and phenol red-negative medium (phenol red -) with 5% dextran-charcoal-treated serum (DCC-FBS). Alfa tubulin was used as loading control (tub-a). Western blot image at the bottom of the figure shows PR protein levels in non-pretreated (no preE2) and 12 hr E2-pretreated (preE2) Ishikawa cells using alfa tubulin as loading control (tub-a). (F) Western blot image showing PR protein levels in Ishikawa cells pretreated with E2 for 12 hr (preE2) and non-pretreated (no preE2). Alfa tubulin was used as loading control (tub-a). (G) mRNA levels of ERalpha, ERbeta and actin in serum-deprived Ishikawa cells. (***) p < 0.001. (H) Representative image of anti-ERalpha detection in Ishikawa cells treated or not with R5020 for 60 min. Quantification was performed as in Figure 1. Nuclei were revealed with DAPI (blue).
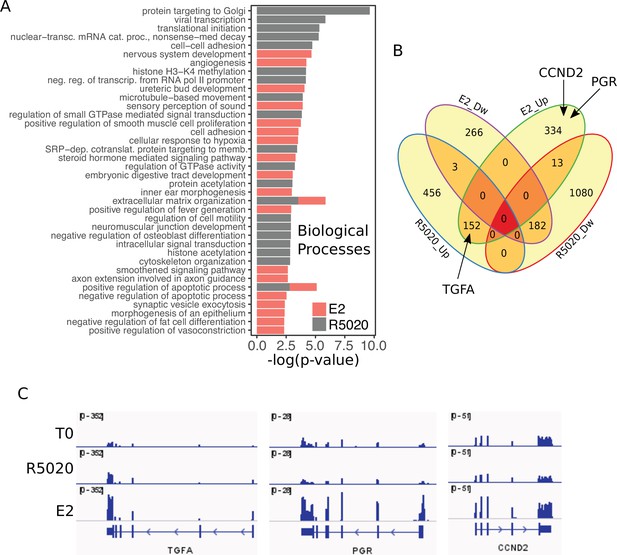
RNAseq results from hormone-treated Ishikawa cells.
(A) Overrepresented Biological Processes in RNAseq results from Ishikawa cells treated with R5020 and E2 for 12h analysed using DAVID web-based tool. Only the top 20 terms with p<0.05 were selected for each hormone. (B) Venn Diagram represents intersection between differentially expressed genes (v.T0; log2FC=±0.8, q < 0.05) in Ishikawa cells treated with R5020 and E2 for 12h. Upregulated and downregulated genes are indicated by Up and Dw, respectively. (C) Normalized expression profiles of genes located in the proximity of steroid receptor binding. RNAseq results expressed in RPKM for genes TGFA, PGR and CCND2 displayed in IGV-based environment. Tracks for untreated (T0) and R5020 10nM (R5020) and E2 10nM (E2) 12 hr treatments were represented in the same vertical viewing range value.
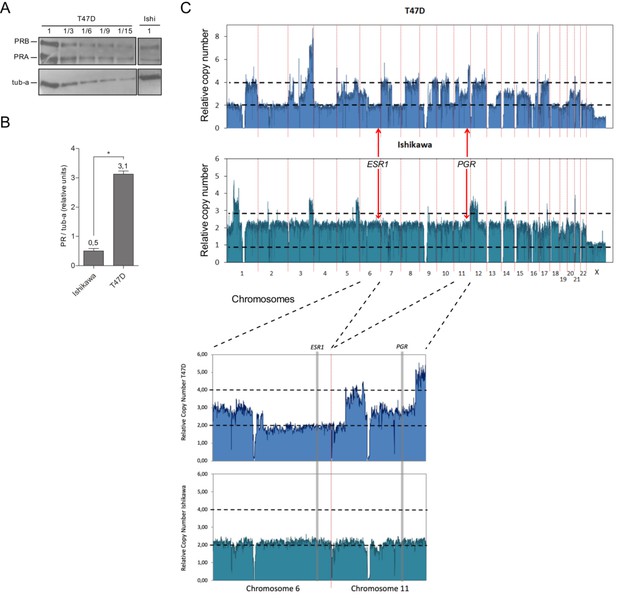
Ishikawa cells express six times less PR than T47D cells.
(A) Representative image of western blot assays using protein extracts from Ishikawa and T47D cells. Thirty µg (1) of T47D extracts were seeded followed by 10µg (1/3), 5µg (1/6), 3.3µg (1/9), and 2µg (1/15) of the same extract. 30µg of protein extract from Ishikawa cells (Ishi) were loaded on the same gel. PR isoforms (PRA and PRB) levels were evaluated and tubulin (tub-a) as control. Image was processed to remove lanes that did not contribute to the present analysis. (B) Quantification of PR protein levels in Ishikawa and T47D cells expressed as mean units of PRA + PRB relative to tubulin protein levels (tub-a) of three independent experiments. Numbers above bars indicate value of quantification relative to tubulin determined by densitometry of western blot assays. (*) pP < 0.05. (C) Copy number variation (CNV) analysis of 100kb regions distributed over all chromosomes (hg19 UCSC) in T47D (upper panel) and Ishikawa (lower panel) cell lines. Black dotted lines limit diploid (2) and tetraploid (4) numbers. Location of estrogen receptor 1 (ESR1) and progesterone receptor (PGR) loci are indicated with red arrows. Enlarged image of regions covering chromosome 6 (ESR1) and chromosome 11 (PGR) is shown below graph.
