Multiplexed mRNA assembly into ribonucleoprotein particles plays an operon-like role in the control of yeast cell physiology
Figures
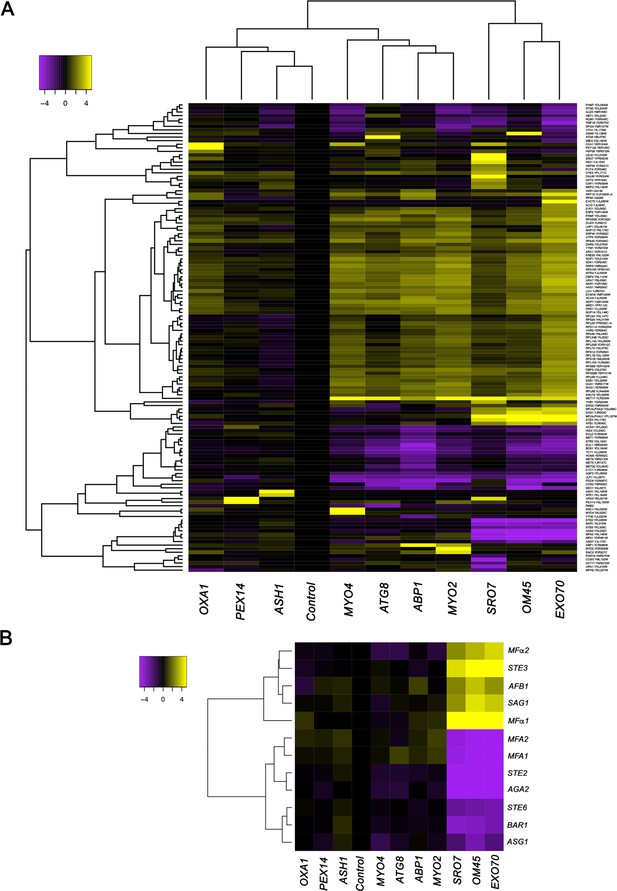
mRNAs encoding secreted and membrane proteins involved in yeast mating undergo multiplexing.
(A) RaPID RNA pulldown and RNA-seq reveal transcript multiplexing in yeast. Different MS2 aptamer-tagged mRNAs (listed on the x axis) expressed from their genomic loci were precipitated from MATα yeast (BY4742) by MS2-CP-GFP-SBP after formaldehyde crosslinking in vivo and cell lysis procedures (RaPID; see 'Materials and methods'). RNA-seq was performed and the reads were plotted to yield a heat map for the relative enrichment of the tagged as well as non-tagged mRNAs (see color key for approximate values). All non-coding RNAs (e.g., Ty elements, tRNAs, snoRNAs, ribosomal RNAs) were removed but are included in the heat map of Figure 1—figure supplement 1. A list of the log2 transformed and control-subtracted expression values, as plotted here, is given in Supplementary file 1. (B) mRNAs encoding proteins involved in mating in MATα cells are enriched in the pulldowns of SRO7, EXO70, and OM45 mRNAs. A selective heat map comprising the mRNAs encoding secreted and membrane proteins involved in yeast cell mating is shown. Positive enrichment of cell type-specific MATα mRNAs (STE3, MFα1, MFα2, SAG1, AFB1) is shown in purple, while negative enrichment of the non-expressed MATa mRNAs (MFA1, MFA2, STE2, AGA2, BAR1, STE6, ASG7) is shown in yellow.
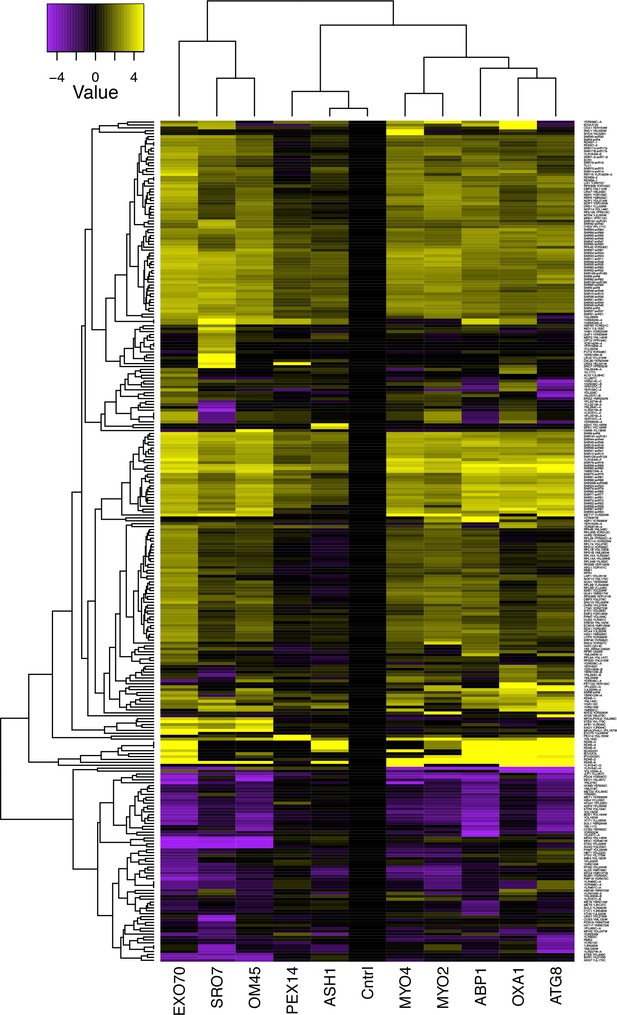
mRNAs encoding secreted and membrane proteins involved in yeast mating multiplex into a single ribonucleoprotein particle.
An expanded version of the figure shown in Figure 1A is given. Different MS2 aptamer-tagged mRNAs (listed on the x axis) expressed from their genomic loci were precipitated from MATα yeast (BY4742) by MS2-CP-GFP-SBP after formaldehyde crosslinking in vivo and cell lysis procedures (RaPID; see 'Materials and methods'). RNA-seq was performed and the reads (including those of non-coding RNAs, including Ty elements, tRNAs, snoRNAs, ribosomal RNAs, etc) were plotted to yield a heat map for the relative enrichment of the tagged as well as non-tagged mRNAs (see color key for approximate values). Red-hatched rectangles indicate the target-tagged mRNAs.
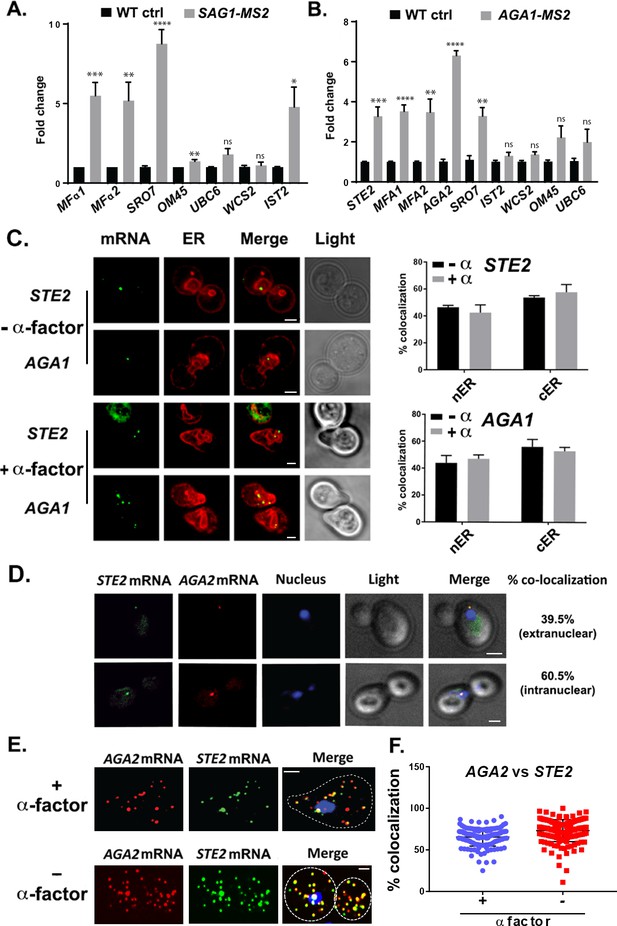
mRNAs encoding secreted and membrane proteins involved in mating multiplex in both MATα and MATa cells and localize to the ER.
