In vivo proximity labeling identifies cardiomyocyte protein networks during zebrafish heart regeneration
Figures
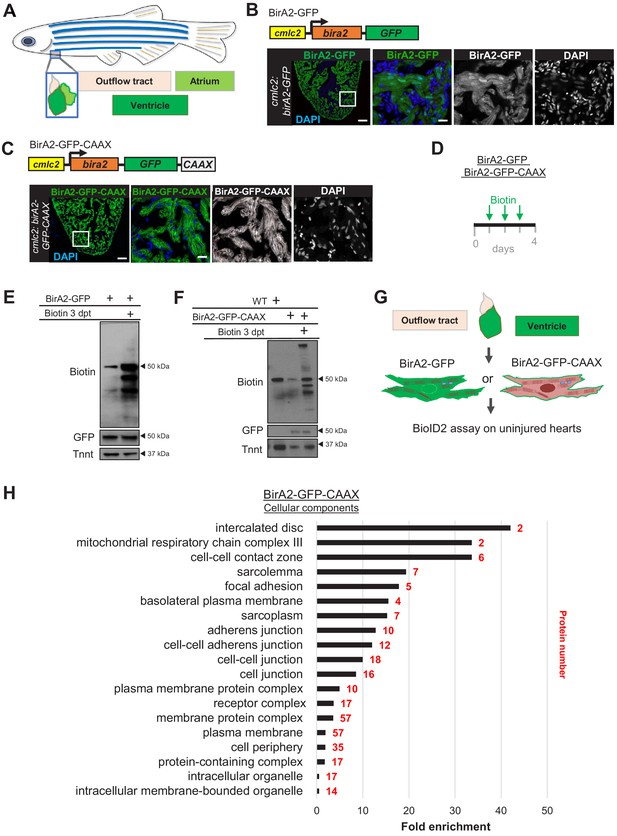
In vivo BioiD2 captures the proteome in cardiomyocytes of adult zebrafish hearts.
(A) Schematic representation of a zebrafish heart. The heart consists of a ventricle, atrium, and outflow tract. (B) BirA2-GFP is expressed in cardiomyocytes via the cmlc2 promoter. Section of ventricle from cmlc2:birA2-GFP transgenic animal. BirA2 is distributed throughout the cardiomyocytes, including the nucleus. DAPI staining and native GFP signal are shown. (C) BirA2-GFP-CAAX is expressed in cardiomyocytes via the cmlc2 promoter. Section of ventricle from cmlc2:bira2-GFP-CAAX transgenic animal. The CAAX-tag localizes BirA2 to the membranes of cardiomyocytes. (D) Timeline of biotin administration by intraperitoneal injections (IP injections). (E) Western blot analysis of biotinylation activity of BirA2-GFP-expressing ventricles. BirA2 is functional and biotinylates sufficiently after three daily biotin injections. Endogenous biotinylated carboxylase was detected in untreated and biotin-treated hearts. (F) Western blot analysis of BirA2-GFP-CAAX-expressing ventricles. BirA2 is functional and biotinylates sufficiently after three daily injections with biotin. WT: wild type. (G) Schematic summary of BioID2 assay on uninjured zebrafish hearts. (H) Over-representation test for cellular components. BioID2 for BirA-GFP-CAAX enriches for membrane-associated proteins. 343 total proteins were gated at a 2.5-fold change when normalized to BirA-GFP. p<0.001, false discovery rate (FDR) < 0.015%. dpt: days post treatment. Scale bar in images, 50 μm; magnification scale bar: 10 μm.
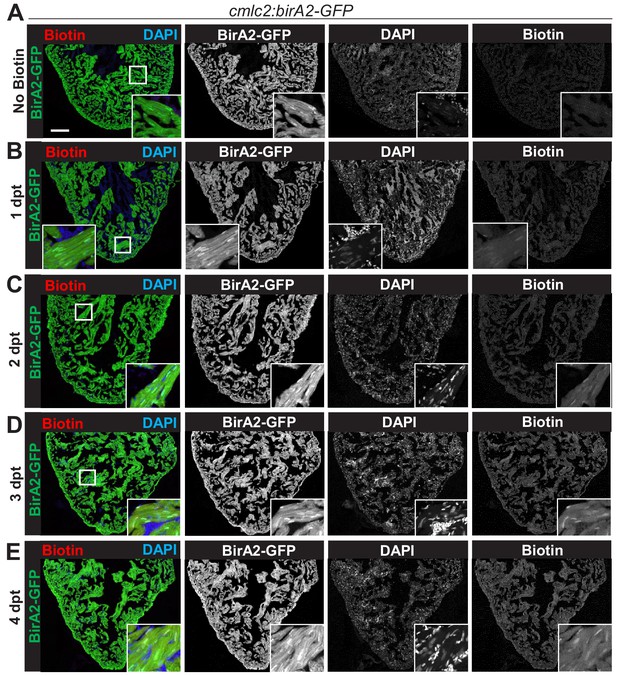
Evaluation of biotinylation in BirA2-GFP-expressing transgenic zebrafish.
(A–E) Immunofluorescence images of sections of ventricles with native BirA2-GFP signal, stained for DAPI (nuclei). Animals were injected intraperitoneally with biotin for 1, 2, 3, or 4 days. dpt: days post treatment. Scale bar: 50 µm.