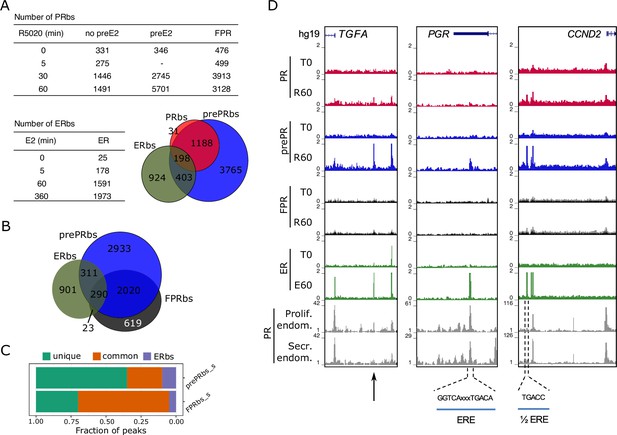
Estradiol induces R5020-dependent PR binding to specific regions in chromatin.
(A) Upper table shows total number of PRbs obtained by ChIPseq for untreated (0 min) and R5020-treated (5, 30 and 60 min) endometrial Ishikawa cells under three different conditions: non-pretreated with E2 (PR), pretreated with E2 for 12 h (prePR) and exogenous expression of PR (FPR). Lower table shows number of ERbs using anti-ERalpha antibody on untreated (0 min) and E2-treated (5, 60 and 360 min) Ishikawa cells. Venn Diagram shows shared binding sites among PRbs (red), prePRbs (blue) and ERbs (green) at 60 min. (B) Venn Diagram shows intersection between ERbs (green), FPRbs (dark grey) and prePRbs (blue) at 60 min. (C) Fraction of peaks in FPR and prePR after substraction of shared PRbs (FPR_s and prePR_s, respectively) that are not shared with each other (unique), that are common to each other (common) and that are common with ER (ERbs). (D) Normalized coverage of PR and ERalpha binding in untreated (T0) and 60 min hormone-treated (R60 and E60) Ishikawa cells and PR binding in proliferative (GSE1327133) and secretory (GSE1327134) endometrium. Black arrow indicates peak of interest. R60: 60min R5020 ; E60: 60 min E2 . The three regions displayed include TGFA, PGR and CCND2 genes (indicated at the top). An estrogen response element (ERE) and a half ERE are indicated below the peaks.
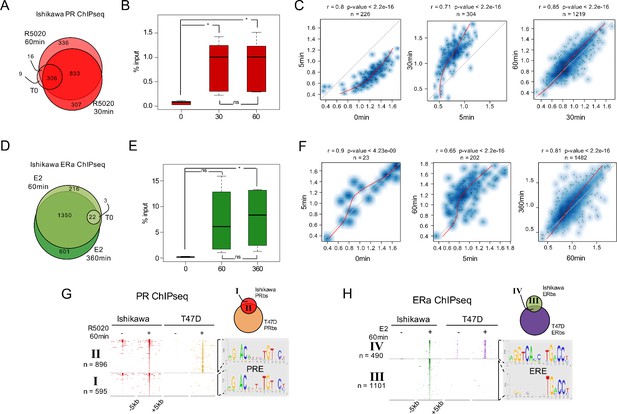
Genome-wide analysis of PR and ERalpha binding in Ishikawa cells.
(A) ChIPseq results using anti-PR antibody in Ishikawa cells. Cells were treated or not (T0) for 5 min (R5), 30 min (R30) and 60 min (R60) with R5020 10 nM. The number of PRbs for T0, R30 and R60 is indicated in the Venn Diagram. (B) Boxplot shows validated PR binding sites expressed as percentage of input (%input) for 0, 30 and 60 min R5020-treated Ishikawa cells. Sites included in this analysis are located in: promoter of ALPP gene, intergenic sequence upstream of ALPP gene, intergenic sequence upstream of EGFR gene, distal promoter of SERPINA3 gene, promoter of RPS6KA1 and intragenic sequence in TGFA gene. Significance was evaluated by ANOVA followed by Tukey's multiple contrasts and significant results are indicated for each individual comparison. (ns) not significant; (*) P < 0,05. (C) Scatterplots depict correlation in peak signal intensity among samples. Number of sites in common (n) as wells as correlation coefficient (r) and p-value are shown over each plot. Non-linear regression curves are indicated as red lines. (D) ChIPseq results using anti-ERalpha antibody in Ishikawa cells treated or not (T0) with E2 10 nM for 5 (E5), 60 (E60) and 360 min (E360). In the Venn Diagram are shown untreated (T0), E60 and E360 as well as the number of ER binding sites (ERbs) for each group. (E) Boxplots shows validated ERalpha binding sites expressed as percentage of input (%input) for 0, 60 and 360 min E2-treated Ishikawa cells. Sites included in this analysis are located in: intergenic region upstream of ALPP gene, intergenic region upstream of TGFA gene, intragenic region upstream of TGFA gene and promoter of TIPARP gene. Significance was evaluated by ANOVA followed by Tukey multiple contrast tests and significant results are indicated for each individual comparison. (ns) not significant; (*) P < 0,05. (F) Scatterplots as in C depict correlation in peak signal intensity among samples immunoprecipitated with anti-ERalpha. (G) Heatmaps of PR ChIPseq data from untreated (-) and 60 min R5020-treated (+) Ishikawa and T47D cells. Regions were defined inside a window of –5 kb to +5 kb centered in peak summit. Intensity of the signal corresponds to the number of reads in the region. ChIPseq data are organized according to the Venn diagram scheme on the top right side of the figure as follows: I, PRbs solely found in Ishikawa cells; II: PRbs shared by both cell lines. De novo discovered motifs in I and II are indicated as sequence logos to the right of the map. PRE: progesterone response element. (H) Heat map constructed as in G of ERalpha ChIPseq data from untreated (-) and 60 min E2-treated (+) Ishikawa and T47D cells. ChIPseq data are organized according to the Venn diagram scheme on the top right site of the figure as follows: III, ERbs solely found in Ishikawa cells; IV: ERbs shared by both cell lines. Discovered motifs in III and IV indicated presented as sequence logos are on the right site of the map. ERE: estrogen response element.