(A) A mating messenger ribonucleoprotein (mRNP) complex is also formed in MATa cells. MATa yeast (BY4741) expressing MS2 aptamer-tagged SAG1 from its genomic locus were grown to mid-log phase (O.D.600 = 0.5) and subjected to RaPID followed by qRT-PCR (RaPID-PCR; see 'Materials and methods'). RNA derived from the total cell extract (Input) or biotin-eluated fraction (RaPID) was analyzed by qRT-PCR using primer pairs corresponding to mRNAs expected to possibly multiplex (based on the results shown in Figure 1). Three biological repeats were performed and an unpaired t-test was used to compare the SAG1 pulldown to the WT control pulldown; ****p<0.0001; ***p<0.0005; **p<0.001; *p<0.01. (B) Analysis of the mating mRNP complex in MATα cells by RaPID-qPCR. MATα yeast expressing MS2 aptamer-tagged STE2 from its genomic locus were grown and processed as in (A). RNA derived from the biotin-eluated fraction (RaPID) was analyzed by qRT-PCR using primer pairs corresponding to mRNAs expected to multiplex (based on the results shown in Figure 1). Three biological repeats were performed and an unpaired t-test was used to compare the AGA1 pulldown to the WT control pulldown; ****p<0.0001; ***p<0.0005; **p<0.001. (C) Mating mRNAs localize to ER in pheromone-treated or -non-treated cells. Wild-type cells expressing MS2 aptamer-tagged STE2 or AGA1 mRNAs (as listed) were transformed with single-copy plasmids expressing MS2-CP-GFP(x3) and either Scs2-mCherry (left panel) or Sec63-RFP (right panel). Cells were grown to mid-log phase and either α-factor (5 µM; +α-factor) or dimethyl sulfoxide (DMSO) (-α-factor) was added for 90 min, followed by fixation with 4% paraformaldehyde in a medium containing 3.5% sucrose and prior to imaging by confocal microscopy. Left panel: fluorescent images: mRNA – MS2-CP-GFP(x3) labeling; ER – Scs2-mCherry labeling; merge – merger of mRNA and ER windows; light – transmitted light. Size bar = 2 μm. Right panel: quantitative analysis of RNA co-localization: nER – nuclear ER; cER – cortical ER. (D) Mating mRNAs STE2 and AGA2 co-localize both inside and outside of the nucleus. Yeast expressing MS2 aptamer-tagged STE2 mRNA and PP7 aptamer-tagged AGA2 mRNA from its genomic locus were transformed with plasmids expressing MS2-CP-GFP(x3) and PP7-PS-tomato, while the nucleus was stained with 4,6-diamidino2-phenylindole (DAPI). STE2 mRNA – MS2-CP-GFP(x3) labeling; AGA2 mRNA – PP7-PS-tomato labeling; nucleus – DAPI labeling; merge – merger of mRNA and nucleus windows; light – transmitted light. Size bar = 2 μm. (E) Single-molecule fluorescence in situ hybridization (smFISH) validation of STE2 and AGA2 mRNA co-localization. Non-transformed cells from (D) were either treated with α-factor (10 µM) or left untreated and processed for smFISH labeling using non-overlapping FISH probes complementary to the STE2 mRNA and PP7 aptamers, prior to labeling with DAPI. The representative image shown is from a single focal plane. AGA2 – Cy3 labeling; STE2 – Alexa488 labeling; merge – merged Cy3/Alexa488 windows with DAPI staining. Size bar = 2 μm. (F) Scatter plot of co-localization data from (E). Co-localization was assessed using the FISHquant algorithm (see 'Materials and methods'). Black lines indicate the avg. ± SEM for each sample. Each data point represents a single cell.
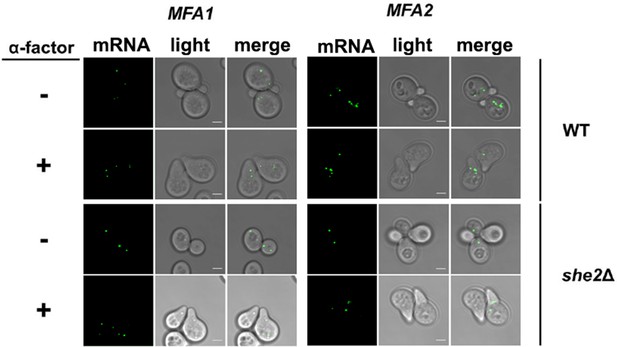
MFA1 and MFA2 mRNAs localize to the cell body.
MFA1 and MFA2 mRNAs localize to the cell body with or without pheromone treatment, and in the presence or absence of She2. MATa wild-type and she2∆ yeast expressing MS2-CP-GFP(x3) from a single-copy plasmid and either MS2 aptamer-tagged MFA1 or MFA2 from their genomic loci were grown to mid-log phase on a selective medium, either treated with or without added pheromone (10 μM; 90 min), and scored for RNA localization by confocal microscopy.
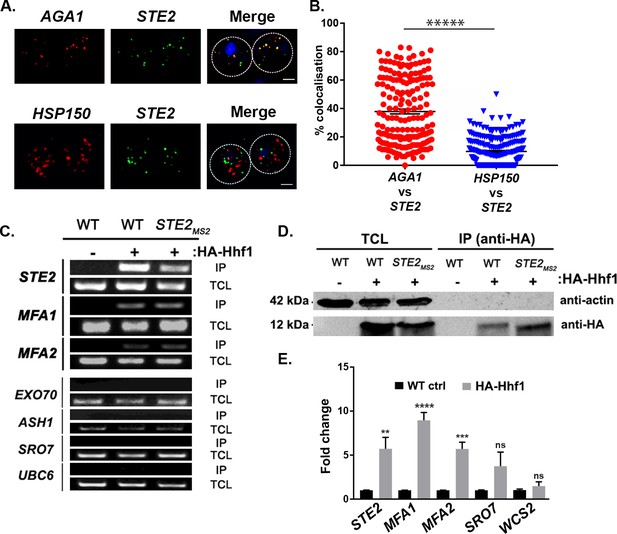
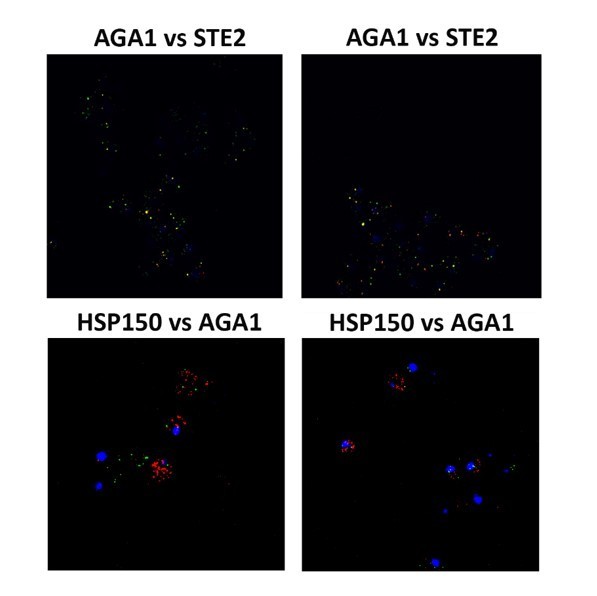
Mating pathway mRNAs co-localize and are co-precipitated by HA-Hhf1.
(A) Endogenous STE2 and AGA1 mRNAs co-localize. Wild-type (WT) BY4741 cells were processed for single-molecule fluorescence in situ hybridization (smFISH) labeling using designated FISH probes complementary to the STE2 and AGA1 mRNAs, and STE2 and HSP150 mRNAs, prior to labeling with 4,6-diamidino2-phenylindole (DAPI). Dotted lines are representative of the cell outline observed in brightfield micrographs. AGA1 – Cy3 labeling; STE2 – Alexa488 labeling; HSP150 – Cy5 labeling; merge – merged Cy3/Cy5 and Alexa-488 windows with DAPI staining. Size bar = 2 μm. The representative image shown is from a single focal plane. (B) Scatter plot of co-localization data from (A). Co-localization was assessed using the FISHquant algorithm (see 'Materials and methods'). Black lines indicate the average + SEM for each sample. Each data point represents a single cell. ***** indicates p-value<0.00001. (C) HA-Hhf1 precipitates mRNAs encoding secreted mating pathway components. MATa WT cells and cells expressing MS2 aptamer-tagged STE2 mRNA from its genomic locus were transformed with a 2 µ plasmid expressing HA-HHF1 (+HA-Hhf1). WT cells were also transformed with an empty vector (–HA-Hhf1) as control. Cells were grown to mid-log phase, lysed to yield the total cell lysate (TCL), and subjected to immunoprecipitation (IP) with anti-HA antibodies. Following IP, the RNA was extracted, DNase-treated, and reverse-transcribed. Transcripts were amplified using specific primers to the genes listed. A representative ethidium-stained 1% agarose gel of electrophoresed samples is shown; n = 3 experiments. (D) HA-Hhf1 migrates as a 12 kDa protein in SDS-PAGE gels. Aliquots (5% of total) of the TCL and IP fractions were resolved on 15% SDS-PAGE gels and transferred to nitrocellulose membranes for detection with anti-HA and actin antibodies to verify the presence of HA-tagged Hhf1 in both fractions. (E) HA-Hhf1 precipitates the mating mRNAs. Same as in (C), except the presence of mating mRNAs was analyzed by qRT-PCR using primer pairs corresponding to mRNAs expected to co-purify with HA-Hhf1, as well as with different controls.
