The EMT transcription factor Snai1 maintains myocardial wall integrity by repressing intermediate filament gene expression
Figures
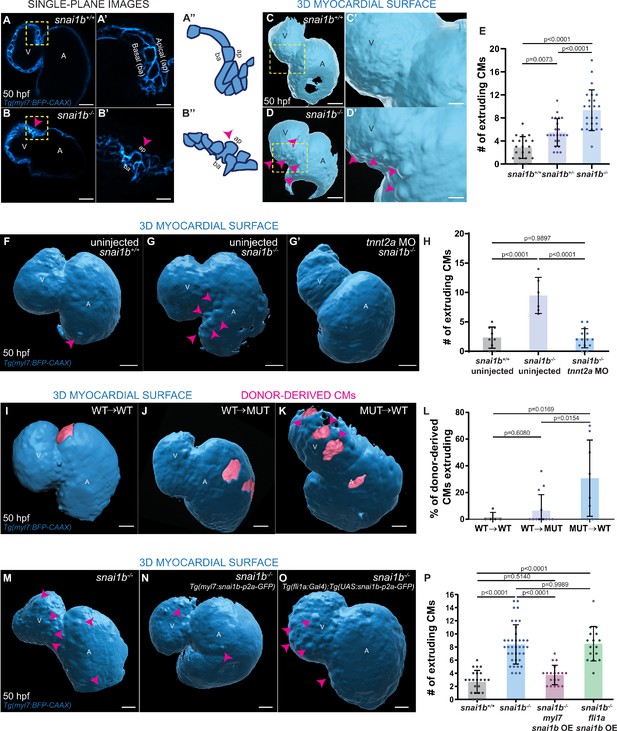
Loss of snai1b leads to cardiomyocyte (CM) extrusion, disrupting myocardial wall integrity.
(A–B”) Single-plane images of Tg(myl7:BFP-CAAX) snai1b+/+ (A) and snai1b-/- (B) hearts at 50 hpf. Close-up of boxed areas (A’, B’) and schematic (A”, B”). (C–D’) 3D surface rendering of the myocardium of Tg(myl7:BFP-CAAX) snai1b+/+ (C, C’) and snai1b-/- (D, D’) embryos at 50 hpf. CM extrusions are clearly observed in snai1b-/- embryos (magenta arrowheads in B, B’, B”, D, D’). (E) More CMs are extruding in Tg(myl7:BFP-CAAX) snai1b-/- embryos compared with snai1b+/+ and snai1b+/- siblings at 50 hpf (snai1b+/+, n = 20; snai1b+/-, n = 23; snai1b-/-, n = 24). (F–H) Blocking cardiac contractions with tnnt2a MO leads to a reduced number of extruding CMs in snai1b-/- embryos, comparable with uninjected snai1b+/+ embryos. (F–G’) 3D surface rendering of the myocardium of snai1b+/+ (F) and snai1b-/- (G) uninjected embryos and snai1b-/- embryos injected with tnnt2a MO (G’). (H) Fewer CMs are extruding (magenta arrowheads in G) in snai1b-/- embryos injected with tnnt2a MO (n = 14) compared with uninjected snai1b-/- (n = 6) and snai1b+/+ (n = 9) embryos at 50 hpf. (I–L) 3D surface rendering of the myocardium showing snai1b+/+ donor-derived CMs in a snai1b+/+ (I) or snai1b-/- (J) heart, and snai1b-/- donor-derived CMs in a snai1b+/+ heart (K). (L) The percentage of donor-derived CMs that extrude is higher when snai1b-/- donor-derived CMs are in snai1b+/+ hearts (n = 8) than when snai1b+/+ donor-derived CMs are in snai1b+/+ (n = 5) or snai1b-/- (n = 14) hearts. (M–P) Overexpression of snai1b specifically in CMs partially rescues the CM extrusion phenotype in snai1b-/- embryos. 3D surface rendering of the myocardium of a snai1b-/- embryo (M), and snai1b-/- embryo overexpressing snai1b under a myl7 (N) or a fli1a (O) promoter. (P) Fewer CMs are extruding (magenta arrowheads) in snai1b-/- embryos (n = 19) overexpressing snai1b in CMs (N, P) compared with snai1b-/- embryos (M, P, n = 38), and this number is comparable to that in snai1+/+ embryos (n = 24). The number of extruding CMs does not change in snai1b-/- embryos (n = 16) when snai1b is overexpressed in endothelial cells (fli1a) (O, P). Plot values represent means ± S.D.; p-values determined by one-way ANOVA followed by multiple comparisons with Dunn test (E, H, L, P). Scale bars: 20 µm. V: ventricle; A: atrium; ap: apical; ba: basal; n: number of embryos.
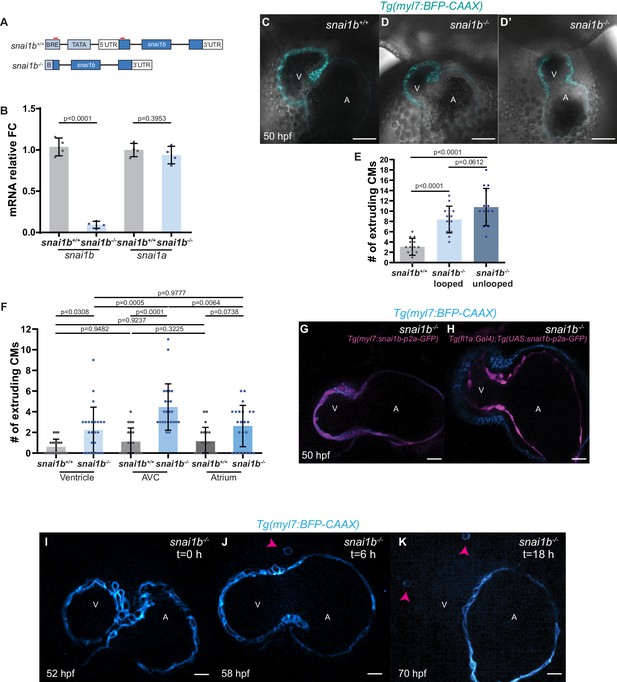
Generation of snai1b mutants.