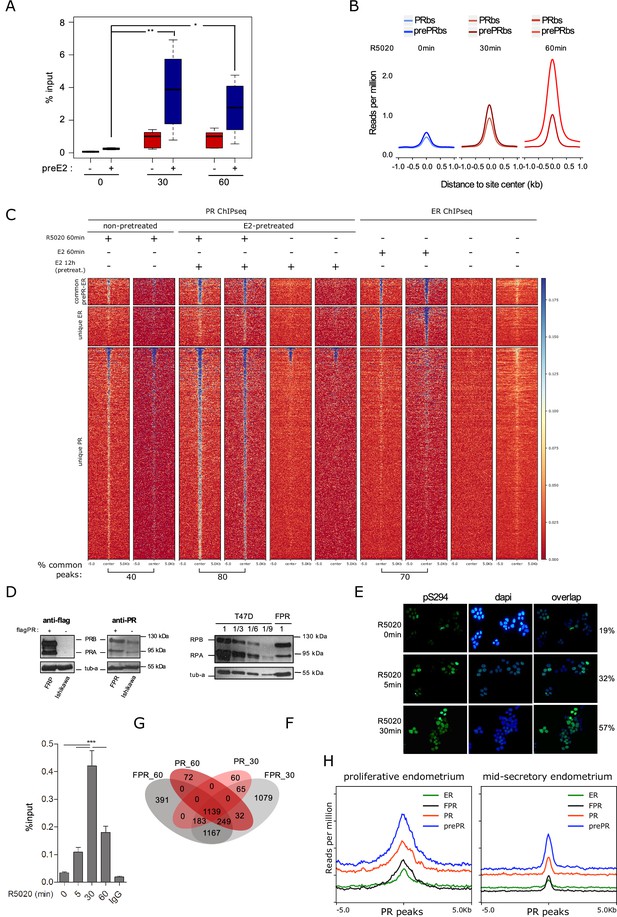
PR overexpression on Ishikawa cells.
(A) Validation of R5020-dependent PR binding in pretreated (+) or not (-) Ishikawa cells expressed as percentage of input (%input). Regions included are described in Figure 2—figure supplement 1B. Significance was evaluated by ANOVA followed by Tukey’s multiple contrast tests. (ns) not significant; (*) P < 0,05; (**) P < 0,01. (B) Signal strength for PR and preE2PR binding of untreated (T0) and R5020-treated cells. Intensity was defined as reads per million relative to distance to site center in kb inside a window of ±1kb. (C) PR and ERalpha ChIPseq replicates from two independent experiments are displayed as a heatmap in a 10kb window centered in peaks from pretreated cells. Labels on the left indicate common peaks between PR and ER (601), unique ER (924) and unique PR (5,064) binding sites. (D) (left side panels) Western blot of PR protein levels on Ishikawa and FPR Ishikawa (exogenous expression of triple flag-PR) cells. (right side panel) Representative image of western blot assays using protein extracts from FPR Ishikawa and T47D cells. 30µ g (1) of T47D extract were seeded followed by 10µ g (1/3), 5µ g (1/6), 3.3µ g (1/9) and 2µ g (1/15) of the same extract. 30µ g of extract from FPR Ishikawa cells (FPR) were loaded on the same gel. Anti-PR antibody was used to reveal PR isoforms (PRA and PRB) and tubulin (tub-a) as control. (E) Immunofluorescence of serine 294 phosphorylated PR (pS294) (green). Percentage of positive nuclei was calculated as green dots x 100/blue dots. (F) Recruitment of PR to EGFR enhancer sequence -described in Figure 1F- in FPR cells treated with R5020 and unspecific IgG control. Results are expressed as in Figure 1F (two independent experiments). (***) p < 0.001. (G) Shared binding sites among 30min- and 60min-treated FPR and parental Ishikawa cells. (H) Binding profiles (coverage signal) of prePRbs, PRbs, ERbs, and FRPbs on PR peaks from normal proliferating and mid-secretory endometrium. Normal endometrium data was obtained from Chi et al., 2020.
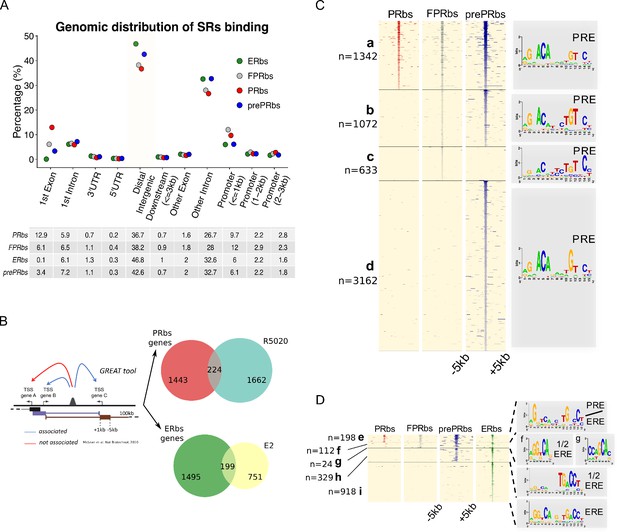
A fraction of E2-induced PRbs localize on ERbs and contain half ERE motifs.