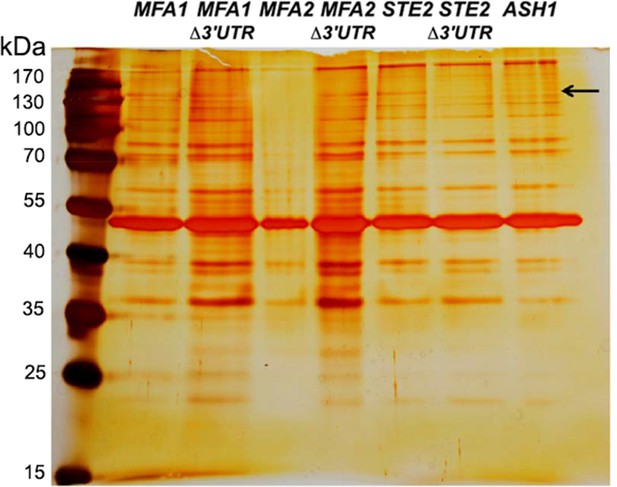
Identification of an RNA-binding protein that binds to the 3’UTR of the mating mRNAs.
Identification of an RNA-binding protein (Hhf1) that binds to the 3’UTR of STE2 mRNA. Yeast strains expressing MS2 aptamer-tagged MFA1, MFA2, and STE2 either with or without their 3’UTRs, and ASH1 were grown and processed for RaPID-MS (see 'Materials and methods'). The eluate after biotin release was resolved by SDS-PAGE on a 10% polyacrylamide gel and silver stained. The arrow indicates a band of ~150 kDa that exists in the STE2 lane (and all MFA1 and MFA2 lanes), but not in the ASH1 or STE2 ∆3’UTR lane. kDa indicates kilodaltons.
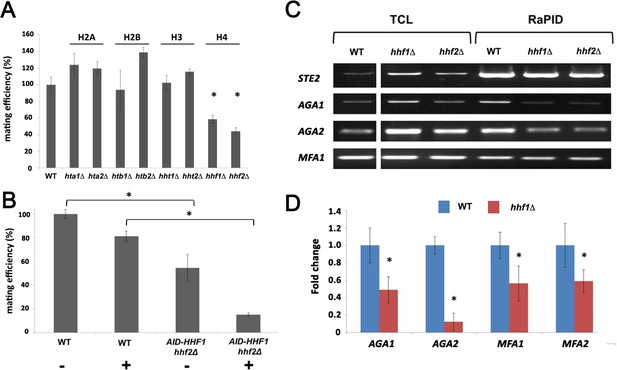
Histone H4 function is required for mating mRNP assembly and mating.
(A) Histone H4 alone is required for mating. MATa yeast (BY4741) bearing mutations in H2A (hta1∆, hta2∆), H2B (htb1∆, htb2∆), H3 (hht1∆, hht2∆), and H4 (hhf1∆, hhf2∆) were grown to mid-log phase, crossed against MATα wild-type (WT) cells, and mating was scored using the quantitative mating assay. n = 3 experiments; *p-value<0.05 between mutant and WT. (B) Combined hhf2∆ and AID-HHF1 alleles result in a near-complete block in mating upon auxin induction. MATa WT and hhf2∆ AID-HHF1 cells were grown to mid-log phase, treated either with or without auxin (3-IAA; 4 mM) for 4 hr, and examined for mating using the quantitative mating assay. n = 3 experiments; p-value<0.05 between mutant and WT. (C) Deletions in H4 histones result in defects in mating mRNP assembly – PCR analysis. MATa WT, hhf1∆, and hhf2∆ cells expressing MS2 aptamer-tagged STE2 from the genome were grown to mid-log phase, processed for RaPID-PCR, and the extracted RNA analyzed using primers against the listed mating mRNAs. A representative ethidium-stained 1% agarose gel of electrophoresed samples is shown; n = 3 experiments. (D) Deletions in an H4 histone result in defects in mating mRNP assembly – qRT-PCR analysis. Same as in (C), except that qRT-PCR was performed instead of PCR on WT and hhf1∆ cells expressing MS2 aptamer-tagged STE2 from the genome. Three experiments were performed and gave similar results; p-values<0.05 between mutant and WT cells.
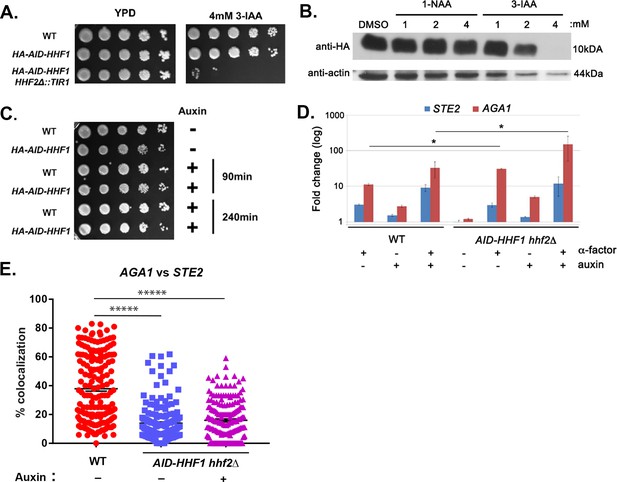
Conditional removal of both histone H4 paralogs leads to synthetic lethality over extended time.
(A) Long-term treatment of HA-AID-HHF1 hhf2∆ cells with auxin leads to lethality. Wild-type (WT; BY4741) yeast and HA-AID-HHF1 strains, either bearing or lacking TIR1 integration into the HHF2 locus (i.e., the hhf2∆ mutation), were grown on liquid YPD (yeast extract - peptone - dextrose medium) to mid-log phase (O.D.600 = 0.5). One O.D.600 unit from each strain was removed, washed, and diluted serially five times (tenfold each). Cells were spotted onto pre-warmed solid YPD medium either with or without 4 mM of 3-IAA and grown for 48 hr at 26°C prior to photodocumentation. (B) HA-AID-Hhf1 disappeared upon treatment with 4 mM 3-indole acetic acid. HA-AID-HHF1 hhf2∆ cells were grown to mid-log phase prior to treatment with dimethyl sulfoxide (DMSO) or different concentrations of 1-NAA or 3-IAA (auxin) for 2 hr. After harvesting and total cell lysate (TCL) preparation, samples (30 μg) were resolved on 15% SDS-PAGE gels, transferred to nitrocellulose membranes, and probed with antibodies against HA and actin. (C) HA-AID-HHF1 hhf2∆ cells remained viable for at least 4 hr of treatment with 4 mM 3-IAA. The indicated WT (BY4741) and HA-AID-HHF1 hhf2∆ cells were grown on a rich medium containing glucose (YPD) to mid-log phase (O.D.600 = 0.5) and treated for the indicated times with 4 mM 3-IAA. One O.D.600 unit from each strain was removed, washed, and diluted serially five times (tenfold each). Cells were spotted onto the same medium and grown for 48 hr at 26°C, prior to photodocumentation. (D) Neither auxin (3-IAA) treatment nor histone H4 depletion abolished pheromone-induced gene expression. WT and HA-AID-HHF1 hhf2∆ cells were grown to mid-log phase prior to treating cells with either auxin (3-IAA; 4 mM), pheromone (10 uM α-factor), or both for 2 hr. After treatment, TCLs were prepared, RNA extracted, and analyzed by qRT-PCR using primers against STE2 and AGA1. RNA levels derived from WT and HA-AID-HHF1 hhf2∆ cells under the different conditions were compared; p-values<0.05. (E) Reduced co-localization of AGA1 and STE2 mRNA in AID-HHF1 hhf2∆ cells upon histone H4 depletion. WT and HA-AID-HHF1 hhf2∆ cells were grown to mid-log phase prior to treating cells with auxin (3-IAA; 4 mM) and processed for single-molecule fluorescence in situ hybridization (smFISH). A scatter plot showing the level of co-localized smFISH puncta is shown. Co-localization was assessed using the FISHquant algorithm (see 'Materials and methods'). Black lines indicate the avg. ± SEM for each sample. Each data point represents a single cell. *****p-value<0.00001.