(A) Schematic of the promoter-less snai1b allele. Two gRNAs (red lines) were used to generate a snai1b mutant allele lacking 1300 bp upstream of the start codon. (B) Relative mRNA levels of snai1b are significantly reduced in snai1b-/- hearts at 48 hpf, whereas snai1a expression levels appear unchanged, indicating lack of transcriptional adaptation by the paralogue; n = 4 biological replicates, 30 embryos each. (C–D’) Single-plane images of 50 hpf snai1b+/+ (C) and snai1b-/- (D–D’) hearts, with 50% of snai1b-/- hearts exhibiting cardiac looping defects (D’). (E) Unlooped and looped snai1b-/- hearts display a similar number of extruding CMs (snai1b+/+, n = 14; looped snai1b-/-, n = 14; unlooped snai1b-/-, n = 14). (F) A higher proportion of the extruding CMs in snai1b-/- hearts are located in the atrioventricular canal compared with the ventricle and atrium (snai1b+/+, n = 20; snai1b-/-, n = 24). (G, H) Single–plane images of 50 hpf snai1b-/- hearts overexpressing snai1b under a myl7 (G) or fli1a (H) promoter. (I, J) Frames from time-lapse imaging of the beating heart of a snai1b-/- embryo to visualize extruding cardiomyocytes (CMs) at 52 (I), 58 (J), and 70 (K) hpf; extruded CMs are found in the pericardial cavity (arrowheads in J and K). Plot values represent means ± S.D.; p-values determined by Student’s t-test (B) or by one-way ANOVA followed by multiple comparisons with Dunn test (E, F). Scale bars: 20 µm (C–D’, G–K). BRE: transcription factor IIB response element; V: ventricle; A: atrium; n: number of embryos; FC: fold change. All Ct values are listed in Supplementary file 2.
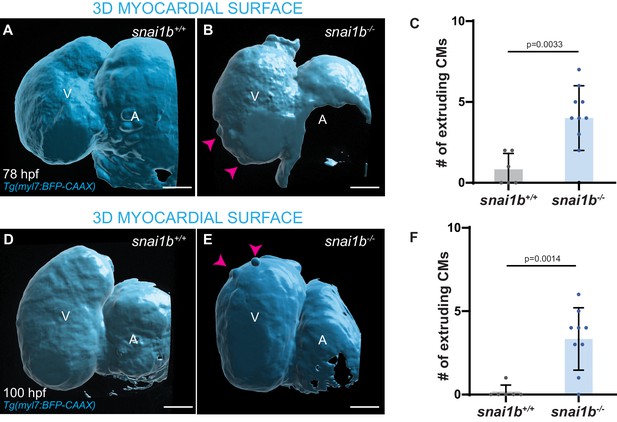
Increased cardiomyocyte (CM) extrusion in snai1b-/- larvae.
(A–F) 3D surface rendering of the heart at 78 and 100 hpf. snai1b-/- larvae (B, E) exhibit more extruding CMs (arrowheads) compared with snai1b+/+ larvae (A, D); quantification (C, F) (C, snai1b+/+, n = 6; snai1b-/-, n = 10; F, snai1b+/+, n = 12; snai1b-/-, n = 10). Plot values represent means ± S.D.; p-values determined by Student’s t-test. Scale bars: 20 µm. V: ventricle; A: atrium; n: number of embryos.
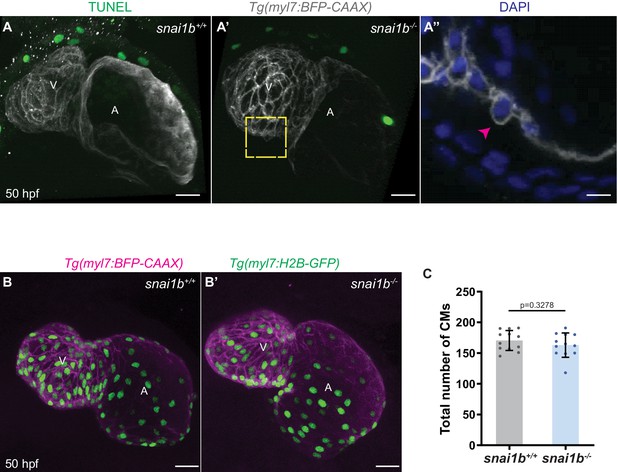
Wild-type like cardiomyocyte (CM) numbers in snai1b mutants.
(A–A”’) TUNEL assay analysis indicates that extruding CMs in snai1b-/- embryos are not apoptotic. Maximum intensity projection of Tg(myl7:BFP-CAAX) snai1b+/+ (A) and snai1b-/- (A’–A”) hearts at 50 hpf. Close-up of extruding CM (arrowhead) labelled with DAPI, but not with TUNEL (A”). (B, B’) Maximum intensity projections of Tg(myl7:H2B-GFP) snai1b+/+ (B) and snai1b-/- (B’) hearts at 52 hpf. (C) The total number of CMs does not change significantly in snai1b-/- hearts (n = 12) compared with snai1b+/+ hearts (n = 12). Plot values represent means ± S.D.; p-value determined by Student’s t-test. Scale bars: 20 µm (A–A’, B–B’); 10 µm (A’’). V: ventricle; A: atrium; n: number of embryos.
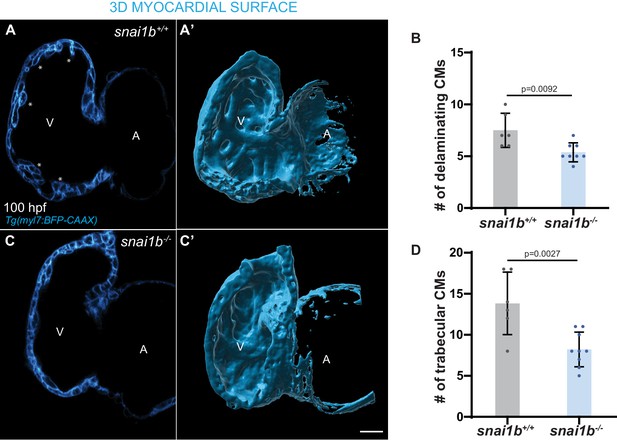
snai1b mutants exhibit reduced cardiac trabeculation.