(A) Classification of steroid receptor binding relative to genomic features expressed as percentage (%) of peaks after 60 min of hormone treatment inside each feature. Legend at the top right corner indicates the color key for ERbs (green dots) and three conditions of PR binding: non-pretreated with E2 (PRbs, red dots), pretreated with E2 for 12 hr (prePRbs, blue dots) and exogenous expression of PR (FPRbs, grey dots). The table below shows percentages represented in the plot. (B) To the left: Representation of GREAT tool association rules adapted with modifications (McLean et al., 2010). To the right: Venn diagrams show intersection between PRbs-associated genes and R5020-regulated genes (top), and ERbs-associated genes and E2-regulated genes (bottom). (C) Peak signals in PRbs, FPRbs and prePRbs from 60 min R5020-treated Ishikawa cells were plotted as heatmaps. Regions were defined inside a window of 10 kb centered in peak summit (±5 kb) and intensity of the signal correspond to number of reads in each region. Heatmap is subdivided into four mutually exclusive groups depending on shared/partly shared/non-shared binding sites: a (n = 1342), sites shared by all three conditions of PR binding; b (n = 1072), sites uniquely found in FPR and prePR; c (n = 633), sites found only in FPR; and d (n = 3162), sites found only in prePR. De novo motif discovery (MEME) was performed on all groups and results are indicated as sequence logos to the right of the map, including the name of the most related known motif. PRE: progesterone response element. (D) Peak signals in PRbs, FPRbs and prePRbs as in (C), and ERbs from 60 min E2-treated Ishikawa cells. Heatmap was subdivided into five mutually exclusive groups: e (n = 198), sites shared by all three conditions of PR binding and ER binding; f (n = 112), sites shared by FPRbs, prePRbs and ERbs; g (n = 24), sites shared by FPRbs and ERbs; h (n = 329), sites shared by prePRbs and ERbs; and i (n = 918), sites uniquely found in ERbs. Motif discovery was performed as in A for all groups and results are shown to the right of the map, including the most related known motif. ERE: estrogen response element; 1/2 ERE: half ERE.
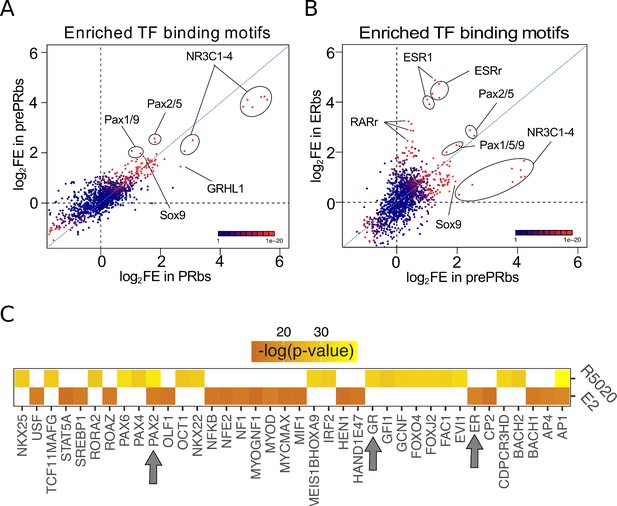
Putative PAX2 binding sites are associated with PR and ER alpha binding and hormone-regulated genes in Ishikawa cells.
(A) Fold enrichment values (log2FE) of 1.395 known TF binding motifs on prePRbs and PRbs. Combined p-values for enrichment analyses are indicated through the color key displayed at the lower right corner of the plot. Relevant motifs pointed on the plot correspond to NR3C1-4, members of the PAX family (1, 2, 5, and 9) and SOX9. (B) Comparison as in (A) between prePRbs and ERbs. Relevant motifs pointed on the plot correspond to NR3C1-4, members of the PAX family (1, 2, 5, and 9), SOX9, ESR1 and estrogen related (ESRr) and retinoic acid receptor (RARr). (C) Predicted UCSC Transcription Factor (TFBS) binding on genes regulated by 12 hr treatments with R5020 and E2 in Ishikawa cells were analyzed using DAVID web-based functional enrichment tool. Heatmap shows the top 20 TFBS predicted (p < 0.05)for R5020- and E2-regulated genes from RNAseq results expressed as -log(p-value). Arrows indicate position of PAX2, GR (PR-like binding motif), and ER.
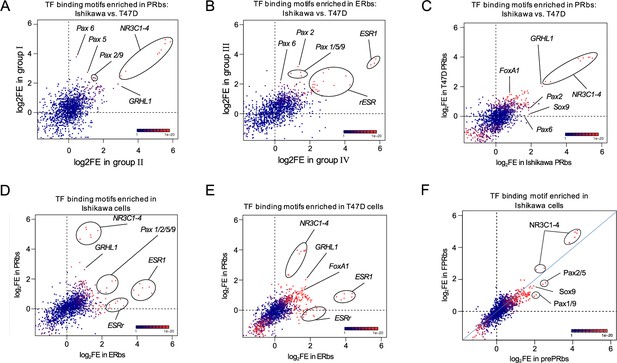
PAX2 binding to chromatin is hormone dependent in Ishikawa cells while it is not localized to nuclei of T47D cells.
Scatter plots showing fold enrichment values (log2FE) of 1,395 known TF binding motifs on (A) PRbs in group I and group II from Figure 2—figure supplement 1G, (B) ERbs in group III and group IV from Figure 2—figure supplement 1H, (C) PRbs from Ishikawa and T47D cells, (D) ERbs and PRbs from Ishikawa cells, (E) ERbs and PRbs from T47D cells and (F) FPRbs and prePRbs. Combined p-values (product of x and y p-value multiplication) for enrichment analyses are indicated through the color key displayed at the lower right corner of the plots. Relevant motifs pointed on the plots correspond to NR3C1-4, ESR1, rESR, members of the Pax family (1, 2, 5, 6, and 9), GRHL1 (TF involved in epithelial development), SOX9, and FOXA1.