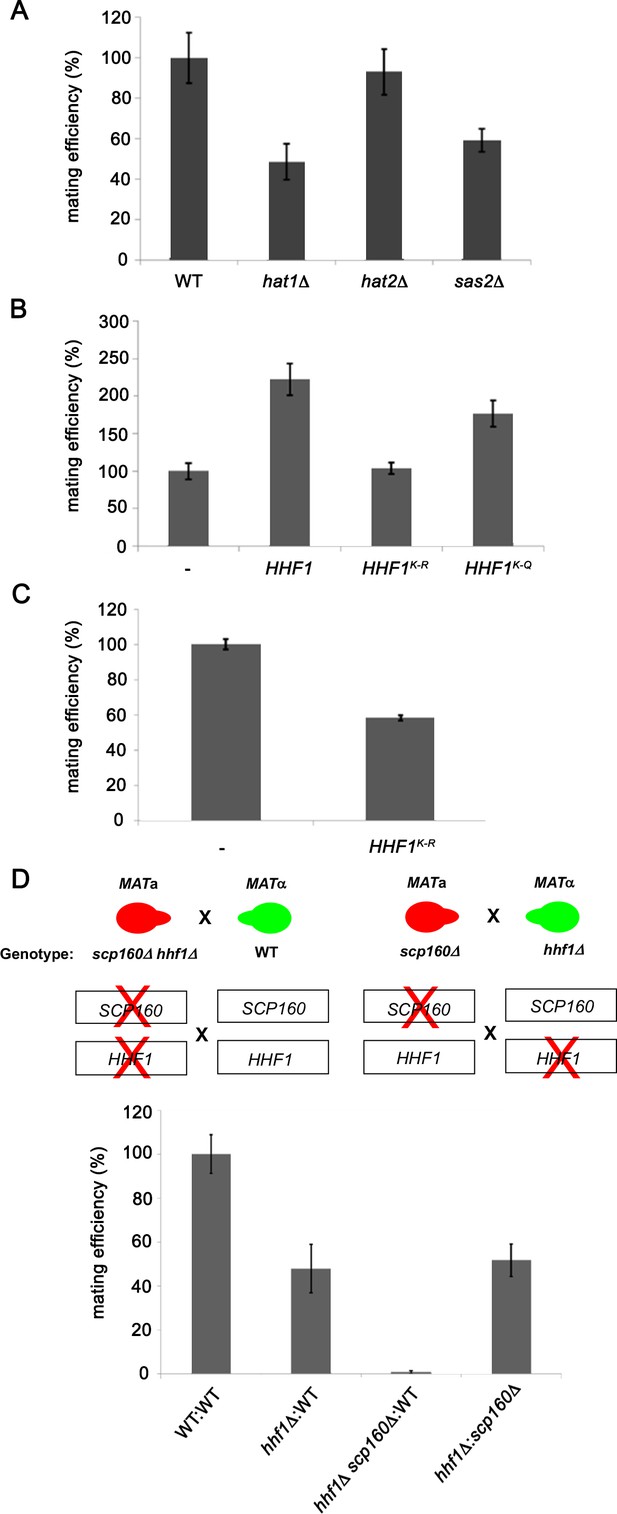
Histone H4 acetylation is required for mating.
(A) Histone acetyltransferases are required for mating. Wild-type (WT; BY4741) and yeast lacking known acetyltransferase genes (hat1∆, hat2∆, and sas2∆) were grown to mid-log phase (O.D.600 = 0.5) and examined for mating with WT MATα cells (BY4742). Three quantitative mating experiments were performed and gave similar results; p-values<0.05 between mutants and WT cells. (B) Mutations that either mimic or block N-terminal histone H4 acetylation increase or decrease mating efficiency, respectively. MATa WT (BY4741) cells alone (-) or overexpressing HHF1 (HHF1), along with cells overexpressing HHF1 bearing K-to-R (HHF1K-R) or K-to-Q (HHF1K-Q) mutations from a 2 um plasmid, were grown to mid-log phase and examined for mating with MATα cells. Three experiments were performed; p-values<0.05 between mutants and WT cells. (C) Expression of the K-to-R mutation in HHF1 in the genome inhibits mating comparative to the deletion of HHF1. MATa WT (BY4741) cells alone (–) or expressing HHF1 bearing the K-to-R (HHF1K-R) mutation from the genome were grown to mid-log phase and examined for mating with MATα cells. Three experiments were performed; p<0.05. (D) HHF1 and SCP160 are not epistatic, and hhf1∆ scp160∆ double mutants are sterile. Crosses between WT, hhf1∆, or hhf1∆ scp160 MATa cells and MATα WT cells, along with an hhf1∆ and scp160∆ cross, were performed. Illustrated are the hhf1∆ scp160 x WT cross (upper left side) and the hhf1∆ x scp160∆ cross (upper right side). Three experiments were performed; p-values<0.05 between crosses.
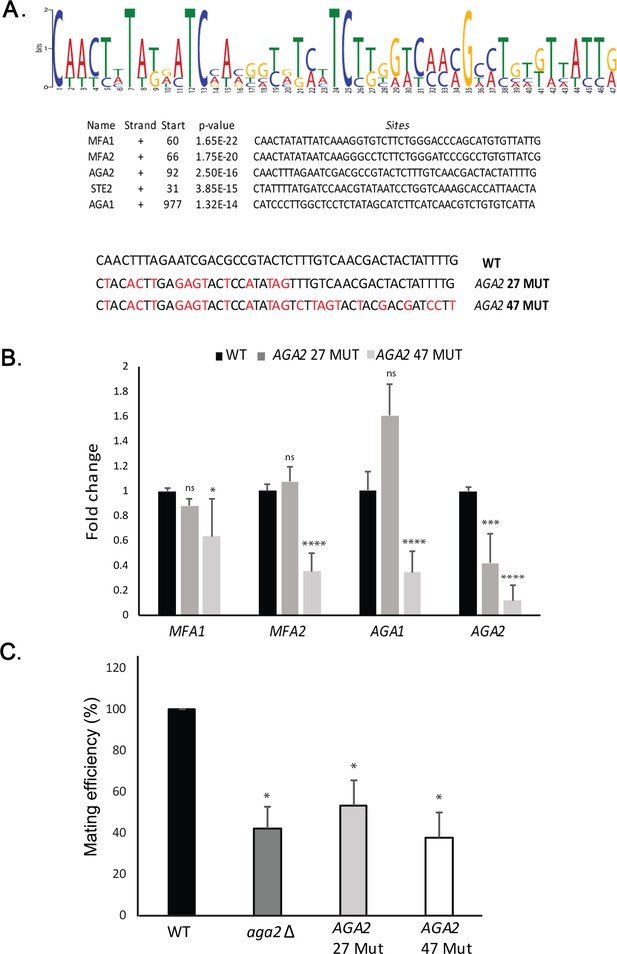
A motif in the mating mRNA coding region facilitates mRNP assembly and mating.
(A) A conserved motif is common to all five MATa mRNAs. MEME-ChIP analysis of the sequences of all five MATa mRNAs was performed and revealed a 47-nucleotide consensus motif in the coding regions, shown schematically as a sequence logo based on nucleotide representation. Also shown are two forms (AGA2 27-MUT and AGA2 47-MUT) in which the motif was altered within the coding region to remove conserved nucleotides without altering the amino acid sequence. (B) Mutation of the consensus motif in AGA2 leads to an inhibition in messenger ribonucleoprotein (mRNP) particle assembly. MS2 aptamer-tagged STE2 cells bearing the aga2Δ deletion were transformed with a single-copy plasmid expressing AGA2 or either of the two AGA2mut forms. Next, WT, AGA2 27-MUT, and AGA2 47-MUT cells expressing MS2-tagged STE2 were subjected to RaPID-qPCR with primers against AGA1, AGA2 (note: not within mutated region), MFA1, and MFA2. The histogram indicates the levels of precipitated mating mRNAs from wild-type (WT) (black) or AGA2 27-MUT and AGA2 47-MUT (dark and light grey, respectively) cells after normalization for the level of STE2 mRNA pulldown. Three biological repeats were performed and an unpaired t-test was performed to compare AGA2 27-MUT and AGA2 47-MUT with WT; ****p<0.0001; ***p<0.0005; *p<0.01. (C) Deletion of AGA2 or mutation of the consensus motif alters mating efficiency. MATa WT cells (WT; BY4741), AGA2 27-MUT or AGA2 47-MUT mutant cells were crossed against WT MATα cells and quantitative mating was assessed. Three biological repeats were performed and an unpaired t-test was performed to compare AGA2 27-MUT and AGA2 47-MUT with WT; *p<0.05.