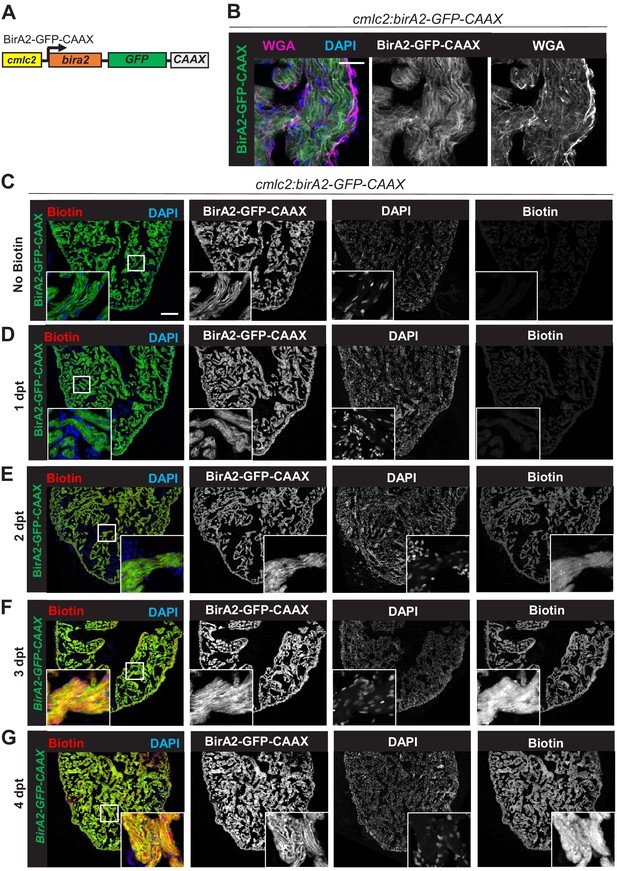
Evaluation of BirA2-GFP-CAAX localization and biotinylation.
(A) BirA2-GFP-CAAX is expressed in cardiomyocytes via the cmlc2 promoter. (B) BirA2-GFP-CAAX is localized to the cell membrane. Membrane staining with wheat germ agglutinin showed colocalization with the BirA2-GFP-CAAX signal. Scale bar: 10 µm. (C–G) Immunofluorescence images of sections of ventricles with native BirA2-GFP-CAAX signal stained for DAPI. Animals were injected intraperitoneally with biotin for 1, 2, 3, or 4 days. dpt: days post treatment. Scale bar: 50 µm.
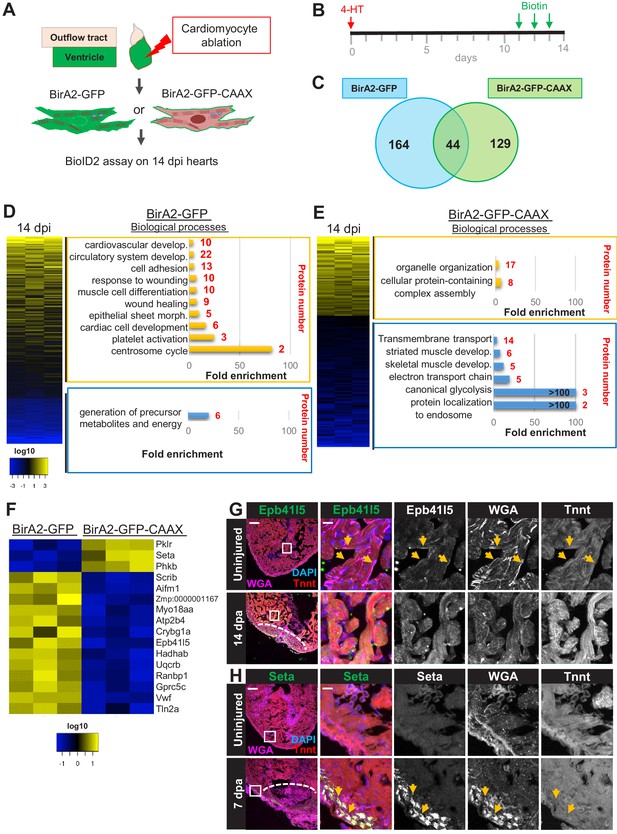
BioID2 identifies changes in membrane protein levels during heart regeneration.