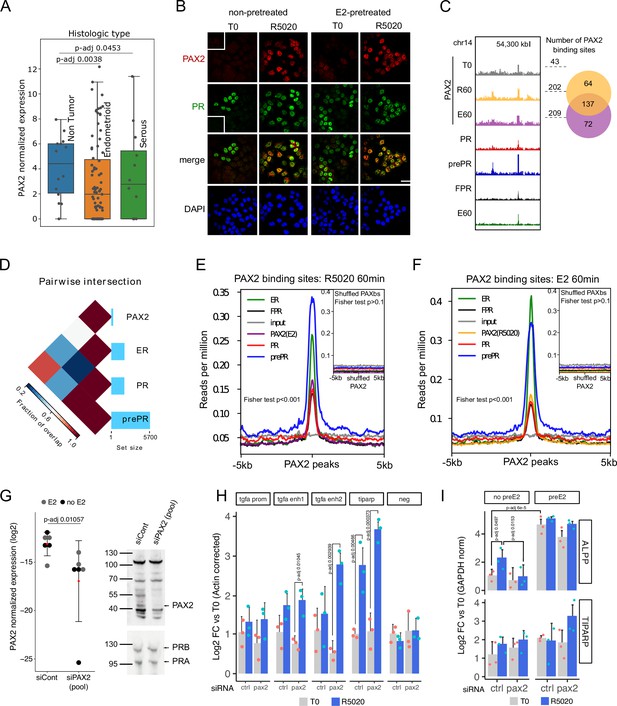
PAX2 co-localizes with PR and ERalpha in the nuclei of Ishikawa cells and it is positioned in the vicinity of PR and ER alpha binding sites.
(A) PAX2 expression in Non Tumor (normal endometrium) and two histologically distinct endometrial cancer samples (Dou et al., 2020). Data stored at the National Cancer Institute’s CPTAC program was accessed through cptac python package developed by Sam Payne lab. (B) Immunofluorescent detection of PR (green) and PAX2 (red) in untreated (T0) and 60min R5020-treated (R5020) Ishikawa cells which were pretreated or not with E2 for 12h (non-pretreated, E2-pretreated). Images were merged for co-localization analysis (merge). Scale bar is shown in the panels and is equivalent to 30 µm. (C) PAX2 binding profile inside a region of 70kb of chromosome 14. Number of PAX2 binding sites for untreated Ishikawa cells and treated with R5020 for 60 min or E2 for 60 min is shown to the right of the profiles as well as the intersection between these last two groups (venn diagram). Tracks for PRbs, prePRbs, FPRbs and ERbs are displayed below the profiles for the same region. (D) Pairwise intersections of the fraction of overlap between PRbs, prePRbs, ERbs and PAXbs. Color key is indicated below the plot and the size of each set is shown as a bar plot to the right side. Intervene software was employed in the analysis (Khan and Mathelier, 2017). (E) Binding profiles of ER (green), PR (red), FPR (black), and prePR (blue) on PAX2 binding sites of 60min R5020-treated Ishikawa cells. PAX2 binding after 60min E2 treatment was included (purple). Inset shows signal profiles centered on shuffled R5020-dependent PAX2 binding sites. P-value for fisher extact test is reported in the plots. (F) Binding profiles as in (E) on PAX2 binding sites of 60min E2-treated Ishikawa cells. PAX2 binding after 60min R5020 treatment was included (orange). As in (E), inset shows signal profiles centered on shuffled E2-dependent PAX2 binding sites. p-Value for fisher extact test is reported in the plots. (G) PAX2 knockdown evaluated by qPCR (left panel) and Western blot (right panel) after treating Ishikawa cells with a control siRNA (siCont) or a mix of specific siRNAs (pool). PCR data from three replicates were normalized by GAPDH expression and shown in the plot together with the mean± SEM. PR was used as control for western blot. (H) Binding of PR in cells treated with siRNA control or siPAX2 followed by 12 hr pretreatment with E2 and treatment with R5020 for 60min. PCR signal was corrected by beta-actin and relativized to untreated control cells. Bars show the mean± SEM of the three replicates displayed as dots. (I) Gene expression levels of hormone responsive genes before and after siPAX2 treatment. Expression was normalized to GAPDH and relativized to T0 using ΔΔCt method and expressed as mean log2 fold change± SEM of the three replicates displayed as dots. For all items, statistically significant comparisons resulting from ANOVA followed by Tukey’s HSD are denoted by the adjusted p-value.
-
Figure 5—source data 1
PR recruitment measured by qPCR in PAX2-knocked down Ishikawa cells.
- https://cdn.elifesciences.org/articles/66034/elife-66034-fig5-data1-v2.csv
-
Figure 5—source data 2
Gene expression measured by qPCR in PAX2-knocked down Ishikawa cells.
- https://cdn.elifesciences.org/articles/66034/elife-66034-fig5-data2-v2.csv
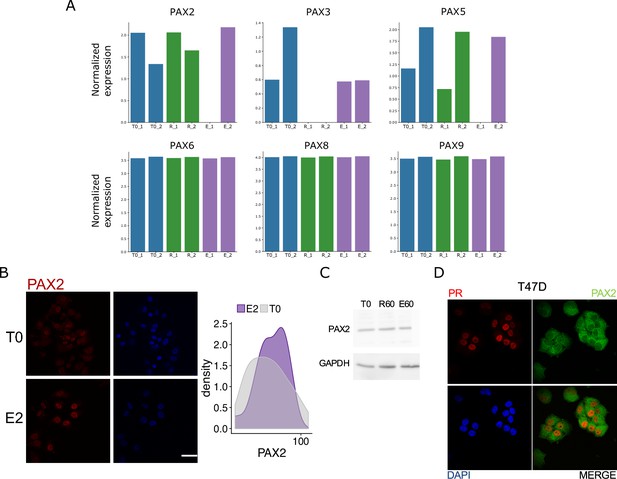
Expression analysis of PAX2 and other PAX family members in Ishikawa cells.
(A) Normalized expression of detected PAX family members in untreated and hormone-treated Ishikawa RNAseq samples. Expression is shown for two independent experimental replicates. (B) Representative images of PAX2 signal (red) in cells treated or not with E2 for 60 min and dapi staining (blue). Scale bar is shown in the figure and equals to 30 µm. Distribution of intensities in nuclear signal for both conditions is shown to the right of the images. (C) Western blot image of PAX2 protein levels in untreated (T0) Ishikawa cells and treated with R5020 for 60 min (R60) or E2 for 60 min (E60). GAPDH was used as normalizing control. (D) PR (red) and PAX2 (green) staining in T47D cells treated with R5020 for 60 min. Dapi was used for revealing nuclei.