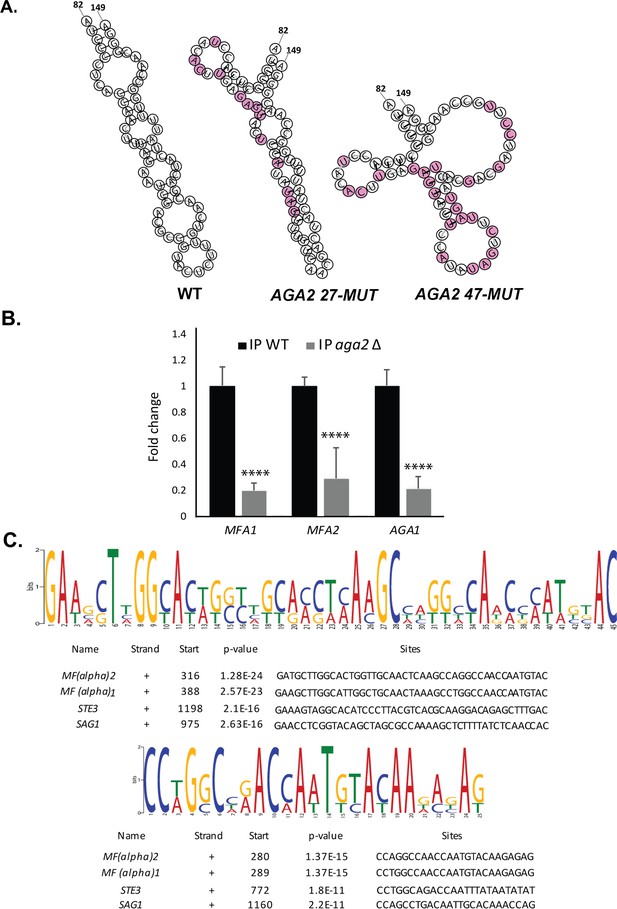
Structure of native and mutated AGA2 mating motifs, effect of AGA2 deletion on mating mRNP assembly, and identification of consensus motifs in MATα mating mRNAs.
(A) AGA2 47-MUT has a distorted structure. RNA-fold was used to model the native and mutant versions of the mating mRNA consensus motif in AGA2. Mutations in AGA2 27-MUT and AGA2 47-MUT are illustrated in pink. (B) Deletion of AGA2 reduces the multiplexing of mating mRNAs. Wild-type (WT) MATa (BY4741) and aga2Δ mutant yeast expressing MS2 aptamer-tagged STE2 from its genomic locus were grown to mid-log phase (O.D.600 = 0.5) and subjected to RaPID followed by RT-PCR (RaPID-PCR; see 'Materials and methods'). RNA derived from the total cell extract or the biotin-eluated fraction was analyzed by qPCR with primers against AGA1, MFA1, and MFA2. The histogram indicates the levels of precipitated mating mRNAs from either WT (black) or aga2Δ (grey) cells after normalization for STE2 pulldown. Three biological replicates were performed and an unpaired t-test was used to compare the pulldown of each gene from WT and aga2Δ mutant cells; ****p<0.0001. (C) Two consensus motifs are common to the four MATα mating RNAs. MEME-ChIP analysis of the sequences of all four MATα mating mRNAs was performed and revealed two conserved motifs (sites) of 45 (upper sequence logo) and 25 (lower sequence logo) nucleotides, as shown schematically for each gene. p-values are listed; '+'=sense strand.
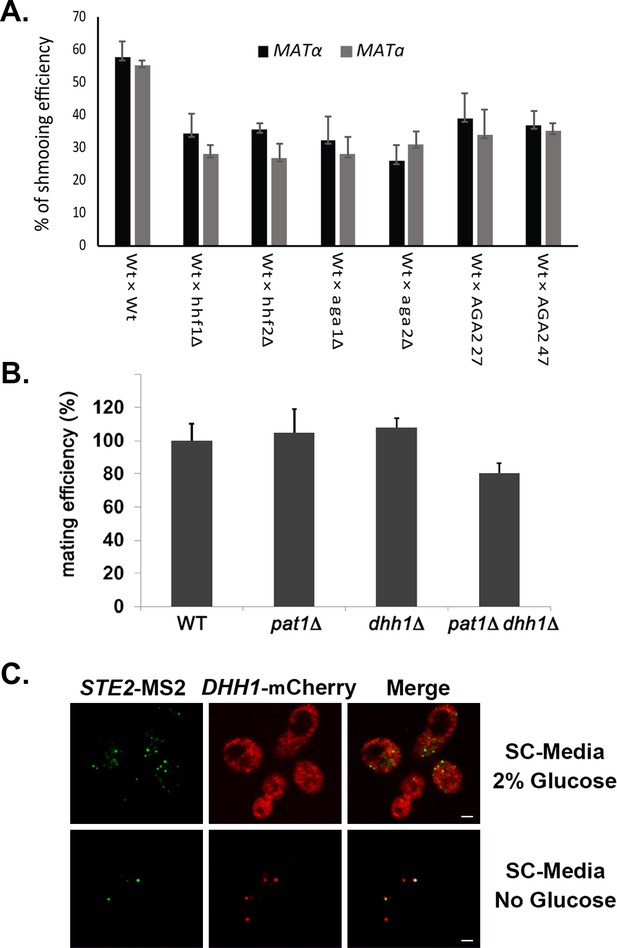
Mutations in AGA2 (in MATa cells) or histone H4 alter both mating partners' responsiveness to pheromone, and P-body components are not essential for mating.
(A) Mutations in the histone H4 paralogs or AGA2 alter MATα cell responsiveness to pheromone, as well as in their WT MATa mating partner. MATa wild-type (WT; BY4741), hhf1∆, hhf2∆, aga1∆, aga2∆, and AGA2mut cells were crossed against WT MATα cells and quantitative mating was assessed. Later, the cells were fixed in 4% paraformaldehyde in a medium containing 3.5% sucrose and analyzed using widefield microscope to determine the percentage of shmooing (i.e., pheromone-responsive) cells. Three biological repeats were performed and gave similar results. A one-way ANOVA with WT vs. mutant pairwise comparisons was performed to calculate the statistical significance of shmooing efficiency between the MATα WT and MATa mutant crosses to the WT MATα and MATa cross; p<0.0001. (B) P-body components are not required for mating. MATa wild-type (WT; BY4741) cells and the pat1∆, dhh1∆, and pat1∆ dhh1∆ deletion mutants were crossed against WT MATα cells and quantitative mating assessed. Three biological repeats were performed and gave non-significant differences between mutant and WT cells. (C) A mating pathway mRNA (STE2) does not co-localize with a P-body marker (Dhh1-mCherry) under normal growth conditions. Yeast expressing MS2 aptamer-tagged STE2 mRNA and DHH1-mCherry from their genomic loci were transformed with MS2-CP-GFP(x3) and grown to mid-log phase (O.D.600 = 0.5). Half of the culture was subjected to growth in a no-glucose medium for 10 min, while the other half was maintained in a medium containing 2% glucose. Next, cells were fixed in 4% paraformaldehyde medium containing 3.5% sucrose and analyzed by confocal microscopy to determine the percentage of STE2 mRNA co-localization with DHH1-mCherry. STE2-MS2 – MS2-CP-GFP(x3) labeling; DHH1-mCherry – mCherry labeling; merge – merger of mRNA and protein windows.
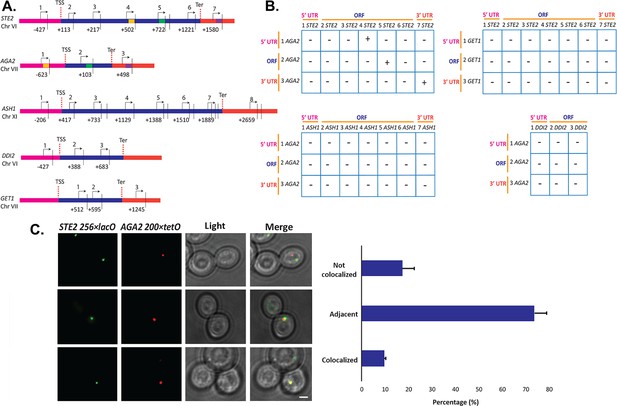
Intergenic association of STE2 and AGA2 genes.