(A–C’) Single-plane images and inner 3D surface rendering of trabecular cardiomyocytes (CMs) (asterisks in A) in 100 hpf snai1-/- (C, C’) and snai1b+/+ (A, A’) larvae. (B–D) Fewer delaminating (B) and trabecular (D) CMs are observed in snai1b-/- larvae compared with snai1b+/+ larvae at 78 (snai1b+/+, n = 6; snai1b-/-, n = 8) and 100 (snai1b+/+, n = 6; snai1b-/-, n = 9) hpf. Plot values represent means ± S.D.; p-values determined by Student’s t-test. Scale bars: 20 µm. V: ventricle; A: atrium; n: number of embryos.
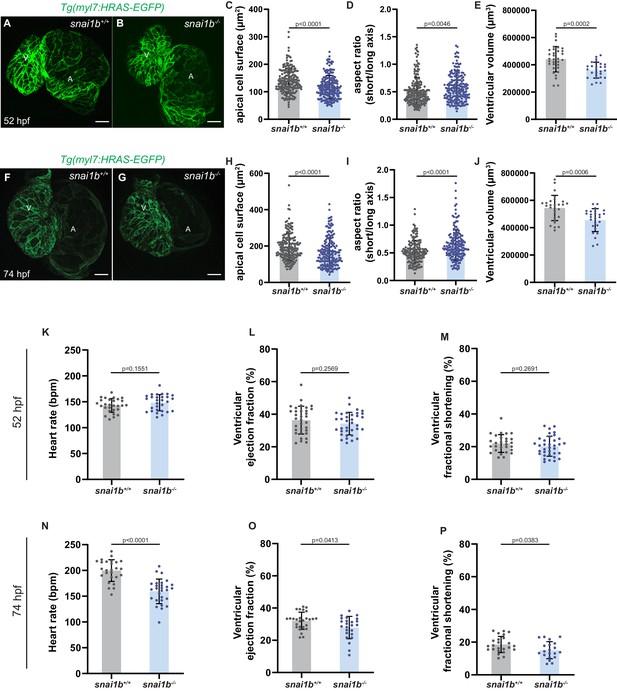
Altered cardiomyocyte (CM) morphology and function in snai1b mutants.
(A, B, F, G) Maximum intensity projection of Tg(myl7:HRAS-EGFP) snai1b+/+ (A, F) and snai1b-/- (B, G) hearts at 52 (A, B) and 74 (F, G) hpf. At both timepoints, snai1b-/- embryos exhibit reduced CM apical cell surface (C, H), increased CM aspect ratio (D, I), and reduced ventricular volume (E, J) compared with snai1b+/+ embryos. (C, H) snai1b+/+, N = 190 at 52 hpf, N = 173 at 74 hpf; snai1b-/-, N = 195 at 52 hpf, N = 178 at 74 hpf. (D, I) snai1b+/+, N = 183 at 52 hpf, N = 162 at 74 hpf; snai1b-/-, N = 188, N = 170 at 74 hpf. (E, J) snai1b+/+, n = 32 at 52 hpf, n = 28 at 74 hpf; snai1b-/-, n = 27 at 52 hpf, n = 26 at 74 hpf. (K–P) snai1b mutants exhibit no differences in heart rate (K), ventricular ejection fraction (L), or ventricular fractional shortening (M) compared with snai1b+/+ embryos at 52 hpf (K, snai1b+/+, n = 28; snai1b-/-, n = 29; L, snai1b+/+, n = 33; snai1b-/-, n = 35; M, snai1b+/+, n = 28; snai1b-/-, n = 35), but exhibit reduced heart rate (N), ventricular ejection fraction (P), and ventricular fractional shortening (O) compared with snai1b+/+ larvae at 74 hpf (N, snai1b+/+, n = 26; snai1b-/-, n = 28; P, snai1b+/+, n = 27; snai1b-/-, n = 24; O, snai1b+/+, n = 24; snai1b-/-, n = 20). Plot values represent means ± S.D.; p-values determined by Mann–Whitney U test. Scale bars: 20 µm. V: ventricle; A: atrium; n: number of embryos; N: number of CMs.
Extruding cardiomyocytes (CMs) in snai1b-/- hearts detach from the myocardium and are visible in the pericardial cavity.
(A) Time-lapse imaging of a Tg(myl7:HRAS-EGFP) snai1b+/+ heart from 52 to 70 hpf. No CMs were observed in the pericardial cavity (three embryos). (B) Time-lapse imaging of a Tg(myl7:HRAS-EGFP) snai1b-/- heart from 52 to 70 hpf. Extruding CMs (arrowheads) detach from the myocardium and can be observed in the pericardial cavity for several hours (three embryos). Scale bars: 20 µm.
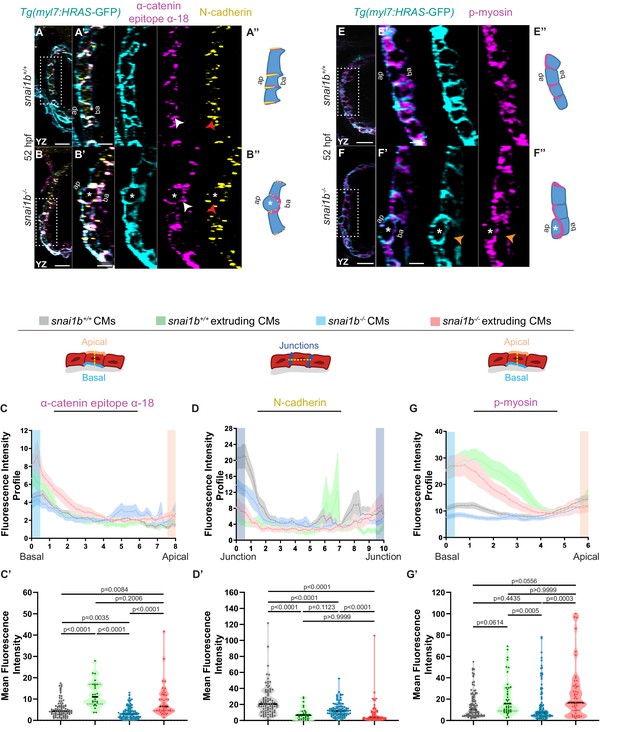
Extruding cardiomyocytes (CMs) exhibit changes in actomyosin components.