(A) Schematic overview of experimental workflow. cmlc2:birA2-GFP or cmlc2:bira2-GFP-CAAX ventricles were collected as uninjured samples or 14 days after induced cardiomyocyte ablation (dpi). (B) Timeline of injury and biotin injections. (C) Venn diagram comparing proteins captured by either the whole-cell BioID2 assay or the membrane BioID2 data set that display at least a 1.5-fold change. (D) Summary of BioID2 analysis of cmlc2:birA2-GFP hearts during regeneration. Left: heat map of proteins found in triplicates of quantitative mass spectrometry analysis. 208 proteins displayed a 1.5-fold change during regeneration when compared to uninjured hearts (p<0.05). Of these, most protein levels increase during heart regeneration. Protein level changes in log10 scale. Right: gene ontology analysis of BirA2-GFP BioID2 data set. Over-representation test of biological processes for at least 1.5-fold increased (yellow) and at least 1.5-fold decreased proteins (blue). Fold enrichment is shown, and protein number is indicated in red. p<0.001, FDR < 0.04%. (E) Summary of BioID2 on cmlc2:bira2-GFP-CAAX hearts. Left: heat map of proteins found in triplicates of quantitative mass spectrometry analysis. 173 proteins displayed an at least 1.5-fold change when compared to uninjured hearts (p<0.05). Of these, most proteins decreased at the membrane during heart regeneration. Right: gene ontology analysis of BirA2-GFP-CAAX BioID2 data set. p<0.001, FDR < 0.05%. (F) Heat map of proteins that have been identified in the BioID2 whole-cell and membrane data sets and that display opposing changes in levels. Those shown changed at least 1.5-fold, p<0.05. Heat map summarizes fold changes measured in three separate pooled samples. (G) Immunofluorescence against indicated proteins in ventricles. Epb41l5 is localized to the plasma membrane (marked by wheat germ agglutinin [WGA] staining) in uninjured hearts, with cytoplasmic fluorescence signals increasing during regeneration of resected tissue (14 days post amputation [dpa]). (H) Seta is poorly detected in uninjured hearts, rising 7 days after resection injury in the epicardium and compact layer of the heart. Seta localizes around the nucleus and colocalizes with the membrane marker WGA. Scale bar in images, 50 μm; magnification scale bar: 10 μm.
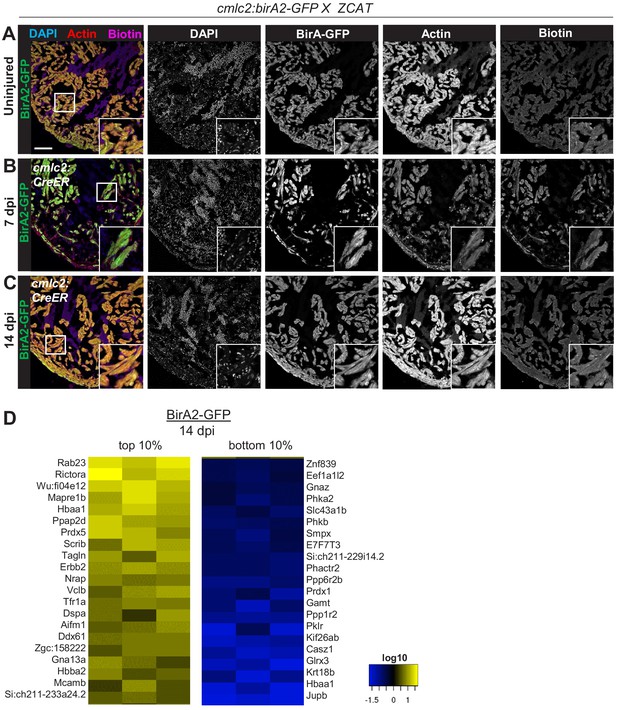
Cardiomyocyte ablation in cmlc2:birA2-GFP fish, and summary of proteins that display largest changes in levels.
(A–C) Immunofluorescence images of sections of ventricles of fish expressing BirA2-GFP crossed to the Z-CAT cardiomyocyte ablation model (Wang et al., 2011). Animals were treated with 4-HT to induce expression of diphtheria toxin (DTA) toxin. Ventricles were stained for DAPI, actin, and biotin. Scale bar: 50 µm. (D) Top 10% and bottom 10% of proteins that were captured in the whole-cell BioID2 assay and show the largest changes in levels after injury in BirA2-GFP; Z-CAT fish. Heat map summarizes fold changes measured in three separate pooled samples.
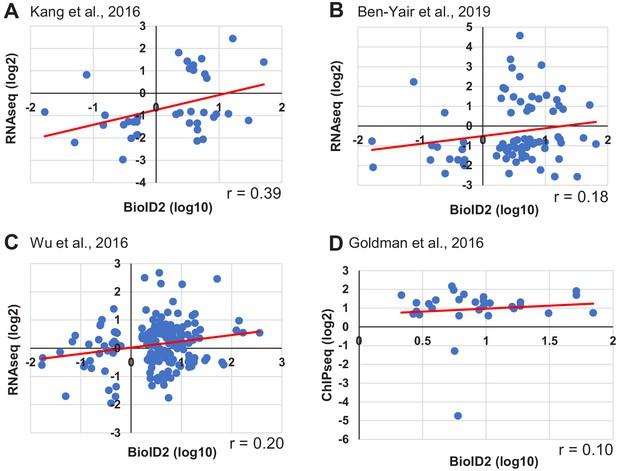
Comparison of the BioID2 data set with published transcriptome or epigenome data sets.
(A–D) The 14 days post incubation (dpi) BirA2-GFP BioID2 data set (normalized to uninjured, p<0.05) was compared to three RNA-seq data sets (Wu et al., 2016; Kang et al., 2016; Ben-Yair et al., 2019) and one ChIP-seq (H3.3) data set (Goldman et al., 2017). For RNA-seq data sets (A–C), expression levels have been normalized to uninjured controls and genes with differences of significance p<0.05 were considered. (A) RNA-seq data set from Kang et al., 2016, where 14 dpi whole hearts were used. 37 genes were identified in both data sets, with a correlation coefficient r = 0.39. (B) RNA-seq data set from Ben-Yair et al., 2019, in which purified gata4-expressing cells were examined for transcriptome changes at 5 dpa. 75 genes were represented in both data sets, with a correlation coefficient r = 0.18. (C) RNA-seq data set from Wu et al., 2016, where 7 days post cryoinjury whole hearts (border zone) were used. 160 genes were identified among both data sets, with a correlation coefficient r = 0.2. (D) ChIP-seq data set from Goldman et al., 2017, in which genes with changes in enrichment for a cardiomyocyte-restricted, tagged H3.3 protein at promoters were used in comparison. Promoters were defined as between 5 kb upstream and 2 kb downstream from the gene start site. FDR < 0.05 for significant differential sites, and p<0.05. 30 genes were identified among both data sets.