(A) Schematic of the STE2, AGA2, ASH1, GET1, and DDI2 genes and the oligonucleotides used for their amplification from 3C DNA samples. Coordinates correspond to TaqI sites (shown as vertical black bars); site numbering is relative to ATG (+1). Forward primers used for 3C analysis were sense-strand identical (arrows) and positioned proximal to TaqI sites as indicated. Primers are numbered to distinguish the pairs used in the PCR reactions shown in (B). 5’UTRs, ORFs, and 3’UTRs are color-coded, as indicated. Transcription start sites (TSS) and termination sites (Ter) are indicated. (B) Matrix summarizing the intergenic association of represented genes as determined by 3C-PCR. Primer pairs corresponding to the different genes listed in (A) were used in PCR reactions. '+' indicates PCR amplification and the interaction between genes. '–' indicates no amplification. See Figure 7—figure supplement 1A for examples of both. 5’UTRs, ORFs, and 3’UTRs are indicated by broken orange lines. (C) Live cell fluorescence microscopy of AGA2−224 × tetR and STE2−256 × lacO yeast grown to mid-log phase prior to imaging. Left panel: STE2−256 × lacO gene is labeled with GFP-lacI; AGA2−224 × tetR is labeled with tetR-tdtomato; merge – merger of STE2 and AGA2 windows; light – transmitted light. Size bar = 2 μm. Right panel: histogram of the data from three biological replicas (avg. ± std.dev.); co-localized – fully overlapping signals; adjacent – partially overlapping signals; not co-localized – no overlap between signals. The representative image shown is from a single focal plane.
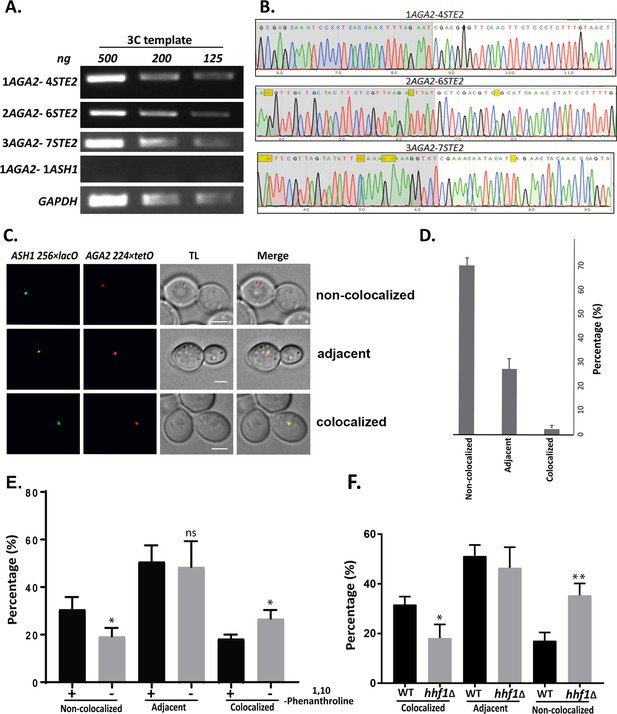
Intergenic association of STE2 and AGA2 genes.
(A) STE2 and AGA2 genes undergo allelic interactions. PCR products derived reactions using the indicated primer pairs (to the genes shown in Figure 7A) and 3C-processed DNA were fractionated on agarose gels (1.5%) and visualized by ethidium bromide staining using an AlphaImager 2000. Lanes represent the 3C-PCR output using the indicated concentrations of DNA template (ng DNA). GAPDH primers were used as a control for the PCR reaction. (B) Sequence confirmation of the STE2-AGA2 gene interaction. Positive PCR products were eluted from the gel and sequenced to confirm the nature of the observed band. A DNA sequencing chromatogram representing the observed sequence of the different STE2 and AGA2 chimeric PCR products is shown. (C) AGA2 and ASH1 genes do not undergo allelic interactions. Live cell fluorescence microscopy of yeast having the AGA2 and ASH1 genes tagged with 224 × tetR and 256 × lacO repeats, respectively. Cells were grown to mid-log phase prior to imaging. ASH1−256 × LacO repeats were visualized by GFP-lacI; AGA2−224 × TetR was visualized by tetR-td tomato; merge – merger of STE2 and AGA2 windows; TL – transmitted light. Size bar = 2 μm. Representative images are shown. (D) Histogram of the data in (C) obtained from three biological replicas (avg. ± SEM); co-localized – fully overlapping signals; adjacent – partially overlapping signals; not co-localized – no overlap between signals. (E) AGA2 and STE2 genes show reduced allelic interactions after treatment with a transcription inhibitor. Cells bearing the AGA2 and STE2 genes tagged with 224 × tetR and 256 × lacO repeats were grown to mid-log phase and treated with or without 200 ug/ml 1,10 phenanthroline for 2 hr prior to fixation and imaging. Histogram of the data from three biological replicas (avg. ± SEM); co-localized – fully overlapping signals; adjacent – partially overlapping signals; non-co-localized – no overlap between signals. A paired t-test was performed to calculate the statistical significance; *p<0.01 between with and without treatment; ns – non-significant. (F) HHF1 deletion reduces allelic interactions between the AGA2 and STE2 genes. Wild-type (WT) cells and hhf1Δ cells bearing AGA2 and STE2 genes tagged with 224 × tetR and 256 × lacO repeats, respectively, were grown to mid-log phase prior to fixation and imaging. A histogram of the data is shown: co-localized – fully overlapping signals; adjacent – partially overlapping signals; non-co-localized – no overlap between signals showing avg. ± SEM. A paired t-test was performed to calculate the statistical significance: *p<0.01; **p<0.001 between the WT and hhf1∆ cells.
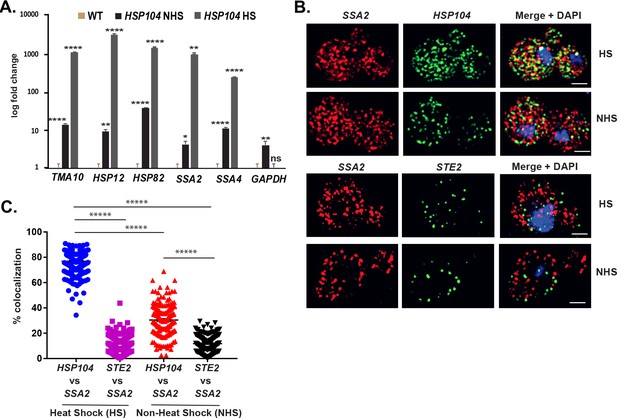
mRNAs encoding heat shock proteins undergo enhanced multiplexing upon heat shock.
MATa yeast strains (BY4741) expressing MS2 aptamer-tagged HSP104, UGO1, or STE5 from their genomic loci or untagged control (WT) cells were grown to mid-log phase (O.D.600 = 0.5) and subjected to RaPID followed by qRT-PCR (RaPID-qRTPCR; see 'Materials and methods'). RNA derived from the total cell extracts or biotin-eluated fractions was analyzed by qRT-PCR using primer pairs corresponding to mRNAs expected to multiplex (see listed genes) or not (e.g., GAPDH mRNA). (A) mRNAs encoding heat shock proteins (HSPs) multiplex upon heat shock. Cells expressing MS2 aptamer-tagged HSP104 were either exposed to heat shock (10 min; 40°C; HS) or maintained at 30°C (NHS) prior to fixation and RaPID-qRT-PCR. Three biological replicates were performed and an unpaired t-test was used to compare each under non-heat and heat shock conditions with WT; ****p<0.0001; **p<0.001; *p<0.01. (B) mRNAs of HSPs co-localize upon heat shock. Representative single-molecule fluorescence in situ hybridization (smFISH) images of BY4741 WT cells that underwent heat shock (HS) or did not (NHS). BY4741 were processed for smFISH labeling using designated FISH probes complementary to the HSP104 and SSA2 and HSP104 and STE2 mRNA pairs prior to labeling with DAPI. SSA2 – Cy3 labeling; HSP104/STE2 – Alexa488 labeling; merge – merged Cy3 and Alexa-488 windows with DAPI staining. Size bar = 2 μm. The representative image shown is from a single focal plane. (C) Scatter plot of data from (B) showing the proportion of co-localized smFISH foci. Black lines represent average ± SEM distribution. Each data point represents a single cell. *****p-value<0.00001.