(A–B”) Orthogonal projections in the YZ plane of a 52 hpf snai1b+/+ heart (A) immunostained for α-catenin epitope α-18, N-cadherin, and GFP compared with a snai1b-/- sibling heart (B). Close-up of boxed areas of snai1b+/+ (A’) and snai1b-/- (B’) CMs. Schematics illustrate the localization of activated α-catenin (magenta) in the basal domain of extruding CMs in snai1b-/- embryos and defects in N-cadherin (yellow) localization in the junctional domain of snai1b-/- CMs (A”–B”). (C–D’) Fluorescence intensity profile (FIP) (C–D) and mean fluorescence intensity (mFI) (C’–D’) of α-catenin epitope α-18 and N-cadherin immunostaining in 52 hpf snai1b+/+ and snai1b-/- CMs, and in snai1b+/+ and snai1b-/- extruding CMs. The α-catenin epitope α-18 is observed in the basal domain (white arrowhead in B’) of extruding CMs (white asterisks in B’) in snai1b-/- embryos, and a reduction in junctional N-cadherin (red arrowhead in B’) is observed in snai1b-/- CMs. (FIP α-catenin epitope α-18: snai1b+/+ CMs, N = 179; snai1b+/+ extruding CMs, N = 60; snai1b-/- CMs, N = 140; snai1b-/- extruding CMs, N = 54; mFI α-catenin epitope α-18: snai1b+/+ CMs, N = 90; snai1b+/+ extruding CMs, N = 24; snai1b-/- CMs, N = 88; snai1b-/- extruding CMs, N = 44. FIP N-cadherin: snai1b+/+ CMs, N = 90; snai1b+/+ extruding CMs, N = 12; snai1b-/- CMs, N = 98; snai1b-/- extruding CMs, N = 49; mFI N-cadherin: snai1b+/+ CMs, N = 90; snai1b+/+ extruding CMs, N = 25; snai1b-/- CMs, N = 92; snai1b-/- extruding CMs, N = 70.) (E–F”) Representative images of a 52 hpf snai1b-/- heart (F) immunostained for p-myosin and GFP compared with a snai1b+/+ sibling heart (E). Schematics illustrate the basal enrichment of p-myosin (magenta) in extruding CMs in snai1b-/- embryos (E”–F”). (G–G’) FIP (G) and mFI (G’) of p-myosin immunostaining in snai1b+/+ and snai1b-/- CMs, and in snai1b+/+ and snai1b-/- extruding CMs. p-myosin is enriched basally (orange arrowheads in F’) in snai1b-/- extruding CMs in (white asterisks in F’). (FIP p-myosin: snai1b+/+ CMs, N = 204; snai1b+/+ extruding CMs, N = 60; snai1b-/- CMs, N = 140; snai1b-/- extruding CMs, N = 49; mFI p-myosin: snai1b+/+ CMs, N = 100; snai1b+/+ extruding CMs, N = 29; snai1b-/- CMs, N = 153; snai1b-/- extruding CMs, N = 48). Plot values represent means ± S.E.M. (C, D, G). In the violin plots (C’, D’, G’), solid black lines indicate median. p-values determined by Kruskal–Wallis test (C’, D’, G’). Scale bars: 20 µm (A, B, E, F); 2 µm (A’, B’, E’, F’). ap: apical; ba: basal; N: number of CMs. See also Figure 2—figure supplement 1.
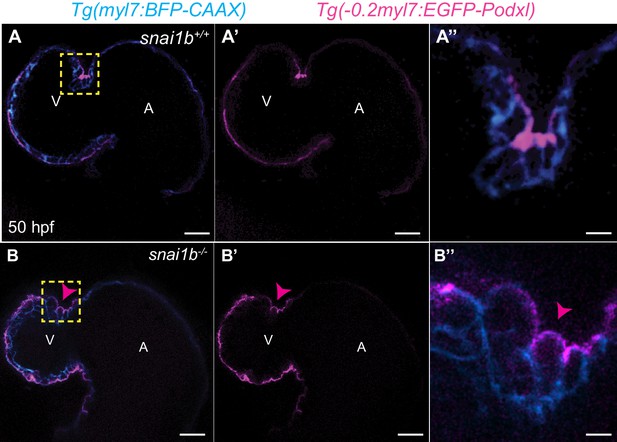
Apicobasal polarity is maintained in extruding cardiomyocytes (CMs) in snai1b mutants.
(A–B’’) Single-plane images of Tg(myl7:BFP-CAAX); Tg(−0.2myl7:EGFP-podocalyxin) snai1b+/+ (A–A’) and snai1b-/- (B–B’) hearts at 50 hpf. Close-up of boxed areas (A’’, B’’). No changes in the localization of the apical marker Podocalyxin are observed in extruding CMs (arrowheads in B–B’’). Scale bars: 20 µm (A–A’, B–B’), 2 µm (A’’, B’’). V: ventricle; A: atrium.
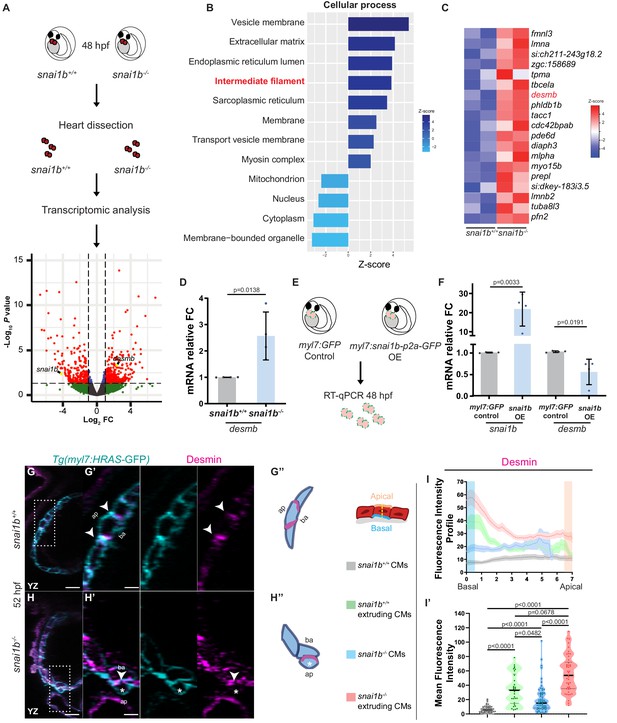
Transcriptomic analysis reveals upregulation of intermediate filament genes in snai1b-/- hearts.