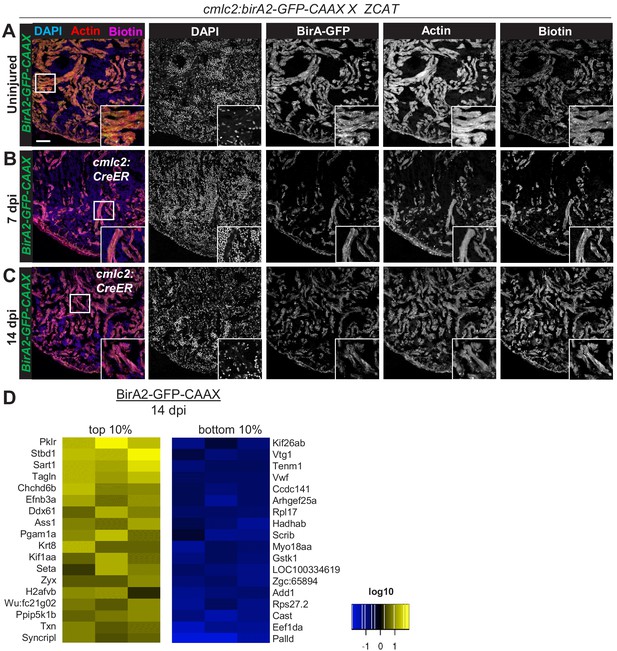
Cardiomyocyte ablation in cmlc2:bira2-GFP-CAAX fish, and summary of proteins that display largest changes levels.
(A–C) Immunofluorescence images of sections of ventricles of fish expressing BirA2-GFP-CAAX crossed to the Z-CAT ablation model (Wang et al., 2011). Animals were treated with tamoxifen to induce expression of DTA toxin. Ventricles were stained for DAPI, actin, and biotin. Scale bar: 50 µm. (D) Top 10% and bottom 10% of proteins that were captured in the whole-cell BioID2 assay and show the largest changes in levels after injury. Heat map summarizes fold changes measured in three separate pooled samples.
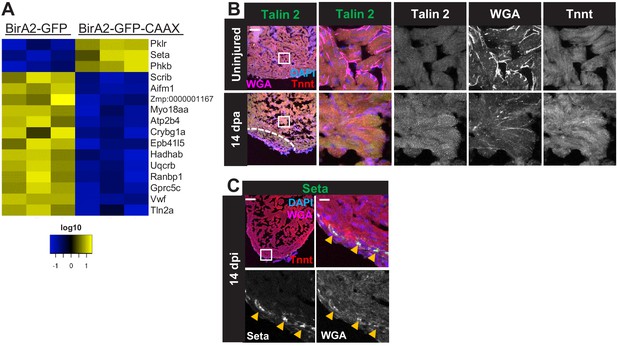
Protein levels and localization from BioID2 comparison of whole cardiomyocytes and membranes.
(A) Heat map of proteins identified in the BioID2 whole-cell and membrane data sets that display opposing changes in levels. Changes 1.5-fold or greater with p<0.05. Log10 scale is used. Heat map summarizes fold changes measured in three separate pooled samples. (B,C) Immunofluorescence for indicated proteins in ventricular sections. (B) Talin 2 levels are low and display a punctate distribution in the uninjured heart. During regeneration, Talin levels rise in the cytoplasm. (C) Seta-positive cells are localized to the compact layer and epicardium in 14 days post incubation hearts injured by the Z-CAT ablation model. Scale bars: 50 μm; magnification scale bar: 10 μm.

BioID2 identifies Rho A as a downstream target of ErbB2.