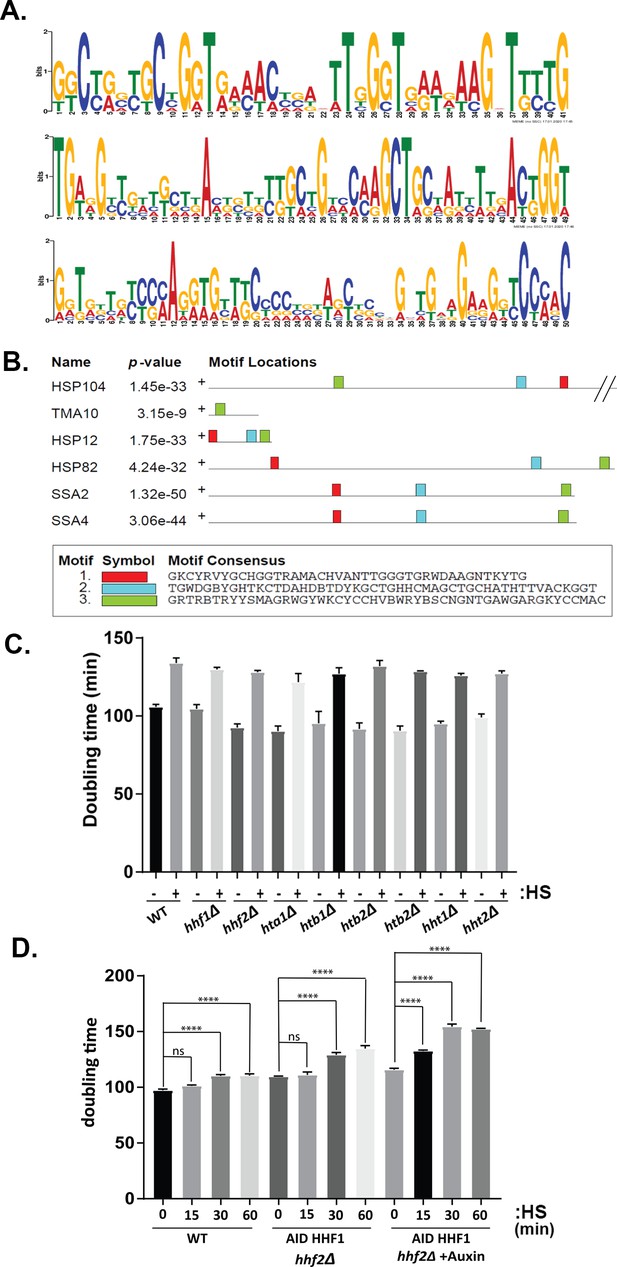
Motifs in HSP mRNA coding regions and role of histone H4 in transperon formation.
(A) A conserved motif is common to all six heat shock proteins (HSPs) mRNAs. MEME analysis of the sequences of all six mRNAs was performed and revealed three conserved motifs in the coding regions, shown schematically as sequence logo based on nucleotide representation. (B) Distribution of the motifs over the HSP genes. Motif location is plotted, along with their p-values over the different genes. Motifs 1–3 correspond to those shown in (A), respectively. (C) Single gene histone deletions do not affect cell growth after heat shock. The doubling times (minutes) of WT, hhf1Δ, hhf2Δ, hta1∆, hta2∆, htb1∆, htb2∆, hht1∆, and hht2∆ strains were assessed for cells grown on a liquid-rich medium (YPD) exposed to heat shock (50°C; 60 min;+HS) or not (-HS) and then allowed to recover for 18 hr at 30°C. Error bars represent the avg. ± sd of four biological repeats. A one-way ANOVA with multiple comparisons with and without heat shock for each strain was performed to calculate the statistical significance; p<0.0001 for all comparisons. (D) Combined hhf2∆ and AID-HHF1 alleles result in increased doubling time upon auxin induction and heat shock. MATa wild-type (WT) and hhf2∆ AID-HHF1 cells were grown to mid-log phase, treated either with or without auxin (3-IAA; 4 mM) for 3.5 hr, and exposed to heat shock (50°C; 0, 15, 30, 60 min; HS). After removal to a fresh YPD medium lacking auxin, strains were grown for 18 hr at 30°C and the doubling times assessed. Three biological replicates were performed and a one-way ANOVA test with multiple comparisons was performed to calculate the statistical significance between time points; ****p-value<0.0001. ns – non-significant.
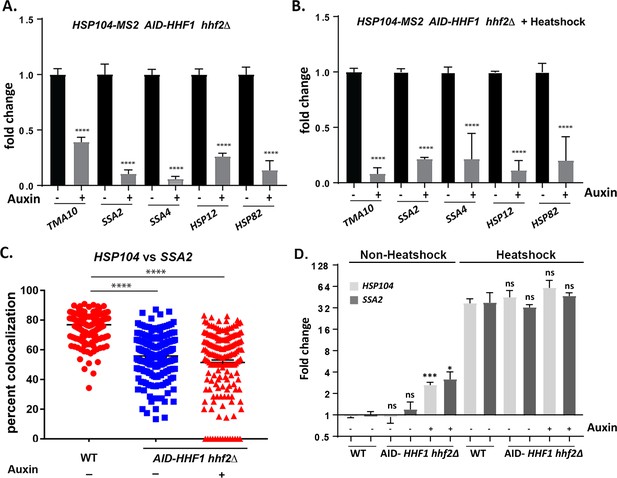
Histone H4 function is required for HSP mRNP assembly.
(A) Depletion of histone H4 inhibits heat shock protein (HSP) mRNA multiplexing. MATa hhf2∆ AID-HHF1 cells expressing MS2 aptamer-tagged HSP104 from its genomic locus were grown to mid-log phase, treated either with or without auxin (3-IAA; 4 mM) for 3.5 hr, and then heat shocked (50°C; 10 min) as shown in (B) or maintained at 30°C, as shown in (A). RaPID pulldown of HSP104 mRNA was performed, followed by qRT-PCR of the co-precipitated mRNAs. Three biological replicates were tested and unpaired t-tests were used to compare the levels of each mRNA either with or without auxin treatment; ****p<0.0001. (C) The co-localization of HSP104 and SSA2 mRNAs is reduced upon histone H4 depletion. Wild-type (WT) and AID-HHF1 hhf2∆ cells were grown to mid-log phase, treated either with or without auxin (3-IAA; 4 mM) for 3.5 hr, and fixed for single-molecule fluorescence in situ hybridization (smFISH). A scatter plot showing the level of co-localized smFISH puncta. Co-localization was assessed using FISHquant (see 'Materials and methods'). Black lines indicate the avg. ± SEM for each sample. Each data point represents a single cell. ****p-value<0.0001. (D) Neither auxin (3-IAA) treatment nor histone H4 depletion abolishes heat shock-induced gene expression. WT and HA-AID-HHF1 hhf2∆ cells were grown to mid-log phase prior to treating cells with or without auxin (3-IAA; 4 mM; 3.5 hr), with or without subsequent heat shock (50°C for 10 min), both auxin treatment and heat shock, or neither. After treatment, total cell lysates were prepared, RNA extracted, and analyzed by qRT-PCR using primers against HSP104 and SSA2. RNA levels derived from WT and HA-AID-HHF1 hhf2∆ cells under the different conditions were compared; ***p-value<0.0005; *p-value<0.01.
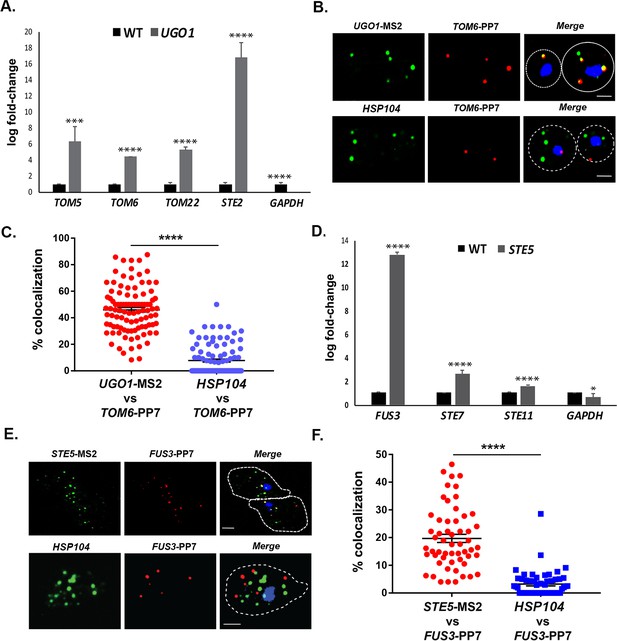
mRNAs encoding mitochondrial outer membrane and MAPK proteins multiplex to form different transperons.