(A) RNA extracted from 48 hpf snai1b+/+ and snai1b-/- hearts was used for transcriptomic analysis. (B) GO analysis of cellular processes shows enrichment of intermediate filament components in snai1b-/- hearts. (C) Heatmap of a list of upregulated cytoskeletal genes, including desmb. (D) Relative mRNA levels of desmb are significantly increased in snai1b-/- hearts at 48 hpf; n = 4 biological replicates, 30 hearts each. (E) Schematic of snai1b overexpression under a myl7 promoter; snai1b and desmb mRNA levels analysed at 48 hpf. (F) Relative mRNA levels of desmb are significantly reduced in snai1b cardiomyocyte (CM)-specific overexpressing hearts at 48 hpf; n = 4 biological replicates, 30 hearts each. (G–H”) Orthogonal projections in the YZ plane of a 52 hpf snai1b-/- heart (H) immunostained for Desmin and membrane GFP compared with a snai1b+/+ heart (G). Close-up of boxed areas of snai1b+/+ (G’) and snai1b-/- (H’) CMs. Schematics (Desmin in magenta) illustrate the basal enrichment of Desmin in extruding CMs in snai1b-/- embryos (G”–H”). (I–I’) Fluorescence intensity profile (FIP) (I) and mean fluorescence intensity (mFI) (I’) of Desmin in snai1b+/+ and snai1b-/- CMs, and in snai1b+/+ and snai1b-/- extruding CMs. Desmin immunostaining is observed throughout the snai1b-/- myocardium, with an enrichment in the basal domain (white arrowheads in H’–G’) in extruding CMs (white asterisks in H’). (FIP: snai1b+/+ CMs, N = 49; snai1b+/+ extruding CMs, N = 41; snai1b-/- CMs, N = 45; snai1b-/- extruding CMs, N = 41; mFI: snai1b+/+ CMs, N = 56; snai1b+/+ extruding CMs, N = 30; snai1b-/- CMs, N = 65; snai1b-/- extruding CMs, N = 46). Plot values represent means ± S.D. (D, F) or mean ± S.E.M. (I). In the violin plot (I’), solid black lines indicate median. p-Values determined by Student’s t-test (D, F) or Kruskal–Wallis test (I’). Scale bars: 20 µm (G, H); 2 µm (G’, H’). ap: apical; ba: basal; n: number of embryos; N: number of CMs; FC: fold change. All Ct values are listed in Supplementary file 2. See also Figure 3—figure supplement 1.
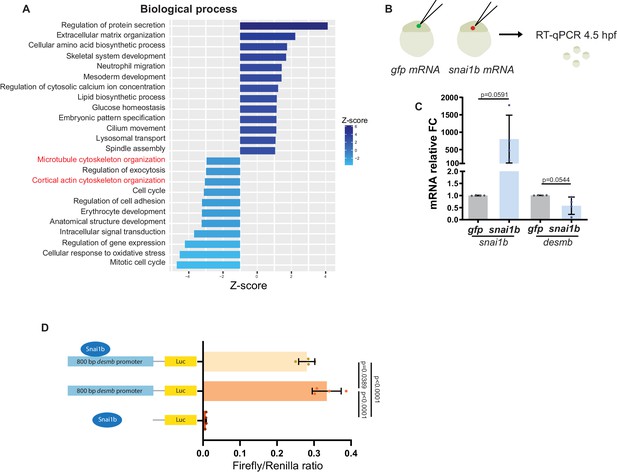
Snai1b regulates desmb expression.
(A) GO analysis of biological processes shows enrichment of cytoskeletal related genes (in red) in snai1b-/- hearts compared with snai1b+/+ hearts. (B, C) Schematic of gfp and snai1b mRNA injections into one-cell stage embryos. snai1b and desmb mRNA levels were analysed in injected embryos at 4.5 hpf. Relative mRNA levels of snai1b are significantly increased, whereas desmb mRNA levels are decreased at 4.5 hpf. n = 4 biological replicates, 30 embryos each. (D) In vitro luciferase assay in HEK293T cells after co-transfection of pGL4.14-luciferase and pGL4.14–800 bp desmb-luciferase with pTol2-CMV-snai1b or alone. Reduced luciferase activity was detected when pTol2-CMW-snai1b was co-transfected with pGL4.14–800 bp desmb-luciferase. Plot values represent means ± S.D.; p-values determined by Student’s t-test (C) or one-way ANOVA followed by multiple comparisons with Dunn test (D). FC: fold change. All Ct values are listed in Supplementary file 2.
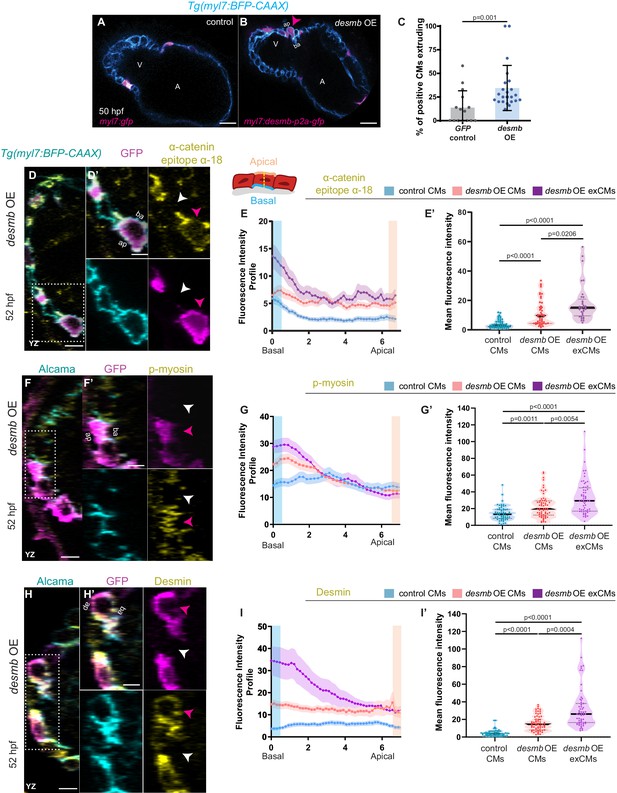
desmb overexpression in cardiomyocytes (CMs) induces their extrusion.