(A) ErbB2-BirA2-HA-P2A-GFP is expressed in cardiomyocytes via the cmlc2 promoter. (B) Summary of experiment and timeline. cmlc2:erbb2-birA2-HA-P2A-GFP ventricles were collected as uninjured samples or 14 days after induced cardiomyocyte ablation (dpi). (C) Known direct interactors of ErbB2 that were captured in the ErbB2 BioID2 assay. 108 proteins showed a change >1.5-fold when normalized to uninjured cmlc2:erbb2-birA2-HA-P2A-GFP ventricles. These data were analyzed for known ErbB2 interactors. Colors and size of interactor correspond to fold changes identified during regeneration. Green: no change; blue: levels decrease; yellow: levels increase. Tns1: tensin 1 (1.57-fold); Hspb2: heat shock protein beta 2 (−2.43-fold); Gapdh: glyceraldehyde 3-phosphate dehydrogenase (p>0.05); Acta1b: actin alpha 1 (p>0.05); Ezrb: ezrin b (p>0.05); Ctnnd1: catenin δ1 (−4.59-fold); RhoA-b: Rho A-b (13-fold); Scrib: scribble (p>0.05); Anxa2a: annexin A2a (7.31-fold). (D) Over-representation test – pathway analysis of proteins increased 1.5-fold or more in ErbB2-BirA data set. p<0.001, FDR < 0.005%. Fold enrichment is shown, and protein number in red. (E) Co-immunoprecipitation of ErbB2-BirA2-HA from uninjured or regenerating cmlc2:erbb2-birA2-HA-P2A-GFP hearts. Rho A association with ErbB2 is increased after injury. Anti-HA antibody was used for ErbB2 detection, and troponin T and IgG were used as loading controls. (F) Analysis of known Rho A interactors in BirA2-GFP BioID2 data set. All known direct interactors were found to be increased when normalized to uninjured hearts. Size of circles indicates fold change; proteins are sorted clockwise after fold change from high to low. Scrib: scribble (90.9-fold); ErbB2: Erb-b2 receptor tyrosine kinase 2 (52.9-fold); Gna13: guanine nucleotide binding protein alpha 13a (25.8-fold); Rtn4a: reticulon 4a (8.82-fold); ROCK2a: Rho-associated, coiled-coil containing protein kinase 2a (5.05-fold); Ilk: integrin-linked kinase (3.91-fold); Pfn1: profilin 1 (3.25-fold); Arhgap21b: Rho GTPase activating protein 21b (2.65-fold); Wasla: WASP-like actin nucleation promoting factor a (2.5-fold); Lrp5: low-density lipoprotein receptor-related protein 5 (2.21-fold); Ctnnd1: catenin δ1 (2.2-fold); Msna: moesin a (1.9-fold). (G) Western blot analysis of ROCK2 levels in uninjured and regenerating hearts. ROCK2 levels are increased during heart regeneration.
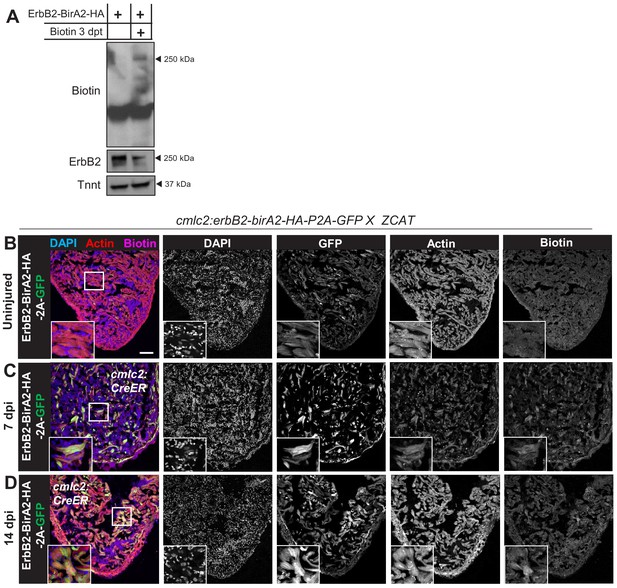
Biotinylation in cmlc2:erbb2-birA2-HA-P2A-GFP ventricles.
(A) Western blot analysis of ErbB2-BirA2-HA assessed for biotin and Human influenza hemagglutinin tag (ammino acids 98-106) (HA) in zebrafish expressing cmlc2:erbb2-birA2-HA-P2A-GFP. Troponin T is used as a loading control. (B–D) Immunofluorescence images of sections of ventricles of fish expressing ErbB2-BirA2-HA crossed to the Z-CAT ablation model. Animals were treated with tamoxifen to induce expression of DTA toxin. Ventricle were stained for DAPI, actin, and biotin. Scale bar: 50 µm.

Alignment of Rho A antibody immunogen with Rho A proteins.
The Rho A antibody immunogen (human Rho A 120aa – 150aa) shows the highest conservation with Rho A-b. aa: amino acids.
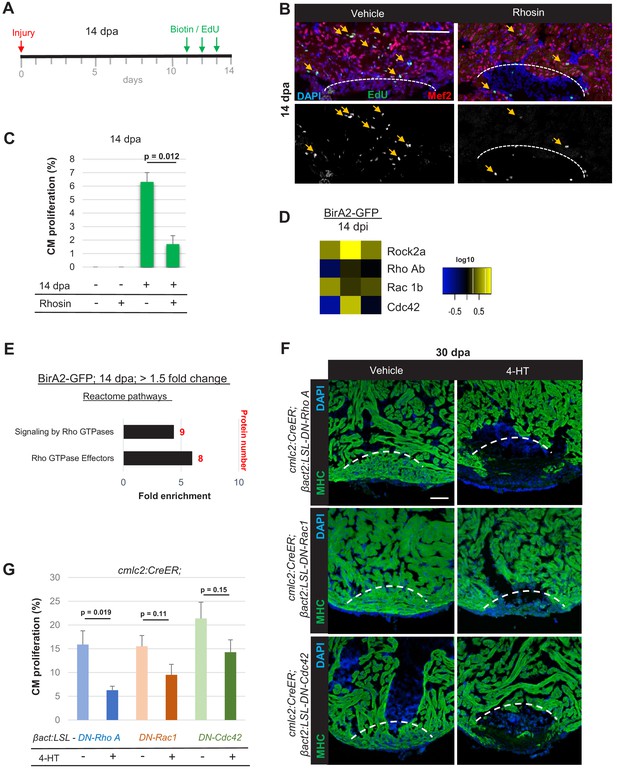
Small GTPases are required for cardiac regeneration.