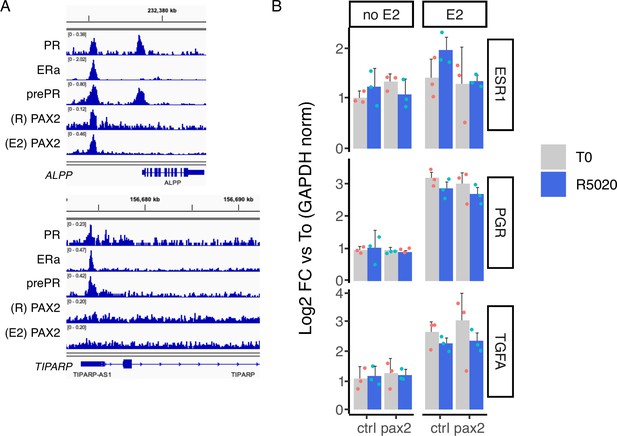
Expression analysis of hormone regulated genes after PAX2 knock down.
(A) IGV-based visualization of treated Ishikawa ChIPseq data. PR, ERalpha, prePR, and PAX2 (R5020 and E2 sites) binding in regulatory regions of ALPP and TIPARP genes is shown in different tracks. (B) Ishikawa cells were incubated with a pool of siRNAs to reduce PAX2 levels followed by 12hr of E2 (pretreatment) and then treated with R5020 for 12hr. RNA was collected and gene expression was analyzed by RT-qPCR. Results are shwon as fold change over untreated cells (T0) after normalizing by GAPDH expression using ΔΔCt method and expressed as mean fold change± SD of three replicates.
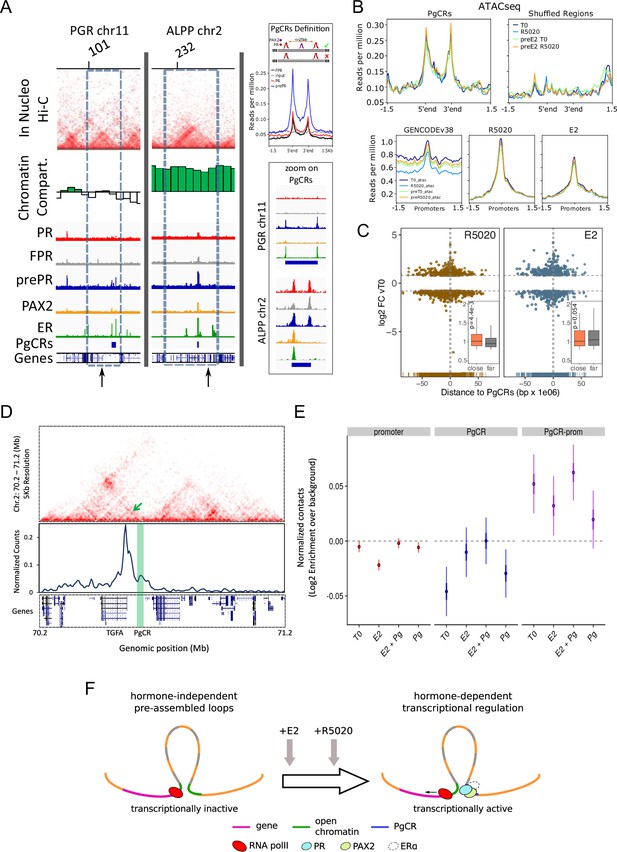
Convergence of PR and PAX2 binding in TADs with regulated genes defines potential endometrial regulatory domains.
(A) Upper panel shows the contact matrices at a resolution of 20kb obtained by In Nucleo Hi-C in PGR and ALPP loci. Middle panel shows the spatial segregation of chromatin as open or closed compartments inside TADs (green bars: A compartment; white bars: B compartment - see Materials and methods section). The bottom panels show ChIPseq signal distribution of PR, FPR, prePR, PAX2 and ERalpha as well as the location of PgCRs and genes over the region. The dashed rectangle restricts the TAD of interest and the vertical arrow marks the TSS of PGR and ALPP. Definition of PgCR: Coverage profiles of PR (red), FPR (black), and prePR (blue) binding on Progesterone Control Regions (PgCRs) delimited by the start and end labels, and flanked upstream and downstream by 1.5Kb regions. Input sample (gray) was included in the plot. Rules for qualifying as a control region are depicted on top of the profile plot. Magnified images over Control Regions are shown to the right (zoom on PgCRs). (B) ATACseq peaks from cells untreated (T0), treated with R5020 for 60 min, 12hr E2-pretreated (preE2 T0) and E2-pretreated followed by 60min treatment with R5020. Signal was plotted over Control Regions, shuffled Control Regions (Shuffled Regions), promoters of all annotated genes from GENCODE database (GENCODEv38) and promoters of genes regulated by 12h treatments with R5020 or E2. (C) Plot shows fold change values of genes regulated by R5020 and E2 (v. untreated cells) relative to Control Regions. Genes located upstream of PgCRs are represented with negative distance values. Dashed horizontal lines mark fold change cut-off points (|log2FC|=0.8) and vertical lines are placed at position –1 and 1Mb. Insets depict comparison of fold change values (absolute values) between genes located beneath (close) and over (far) a 1Mb distance from PgCRs. Statistical significance for this comparison was determined with Welch Two Sample t-test and is represented by a p value on the plot. (D) Top panel: Hi-C contact map at 5kb resolution of chr2 (70,200,000-71,200,000) obtained in Ishikawa cells and showing the organization around TGFA gene locus. Middle panel: Virtual 4C profile at 5kb resolution (expressed as normalized counts per thousands within the region depicted above) using the TGFA promoter as bait and showing the contacts engaged between TGFA promoter and the PgCR detected in this region (highlighted in green). Arrow on top panel highlights the position of the loop in the map. Bottom panel shows the positions of genes in the region depicted. (E) Distributions of observed versus expected interactions established between promoters (red - left), between PgCRs (blue - middle) and between Promoters and PgCRs (purple - right) located within a same TAD in Ishikawa cells treated as indicated below. (F) Representation of a chromatin loop involving a PgCR and the promoter of a regulated gene. Initially, the gene is transcriptionally inactive even though the loop is already formed. After hormone induction (E2 pretreatment followed by R5020), PR, PAX2 and in some cases ER alpha occupy open chromatin compartments in contact with promoters resulting in transcriptional activation.