(A, B) Single-plane images of snai1b+/+ embryos injected with myl7:GFP (A) or with myl7:desmb-p2a-GFP (B) at 50 hpf. (C) A higher percentage of CMs extrude when overexpressing desmb (n = 23) compared with control (n = 15) (magenta arrowheads in B, B’). (D–D’, F–F’, H–H’) Orthogonal projections in the YZ plane of hearts from 52 hpf embryos injected with myl7:desmb-p2a-GFP and immunostained for α-catenin epitope α-18, GFP, and BFP (D–D’), p-myosin, GFP, and Alcama (F–F’), or Desmin, GFP, and Alcama (H–H’). Close-up of boxed areas of desmb-overexpressing and adjacent wild-type CMs (D’, F’, H’). (E–E’, G–G’, I–I’) Fluorescence intensity profile (FIP) (E, G, I) and mean fluorescence intensity (mFI) (E’, G’, I’) of α-catenin epitope α-18 (E–E’), p-myosin (G–G’), and Desmin (I–I’) in CMs that overexpress desmb (magenta arrowheads) and CMs that do not overexpress desmb (white arrowheads). α-catenin epitope α-18, p-myosin, and Desmin immunostaining signals are enriched in the basal domain in desmb overexpressing CMs. FIP: desmb OE CMs, (E) N = 132, (G) N = 120, (I) N = 88; desmb OE extruding CMs, (E) N = 47, (G) N = 54, (I) N = 49; control CMs, (E) N = 153, (G) N = 133, (I) N = 86. mFI: desmb OE CMs, (E’) N = 49, (G’) N = 65, (I’) N = 63; desmb OE extruding CMs, (E’) N = 33, (G’) N = 60, (I’) N = 59; control CMs, (E’) N = 55, (G’) N = 62, (I’) N = 64. Plot values represent means ± S.D. (C) or means ± S.E.M. (E, G, I). In the violin plots (E’, G’, I’), solid black lines indicate median. p-values determined by Mann–Whitney U (C) or Kruskal–Wallis (E’, G’, I’) test. Scale bars: 20 µm (A–B’, D, F, H); 2 µm (D’, F’, H’). V: ventricle; A: atrium; ap: apical; ba: basal; n: number of embryos; N: number of CMs. See also Figure 4—figure supplements 1–3.
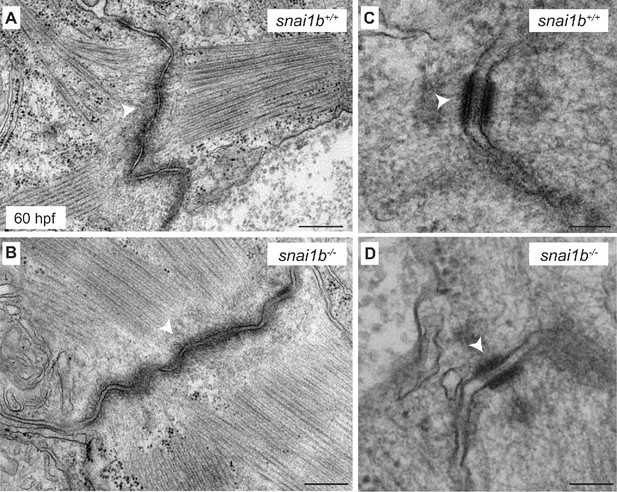
Desmosome organization is maintained in snai1b mutants.
(A–D) Transmission electron microscopy (TEM) images showing the ultrastructure of fascia adherens (white arrowheads in A, B) and desmosomes (white arrowheads in C, D) in 60 hpf snai1b+/+ and snai1b-/- cardiomyocytes. Scale bars: 200 nm (A, B); 500 nm (C, D).
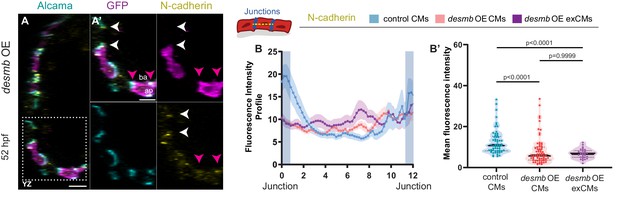
desmb overexpression in cardiomyocytes (CMs) leads to reduced junctional N-cadherin.
(A, A’) Orthogonal projection in the YZ plane of the heart of an embryo injected with myl7:desmb-p2a-GFP and immunostained for N-cadherin, GFP, and Alcama at 52 hpf. Close-up of boxed area of desmb-overexpressing and adjacent control CMs (A’). (B, B’) Fluorescence intensity profile (FIP) (B) and mean fluorescence intensity (mFI) (B’) of N-cadherin in CMs that overexpress desmb (magenta arrowheads) and CMs that do not overexpress desmb (white arrowheads) CMs. N-cadherin immunostaining is reduced in the junctional domains in desmb overexpressing CMs. (FIP: desmb OE CMs, N = 118; desmb OE extruding CMs, N = 44; control CMs, N = 95; mFI: desmb OE CMs, N = 57; desmb OE extruding CMs, N = 30; control CMs, N = 61). Plot values represent means ± S.E.M. In the violin plot, solid black lines indicate median. p-Values determined by Kruskal–Wallis test. Scale bars: 20 µm (A); 2 µm (A’). ap: apical; ba: basal; N: number of CMs.
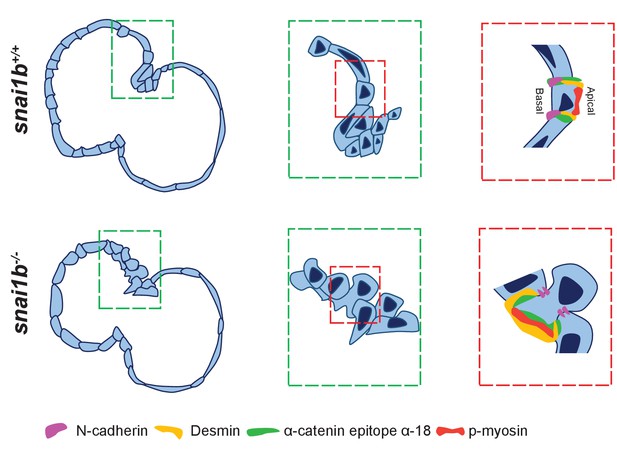
Model.