(A) Timeline of experiment for inhibiting Rho A function in 14 dpa hearts. (B) Immunofluorescence images of sections of 14 dpa ventricles vehicle or Rhosin treated and stained for EdU incorporation, an indicator of cardiomyocyte proliferation. Mef2 staining marks cardiomyocyte nuclei. Dashed line, approximate resection plane. Arrowheads, Mef2+/EdU+ cardiomyocytes. (C) Quantification of Mef2/EdU assays. Inhibition of Rho A by Rhosin reduces cardiomyocyte (CM) proliferation. n = 5 animals for each condition, two independent experiments. Data show mean ± SEM. (Mann–Whitney U test). (D) Heat map of indicated proteins from the whole-cell BioID2 data set. Of these, only levels of ROCK2a changed consistently (5.1-fold; p<0.05) during regeneration. (E) Over-representation test of reactome pathways. Signaling and effectors of Rho GTPases were found to be over-represented. Fold enrichment as indicated, and protein number in red. p<0.001, FDR < 0.02%. (F) Myosin heavy chain (MHC) (green) staining ventricles from animals with induced myocardial-specific dominant-negative Rac, Rho, or Cdc42, along with vehicle-treated ventricles, at 30 dpa. Five to eight animals were assessed in each group treated with vehicle, with none of these animals displaying regeneration defects. Four of five ventricles with induced dominant-negative Rac1, and all ventricles with induced dominant-negative Rho A (n = 7/7), or Cdc42 (n = 9/9), showed obvious areas of missing myocardium. Fisher–Irwin exact test, p<0.05. Dashed line, approximate resection plane. Scale bars, 50 µm. (G) Quantification of cardiomyocyte proliferation by Mef2/Proliferating cell nuclear antigen (PCNA) staining of n = 4 animals for each condition. Data show mean ± SEM. (Mann–Whitney U test).
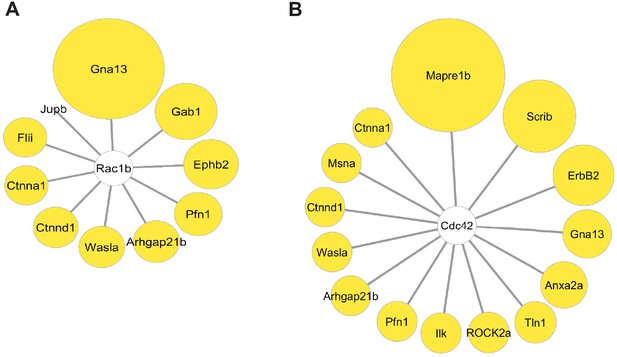
Interactome map of Rac1 and Cdc42.
(A) Analysis of known Rac1b interactors in cmlc2:birA2-GFP BioID2 data set. Except for Jupb, all known interactors were found to be increased during heart regeneration. Size of circles indicate fold change. Proteins are sorted clockwise after fold change from high to low. Gna13: guanine nucleotide binding protein alpha 13a (25.8-fold); Gab1: GRB2-associated binding protein 1 (8.34-fold); Ephb2: ephrin receptor B2 (5.72-fold); Pfn1: profilin 1 (3.25-fold); Arhgap21b: Rho GTPase activating protein 21b (2.65-fold); Wasla: WASP-like actin nucleation promoting factor a (2.5-fold); Ctnnd1: catenin δ1 (2.2-fold); Ctnna1: catenin α1 (1.68-fold); Flii: flightless I actin remodeling protein (1.72-fold); Jupb: junctional plakoglobin b (−192-fold). (B) Analysis of known Cdc42 interactors in cmlc2:birA2-GFP BioID2 data set. See (A) for fold changes of proteins found also in Rac1b interactome. Mapre1b: microtubule-associated protein EB 1b (200-fold); Anxa2a: annexin A2a (20.5-fold); Tln1: Talin 1 (7.56-fold); ROCK2a: Rho-associated, coiled-coil containing protein kinase 2a (5.05-fold); Ilk: integrin-linked kinase (3.91-fold); Msna: moesin a (1.9-fold).
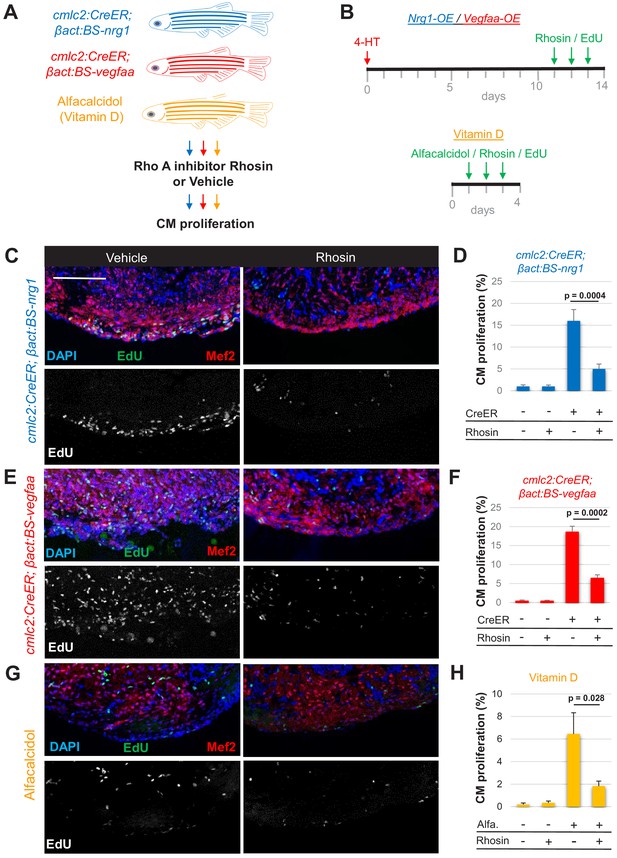
Rho A activity is required for cardiogenic responses to mitogens.