-
Figure 6—source data 1
TAD coordinates (hg38).
- https://cdn.elifesciences.org/articles/66034/elife-66034-fig6-data1-v2.txt
-
Figure 6—source data 2
PgCR coordinates (hg38).
- https://cdn.elifesciences.org/articles/66034/elife-66034-fig6-data2-v2.txt
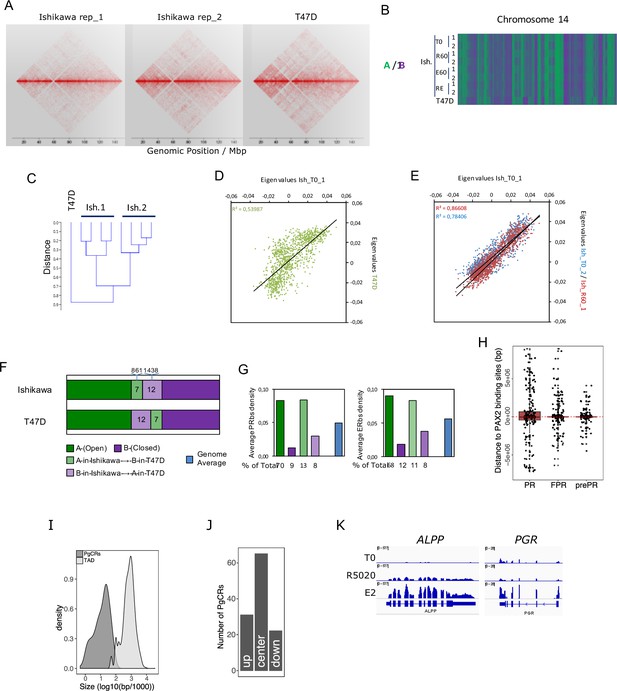
Chromosome compartments analysis and definition of progesterone control regions.
(A) Contact matrices of chr12 region at 100kb resolution obtained by In Nucleo Hi-C performed in two replicates of Ishikawa (Ishikawa_rep1 and Ishikawa_rep2) and T47D cells. (B) Hi-C datasets of untreated (T0) Ishikawa cells or treated with R5020 (R60), E2 (E60) and both hormones combined (RE60) were used to determine the spatial segregation of chromatin in open (A: green) or closed (B: purple) chromatin compartments compared to T47D cells. A fraction of chr14 is shown. (C) Relationship among samples was evaluated after clustering Hi-C datasets. Plot reflects differences as a function of distance. (D and E) Correlation analysis between Ishikawa replicates and T47D samples using eigen values determined from Hi-C datasets. Correlation coefficients are indicated in the figures. (F) Genome-wide proportions of 100kb windows of the genome found commonly in A or B in the two cell lines or showing divergent chromatin states (light green and light purple). (G) Histograms showing the average density of PR and ER binding sites (per 100kb) in the different types of regions mentioned in F. Below are the proportions of total PR and ERalpha binding sites found in those distinct regions. (H) Relative distance in bp from PRbs, FPRbs and prePRbs to PAXbs (both E2 and R5020 originated sites). Red dashed line marks overlapping events (distance = 0). Peaks located upstream of PAXbs are noted as negative distance values, while downstream peaks are positive. (I) Size distribution of PgCRs and TADs, expressed as log10. (J) Distribution of PgCRs in harboring TADs divided into center (center), upstream end (up) and downstream end (down). (K) RNAseq results for genes ALPP and PGR displayed in a genome browser-based environment. Tracks for untreated (T0) and R5020 10nM (R5020) and E2 10nM (E2) 12hr treatments were represented in the same vertical viewing range value (vvrv). A different vvrv was used for each gene in display.
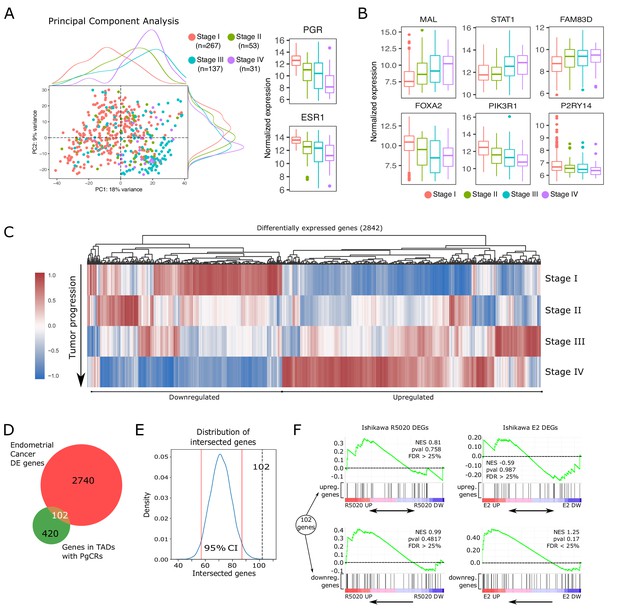
Altered expression of PgCR-genes correlates with drivers of endometrial tumor progression.