Loss of snai1b leads to basal enrichment of Desmin and the actomyosin machinery, and causes an increased incidence of cardiomyocyte extrusion away from the lumen.
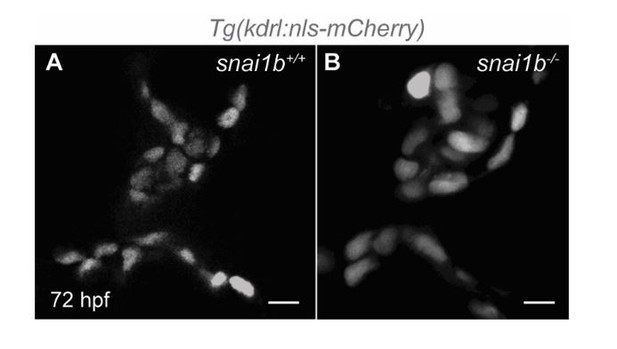
Single-plane images of Tg(kdrl:nls-mCherry)snai1b+/+ (A) and snai1b-/- (B) valve leaflets at 72 hpf.
(snai1b+/+, n=9; snai1b-/-, n=11). Scale bars: 20 µm. n, number of embryos.
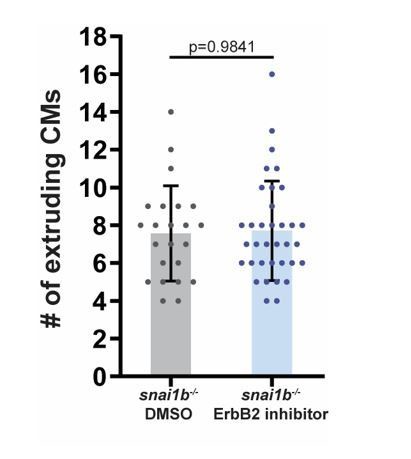
The number of extruding CMs is not significantly different between DMSO and ErbB2 inhibitor treated snai1b-/- embryos at 52 hpf.
(snai1b-/- DMSO, n=23; snai1b-/- ErbB2 inhibitor, n=35). Plot values represent means ± S.D.; p-values determined by Mann-Whitney U test. n, number of embryos.
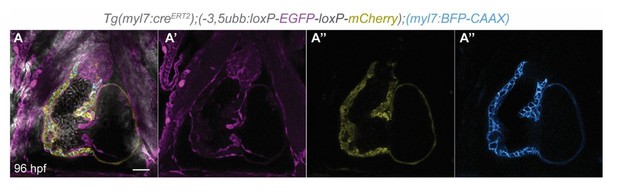
Single-plane images of Tg(myl7:creERT2);(-3.5ubb:loxP-EGFP-loxP-mCherry);(myl7:BFP-CAAX) larvae at 96 hpf.
No switched cells are found in the pericardial cavity. n=23. Scale bar: 20 µm. n, number of embryos.
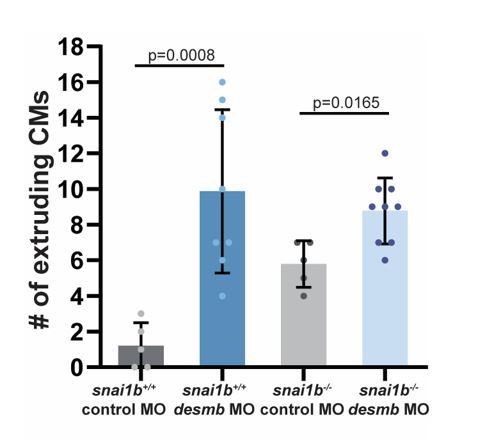
The number of extruding CMs increases in snai1b+/+ and snai1b-/-desmb morphants compared with the respective control morphants at 52 hpf.
(snai1b+/+ control MO, n=5; snai1b+/+ desmb MO, n=8; snai1b-/- Control MO, n=5; snai1b-/- desmb MO, n=9). Plot values represent means ± S.D.; p-values determined by Mann-Whitney U test. n, number of embryos..