(A) Summary of transgenic animals used in Rho A inhibition experiments. (B) Timeline of experiments for Nrg1-overexpression (OE, blue), Vegfaa-OE (red), and alfacalcidol treatment (orange). (C, E, G) Immunofluorescence images of ventricles stained for Mef2/EdU from animals overexpressing Nrg1 (C) or Vegfaa (E) in cardiomyocytes, or injected with alfacalcidol (G). Hearts were treated with either vehicle or Rhosin. Scale bar, 50 μm. (D, F, H) Inhibition of Rho A by Rhosin reduces cardiomyocyte (CM) proliferation. Quantification of Mef2/EdU staining. Five to six animals were assessed for each group in two independent experiments. Data show mean ± SEM (Mann–Whitney U test).
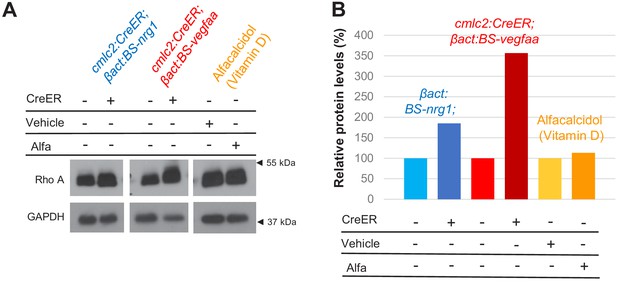
Rho A levels in hearts of cardiac mitogen-treated animals.
(A) Western blot analysis of Rho A levels in ventricles of zebrafish with Nrg1-overexpression (OE), Vegfaa-OE, or vitamin D receptor activation via alfacalcidol. (B) Quantification of western blot shown in (A). In untreated hearts, Rho A levels were set to 100%. Levels were normalized to loading control GAPDH.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Danio rerio) | BirA2-GFP | This paper | Tg(cmlc2:birA2-GFP)pd335 | Transgenic line maintained in K. Poss lab |
| Genetic reagent (Danio rerio) | BirA2-GFP-CAAX | This paper | Tg(cmlc2:birA2-GFP-CAAX)pd336 | Transgenic line maintained in K. Poss lab |
| Genetic reagent (Danio rerio) | ErbB2-BirA2-HA-P2A-GFP | This paper | Tg(erbb2-birA2-HA-P2A-GFP)pd337 | Transgenic line maintained in K. Poss lab |
| Genetic reagent (Danio rerio) | DN-RhoA | This paper | Tg(βactin2:loxp-BFP-STOP-loxp-DN-Rho)pd68 | Transgenic line no longer maintained |
| Genetic reagent (Danio rerio) | DN-Rac1 | This paper | Tg(βactin2:loxp-BFP-STOP-loxp-DN-Rac1)pd69 | Transgenic line no longer maintained |
| Genetic reagent (Danio rerio) | DN-Cdc42 | This paper | Tg(βactin2:loxp-BFP-STOP-loxp-DN-Cdc42)pd70 | Transgenic line no longer maintained |
| Genetic reagent (Danio rerio) | CreER | Kikuchi et al., 2010 | Tg(cmlc2:CreER)pd10 | Transgenic line, K. Poss lab |
| Genetic reagent (Danio rerio) | Nrg1 | Gemberling et al., 2015 | Tg(actb2:loxp-TagBFP-STOP-loxp-nrg1)pd107 | Transgenic line, K. Poss lab |
| Genetic reagent (Danio rerio) | Vegfaa | Karra et al., 2018 | Tg(actb2:loxP-TagBFP-loxp-vegfaa)pd262 | Transgenic line, K. Poss lab |
| Genetic reagent (Danio rerio) | CM ablation | Wang et al., 2011 | Tg(actb2:loxp-mCherry-loxp-DipTox)pd36 | Z-CAT cardiac myocyte ablation model, K. Poss lab |
| Antibody | Anti-Mef2 (rabbit polyclonal) | Abcam | Cat#: ab197070 | IF: 1:100 |
| Antibody | Anti-Rho A (rabbit polyclonal) | Santa Cruz | Cat#: 26C4 | IF: 1:1000 |
| Antibody | anti-epb41L5 (rabbit polyclonal) | Proteintech | Cat#: RPL39 | IF: 1:100 |
| Antibody | Anti-Talin 2 (rabbit polyclonal) | Boster Biological Technology | Cat#: PB9961 | IF: 1:100 |
| Antibody | Anti-Seta (rabbit polyclonal) | Antibodies online | Cat#: ABIN2786762 | IF: 1:100 |
| Antibody | Anti-ROCK2 (rabbit polyclonal) | Bioss Antibodies | Cat#: BS-1205R | IF: 1:500 |
| Antibody | Anti-HA (rabbit polyclonal) | Abcam | Cat#: ab9110 | IF: 1:1000 |
| Antibody | Anti-CT3 (mouse monoclonal) | DSHB | Cat#: CT3 | IF: 1:200 |
| Antibody | Anti-PCNA (mouse monoclonal) | Sigma | Cat#: P8825 | IF: 1:200 |
| Antibody | Anti-MHC (mouse monoclonal) | DSHB | Cat#: F59-S | IF: 1:200 |
| Chemical compound, drug | Rhosin | Calbiochem | Cat#: 555460 | 250 μM in 10 μl IP injections |
| Chemical compound, drug | Biotin | Sigma-Aldrich | Cat#: B4501 | 0.5 mM in 10 μl IP injections |
| Chemical compound, drug | EDU | Life Technology | Cat#: A10044 | 100 nM in 10 μl IP injections |