(A) Mitochondrial outer membrane protein (MOMP) mRNAs multiplex. Cells expressing MS2 aptamer-tagged UGO1 were grown in a medium containing glycerol as a carbon source, prior to fixation and RaPID-qRT-PCR. Three biological replicates were performed and an unpaired t-test was used to compare the UGO1 pulldown to the wild-type (WT) control pulldown. ****p<0.0001; ***p<0.0005. (B) mRNAs encoding MOMP components co-localize. MS2- and PP7 aptamer-tagged UGO1 and TOM6 cells, respectively, were grown in a medium containing glycerol as a carbon source and processed for single-molecule fluorescence in situ hybridization (smFISH) labeling with probes complementary to the aptamers or to native HSP104 mRNA, prior to labeling with DAPI. Dotted lines represent the cell outline observed in brightfield micrographs. UGO1 – Cy3 labeling; TOM6 – Cy5 labeling; HSP104 – Alexa488 labeling; merge – merged Cy3/Alexa-488 and Cy5 windows with DAPI staining. Size bar = 2 μm. The representative image shown is from a single focal plane. (C) Scatter plot of data from (B) showing the proportion of co-localized smFISH foci. RNA localization was scored using the FISHquant algorithm (see 'Materials and methods'). Each dot represents a single cell. Black lines indicate avg. ± SEM distribution. (D) mRNAs encoding MAPK pathway components multiplex. Cells expressing MS2 aptamer-tagged STE5 were treated with α-factor (0.5 μM) prior to fixation and RaPID-qRT-PCR. Three biological replicates were performed and an unpaired t-test was used to compare the STE5 pulldown to the WT control pulldown. ****p<0.0001; *p<0.01. (E) mRNAs encoding pheromone-activated MAPK pathway components co-localize. MS2- and PP7 aptamer-tagged STE5 and FUS3 cells, respectively, were treated with α-factor (0.5 μM) and processed for smFISH labeling with probes complementary to the aptamers for STE5 and FUS3 (MAPK pathway mRNAs) or native HSP104 mRNA prior to labeling with DAPI. Dotted lines represent the cell outline observed in brightfield micrographs. STE5 – Cy3 labeling; FUS3 – Cy5 labeling; HSP104 – Alexa488 labeling; merge – merged Cy3/Alexa-488 and Cy5 windows with DAPI staining. Size bar = 2 μm. The representative image shown is from a single focal plane. (F) Scatter plot of data from (E) showing the proportion of co-localized smFISH foci. RNA localization was scored using the FISHquant algorithm (see 'Materials and methods'). Each dot represents a single cell. Black lines indicate avg. ± SEM distribution.
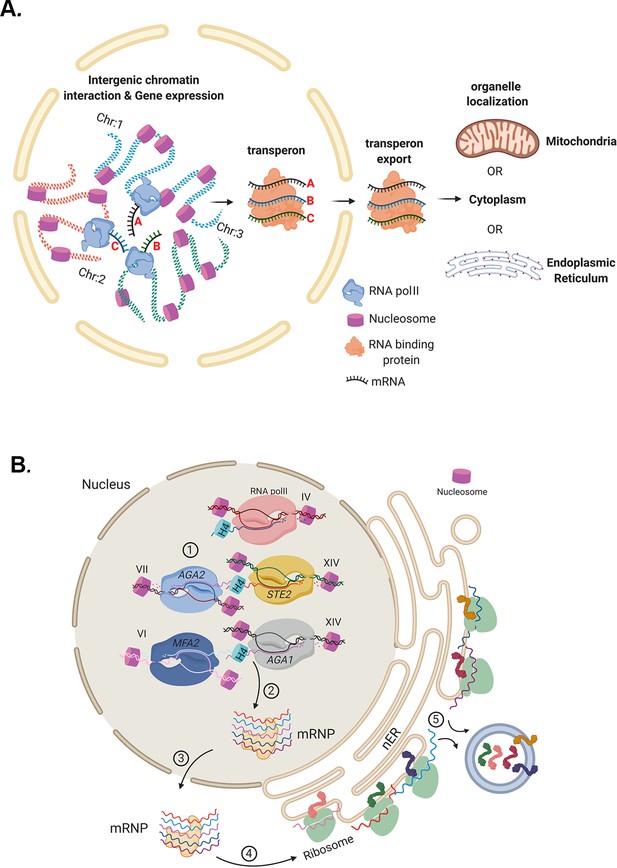
Models for mRNP transperon formation and its role in the co-translation of proteins involved in cell physiology.
(A) A schematic illustrating the mechanism of mRNA multiplexing and its role in cell physiology. Gene loci located on different chromosomes undergo allelic coupling under different growth conditions to form intergenic associations. Upon transcription, intergenic association allows for the mRNAs to assemble into a mRNA ribonucleoprotein (mRNP) complex (transperon) that is exported to the cytoplasm and confers mRNA co-localization at different organelles (mitochondria, ER, cytoplasm). RNA co-localization and co-translation allow for coordination of the pathways responsible for the control of cell physiology. (B) mRNA multiplexing and transperon formation control the mating pathway in yeast. Yeast (MATa cells, as shown) express mRNAs encoding soluble secreted and membrane proteins (e.g., Mfa1, Mfa2, Aga1, Aga2, and Ste2, respectively) from genomic loci located on different chromosomes (as listed) that undergo allelic coupling to form intergenic associations. Histone H4 paralogs (Hhf1 and -2) are required for the mRNAs to assemble into a mating mRNA transperon that is exported to the cytoplasm and confers RNA co-localization at the endoplasmic reticulum peripheral to the nucleus (nER). Co-translational translocation of these proteins at the ER may coordinate the pathways responsible for the cellular responsiveness to pheromone of the opposite mating type (i.e., via both Ste2 signaling and Aga1 and Aga2 function in cell adhesion) and transmission of the cell’s own mating signal to cells of the opposite mating type (i.e., via Mfa1 and Mfa2 secretion).
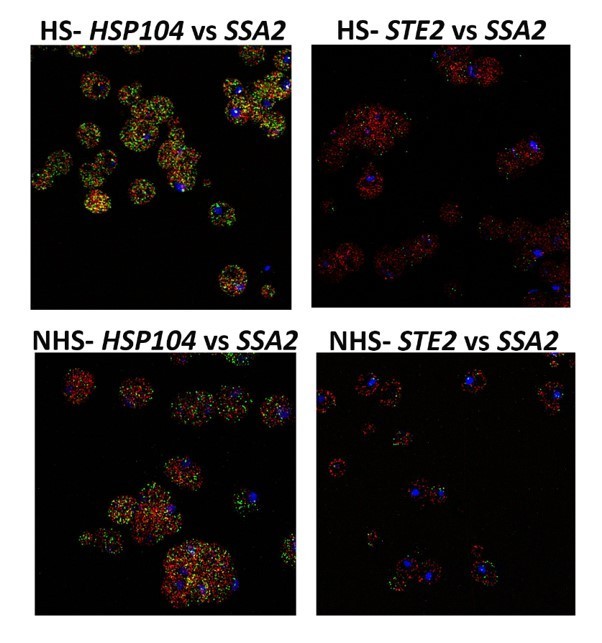
HS – heat shocked cells; NHS – non-heat shocked cells.
Note colocalization of HSP104 mRNA with SSA2 mRNA, but not with STE2 mRNA.
Additional files
-
Supplementary file 1
RNA-sequencing data (Excel table).
Different MS2 aptamer-tagged mRNAs (listed on column heads) expressed from their genomic loci or control cells (Cntrl) lacking a tagged mRNA were precipitated from MATα yeast (BY4742) by MS2-CP-GFP-SBP after formaldehyde crosslinking in vivo and cell lysis procedures (RaPID; see 'Materials and methods'). RNA-seq was performed, and the results for each gene are listed on the following sheets. Sheet 1 (Counts.matrix) represents the number of reads assigned to each gene by Kallisto. Sheet 2 (edgeR_DE_results) represents all genes identified by edgeR, as being differentially expressed in the corresponding pairwise comparison to the control, requiring a false discovery rate (FDR) <0.1. Sheet 3 (DE_expr_sub_cntrl) represents gene length-normalized expression values, log2 transformed followed by subtraction of the control expression value. Sheet 4 (Expr_targets_sub_cntrl) represents the sheet three values restricted to the baits (target mRNAs).
- https://cdn.elifesciences.org/articles/66050/elife-66050-supp1-v3.xlsx
-
Supplementary file 2
Yeast strains used in this study.
In this table, we include the strain name, genotype, and source of origin.
- https://cdn.elifesciences.org/articles/66050/elife-66050-supp2-v3.docx
-
Supplementary file 3
Plasmids used in this study.
In this table, we include the plasmid name, gene expressed, vector backbone used, copy number of the plasmid, selection marker, and source of origin.
- https://cdn.elifesciences.org/articles/66050/elife-66050-supp3-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66050/elife-66050-transrepform-v3.docx