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-tRFP (rabbit polyclonal) | Evrogen | RRID:AB_2571743 | IF(1:200) |
| Antibody | Anti-GFP (chicken polyclonal) | AvesLab | RRID:AB_10000240 | IF(1:800) |
| Antibody | Anti-N-cadherin (rabbit polyclonal) | Abcam | RRID:AB_444317 | IF(1:250) |
| Antibody | Anti-p-myosin (rabbit polyclonal) | Abcam | RRID:AB_303094 | IF(1:200) |
| Antibody | Anti-Desmin (rabbit polyclonal) | Sigma | RRID:AB_476910 | IF(1:100) |
| Antibody | Anti-α-catenin epitope α-18 (rat monoclonal) | Gift from Prof. Akira Nagafuchi | IF(1:300) | |
| Antibody | Anti-Alcama (mouse monoclonal) | DSHB | RRID:AB_531904 | IF(1:50) |
| Antibody | Alexa Fluor 488 Goat anti Chicken IgG (H + L) | Thermo Fisher Scientific | RRID:AB_142924 | IF(1:500) |
| Antibody | Alexa Fluor 647 Goat anti Rabbit IgG (H + L) | Thermo Fisher Scientific | RRID:AB_141663 | IF(1:500) |
| Antibody | Alexa Fluor 647 Goat anti Rat IgG (H + L) | Thermo Fisher Scientific | RRID:AB_141778 | IF(1:500) |
| Antibody | Alexa Fluor 568 Goat anti Rabbit IgG (H + L) | Thermo Fisher Scientific | RRID:AB_2534123 | IF(1:500) |
| Antibody | Alexa Fluor 568 Goat anti Rat IgG (H + L) | Thermo Fisher Scientific | RRID:AB_2534121 | IF(1:500) |
| Chemical compound, drug | Agarose, low gelling temperature | Sigma | Cat# A9414-25g | |
| Chemical compound, drug | Bovine serum albumin | Sigma | Cat# A-9418 | |
| Chemical compound, drug | Chloroform | Merck | Cat# 102445 | |
| Other | DAPI | Sigma | Cat# D954 | (1 µg/mL) |
| Chemical compound, drug | Dimethyl sulfoxide (DMSO) | Sigma | Cat# D8418 | |
| Chemical compound, drug | DMEM(1X)+Glutamax | Thermo Fisher Scientific | Cat# 31966-021 | |
| Chemical compound, drug | DyNAmo ColorFlash SYBR Green qPCR Mix | Thermo Fisher Scientific | Cat# F416S | |
| Chemical compound, drug | Ethanol, undenatured, absolute | Serva | Cat# 11093.01 | |
| Chemical compound, drug | FBS superior | Biochrom | Cat# S0615 | |
| Chemical compound, drug | Glycine | Sigma | Cat# 50046 | |
| Chemical compound, drug | 2-Propanol | Roth | Cat# 6752.4 | |
| Chemical compound, drug | Lipofectamine 3000 Transfection Reagent | Thermo Fisher Scientific | L3000001 | |
| Chemical compound, drug | Methanol | Roth | Cat# 4627.5 | |
| Chemical compound, drug | Normal Goat Serum | Thermo Fisher Scientific | Cat# 16210072 | |
| Chemical compound, drug | Paraformaldehyde | Sigma | Cat# P6148 | |
| Chemical compound, drug | Phosphate buffered saline (PBS) | Sigma | Cat# P4417 | |
| Recombinant DNA reagent | pT3TS-nCas9n (plasmid) | Addgene | Cat# 46757 | |
| Recombinant DNA reagent | pCS2z vector (plasmid) | Addgene | Cat# 62214 | |
| Recombinant DNA reagent | pCMV-Tol2 (plasmid) | Addgene | Cat# 31823 | |
| Recombinant DNA reagent | pGl4.14-luc; SV40:hRLuc (plasmid) | Bensimon-Brito et al., 2020 | ||
| Chemical compound, drug | Sodium citrate monobasic | Sigma | Cat# 71497-1KG | |
| Chemical compound, drug | Triton X-100 | Sigma | Cat# X-100 | |
| Chemical compound, drug | TRIzol Reagent | Thermo Fisher Scientific | Cat# 15596026 | |
| Chemical compound, drug | Tween 20 | Sigma | Cat# P1379 | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay System | Promega | Cat# E1910 | |
| Commercial assay or kit | In Situ Cell Death Detection Kit, Fluorescein | Roche | 11684795910 | |
| Commercial assay or kit | Maxima First Strand cDNA kit | Thermo Fisher Scientific | Cat# K1641 | |
| Commercial assay or kit | MegaShortScript T7 Transcription Kit | Thermo Fisher Scientific | Cat# AM1354 | |
| Commercial assay or kit | MegaScript T3 Transcription Kit | Thermo Fisher Scientific | Cat# AM1348 | |
| Commercial assay or kit | mMESSAGE mMACHINE T7 Transcription Kit | Thermo Fisher Scientific | Cat# AM1344 | |
| Commercial assay or kit | mMESSAGE mMACHINE T3 Transcription Kit | Thermo Fisher Scientific | Cat# AM1348 | |
| Commercial assay or kit | RNA Clean and Concentrator Kit | Zymo Research | Cat# R1013 | |
| Cell line (Homo sapiens) | HEK-293T | ATCC | Cat# CRL-3216 | RRID:CVCL_0063 |
| Strain, strain background (Danio rerio) | Tg(myl7:BFP- CAAX)bns193 | Guerra et al., 2018 | ZFIN:bns193 | |
| Strain, strain background (Danio rerio) | Tg(myl7:H2B-EGFP)zf521Tg | Mickoleit et al., 2014 | ZFIN:zf521Tg | |
| Strain, strain background (Danio rerio) | Tg(myl7:mVenus-gmnn)ncv43Tg | Jiménez-Amilburu et al., 2016 | ZFIN:ncv43Tg | |
| Strain, strain background (Danio rerio) | Tg(−0.2myl7:snai1b-p2a-GFP) bns555 | This paper | ZFIN:bns555 | |
| Strain, strain background (Danio rerio) | Tg(−0.2myl7:EGFP-podocalyxin) bns10 | Jiménez-Amilburu et al., 2016 | ZFIN:bns10 | |
| Strain, strain background (Danio rerio) | Tg(fli1a:Gal4)ubs4 | Zygmunt et al., 2011 | ZFIN:ubs4 | |
| Strain, strain background (Danio rerio) | Tg(UAS:snai1b-p2a-GFP) bns442 | This paper | ZFIN:bns442 | |
| Strain, strain background (Danio rerio) | Tg(myl7:EGFP-Hsa.HRAS)s883Tg | D'Amico et al., 2007 | ZFIN:s883Tg | |
| Strain, strain background (Danio rerio) | snai1bbn351 mutant | This paper | ZFIN:bns351 | |
| Sequence-based reagent | qPCR | This paper | Table S1 | |
| Sequence-based reagent | Genotyping | This paper | Table S1 | |
| Sequence-based reagent | PCR | This paper | Table S1 | |
| Software, algorithm | FiJi ImageJ 1.53 c | Schindelin et al., 2012 | RRID:SCR_002285 | |
| Software, algorithm | GraphPad Prism 6 | GraphPad | RRID:SCR_002798 | |
| Software, algorithm | Imaris, version 8.4.0 | Bitplane | RRID:SCR_007370 | |
| Software, algorithm | Zen Digital Imaging | Carl Zeiss Microscopy | RRID:SCR_013672 |
Additional files
-
Supplementary file 1
Table of primers.
- https://cdn.elifesciences.org/articles/66143/elife-66143-supp1-v1.docx
-
Supplementary file 2
Table of Ct values from RT-qPCR experiments.
- https://cdn.elifesciences.org/articles/66143/elife-66143-supp2-v1.xlsx
-
Supplementary file 3
Luciferase assay raw values.
- https://cdn.elifesciences.org/articles/66143/elife-66143-supp3-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66143/elife-66143-transrepform-v1.pdf