| Chemical compound, drug | Alfacalcidol | Selleck Chemicals | Cat#: S1466 | 200 μM in 10 μl IP injections |
| Recombinant DNA reagent | BirA2-GFP | This paper | cmlc2:birA2-GFP | Plasmid used to generate transgenic line |
| Recombinant DNA reagent | BirA2-GFP-CAAX | This paper | cmlc2:birA2-GFP-CAAX | Plasmid used to generate transgenic line |
| Recombinant DNA reagent | ErbB2-BirA2-HA-P2A-GFP | This paper | cmlc2:erbb2-birA2-HA-P2A-GFP | Plasmid used to generate transgenic line |
Additional files
-
Supplementary file 1
Normalized levels of BioID2 proteins from uninjured cmlc2:birA2-GFP-CAAX hearts.
List of proteins from quantitative mass spectrometry analysis of cmlc2:birA2-GFP-CAAX ventricles. Uninjured BirA2-GFP-CAAX protein levels were normalized to uninjured BirA2-GFP levels. Data are sorted by fold change. Accession number, gene name, description of gene, total unique peptide count, and fold change with p-values are shown.
- https://cdn.elifesciences.org/articles/66079/elife-66079-supp1-v2.xlsx
-
Supplementary file 2
Normalized levels of BioID2 proteins from regenerating cmlc2:birA2-GFP hearts.
List of proteins from quantitative mass spectrometry analysis of cmlc2:birA2-GFP ventricles. Regenerating BirA2-GFP protein levels were normalized to uninjured BirA2-GFP levels. Data are sorted by fold change. Accession number, gene name, description of gene, total unique peptide count, and fold change with p-values are shown.
- https://cdn.elifesciences.org/articles/66079/elife-66079-supp2-v2.xlsx
-
Supplementary file 3
Normalized levels of BioID2 proteins from regenerating cmlc2:birA2-GFP-CAAX hearts.
List of proteins from quantitative mass spectrometry analysis of cmlc2:birA2-GFP-CAAX ventricles. Regenerating BirA2-GFP-CAAX protein levels were normalized to uninjured BirA2-GFP-CAAX levels. Data are sorted by fold change. Accession number, gene name, description of gene, total unique peptide count, and fold change with p-values are shown.
- https://cdn.elifesciences.org/articles/66079/elife-66079-supp3-v2.xlsx
-
Supplementary file 4
Normalized levels of BioID2 proteins from regenerating cmlc2:erbb2-birA2-HA-P2A-GFP hearts.
List of proteins from quantitative mass spectrometry analysis of cmlc2:erbb2-birA2-HA ventricles. Regenerating ErbB2-BirA2-HA protein levels were normalized to uninjured ErbB2-BirA2-HA levels. Data are sorted by fold change. Accession number, gene name, description of gene, total unique peptide count, and fold change with p-values are shown.
- https://cdn.elifesciences.org/articles/66079/elife-66079-supp4-v2.xlsx
-
Supplementary file 5
ErbB2-associated proteins represented in published transcriptome and epigenome data sets.
Association of ErbB2 after injury is color coded as in Figure 3, with yellow denoting increased association with ErbB2, green denoting no change in association, and blue denoting decreased association.
- https://cdn.elifesciences.org/articles/66079/elife-66079-supp5-v2.xlsx
-
Supplementary file 6
rhoa gene expression levels represented in published transcriptome and epigenome data sets after cardiac injury or during Nrg1 overexpression.
For Nrg1 experiments, normalized RNA-seq read counts are shown.
- https://cdn.elifesciences.org/articles/66079/elife-66079-supp6-v2.xlsx
-
Supplementary file 7
Raw data from BioID2 assays used to generate Supplementary files 1–3.
- https://cdn.elifesciences.org/articles/66079/elife-66079-supp7-v2.xlsx
-
Supplementary file 8
Raw data from Erbb2-HA-BirA2-P2A-GFP BioID2 assays used to generate Supplementary file 4.
- https://cdn.elifesciences.org/articles/66079/elife-66079-supp8-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66079/elife-66079-transrepform-v2.pdf