(A) Scatter plot of PCA results showing scores of components 1 and 2 (PC1 and PC2) using transcriptomic data of endometrial cancer samples (protein coding genes) obtained from TCGA-UCEC database (n=488). Samples are classiffied by FIGO stage (I to IV) and identified by color. Marginal density plots represent distribution of scores for each stage. Normalized expression values of PGR and ESR1 genes in these samples is shown to the right of the scatter plot. (B) Expression of genes positively (top row) and negatively (bottom row) regulated during endometrial cancer progression (FIGO stage) from TCGA-UCEC samples. (C) Heatmap shows scaled normalized counts of 2842 differentially expressed genes (DEGs) between Stage IV and the other three stages. (D) Intesection between DEGs presented in C and protein coding genes contained in TADs with PgCRs (522) resulted in 102 gene identities detailed in the box to the left of the Venn diagram. Genes were arranged as Upregulated or Downregulated with respect to Stage IV samples. (E) Bootstrapping statistical approach to test whether intersection described in D could result from randomly picking any set of 2842 genes among all known protein coding genes (GENCODE v38). Vertical red lines mark the 2.5 and 97.5 percentile, which denote the 95% confidence interval of the distribution. Intersection from D is indicated as a vertical black dashed line (102). (F) GSEA plot results using the 102 genes from intersection in D as gene set to match against an expression dataset from Ishikawa cells treated or not with R5020 or E2 for 12h. Normalized enrichment scores and FDR values are reported on the plots.
-
Figure 7—source data 1
Intersection between tumor DEGs and PgCR-genes.
- https://cdn.elifesciences.org/articles/66034/elife-66034-fig7-data1-v2.txt
-
Figure 7—source data 2
Protein coding genes in TADs with PgCRs (PgCR-genes).
- https://cdn.elifesciences.org/articles/66034/elife-66034-fig7-data2-v2.txt
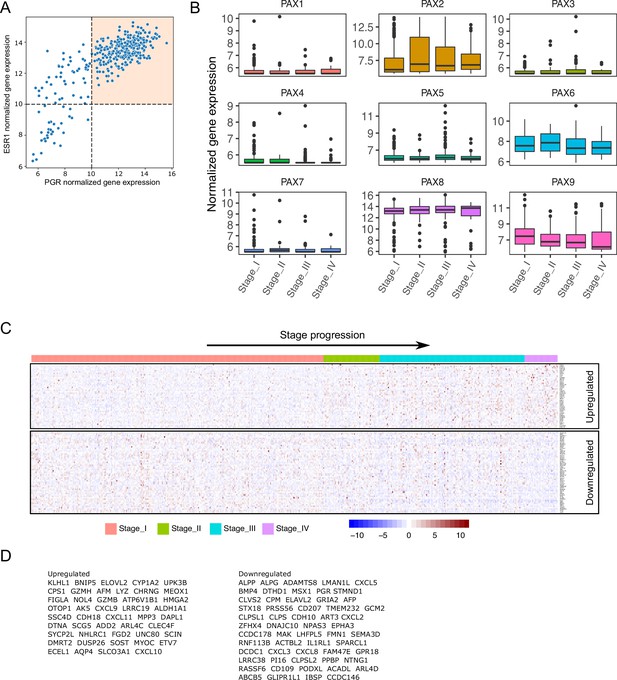
Exploratory analysis and processing of TCGA-UCEC endometrial cancer samples.
(A) Scatter plot of ESR1 and PGR expression levels for Stage I endometrial cancer samples. Orthogonal black dashed lines denote denote the quadrants for sample selection (colored quadrant). (B) Normalized expression levels of PAX genes detected in TCGA-UCEC samples classified by FIGO stage (I to IV). (C) Normalized expression of genes obtained from intersection of endometrial cancer DEGs and PgCR-genes. Samples were arranged by FIGO stage (I to IV). (D) List of genes from heatmap in C classified as up and downregulated relative to Stage IV.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | Edometrial adenocarcinoma (Epithelial) Ishikawa | Dr. Rochefort Unità Hormones and Cancer INSERM, France | Female | |
| Cell line (Homo sapiens) | FPR Ishikawa | This paper | Flag-tagged PR overexpression; Female | |
| Cell line (Homo sapiens) | Breast cancer (Epithelial) T47D | Dr. Beato (Center for Genomic Regulation) | Female | |
| Recombinant DNA reagent | p3xFLAG-CMV-14 (plasmid) | Dr. Beato (Center for Genomic Regulation) | Flag-tagged human PR | |
| Sequence-based reagent | Human PAX2 siRNA | Dharmacon | Pool of 4 on-target oligos | |
| Sequence-based reagent | Human control siRNA | QIAGEN 1027310 | Scramble non-target oligo | |
| Antibody | Anti-PR (H-190 Rabbit polyclonal) | Santa Cruz Bio. sc-7208 | RRID:AB_2164331 | ChIP:30µ l xIP Western (1:200) |
| Antibody | Anti-ERalpha (HC-20 Rabbit polyclonal) | Santa Cruz Bio. sc-543 | RRID:AB_631471 | Western (1:200) |
| Antibody | Anti-ERalpha (HC-20X Rabbit polyclonal) | Santa Cruz Bio. sc-543X | ChIP (25µ l xIP) | |
| Antibody | Anti-ERalpha (H-184X Rabbit polyclonal) | Santa Cruz Bio. sc-7204 | ChIP (25µ l xIP) | |
| Antibody | Anti-phosphoserine 294 PR (Rabbit polyclonal) | Cell Signaling 13736 | RRID:AB_2798307 | IF (1:100) |
| Antibody | Anti-PAX2 (Rabbit polyclonal) | BioLegend PRB-276P (Covance) | RRID:AB_291611 | ChIP:6µl xIP Western (1:200) |
| Antibody | Normal rabbit IgG | Santa Cruz Bio. sc-2027 | RRID:AB_737197 | ChIP (12µ l xIP) |
| Antibody | Anti-alphatubulin (Mouse monoclonal) | Merck T5168 (Sigma-Aldrich) | RRID:AB_477579 | Western (1:1000) |





