Evolutionarily conserved sperm factors, DCST1 and DCST2, are required for gamete fusion
Figures
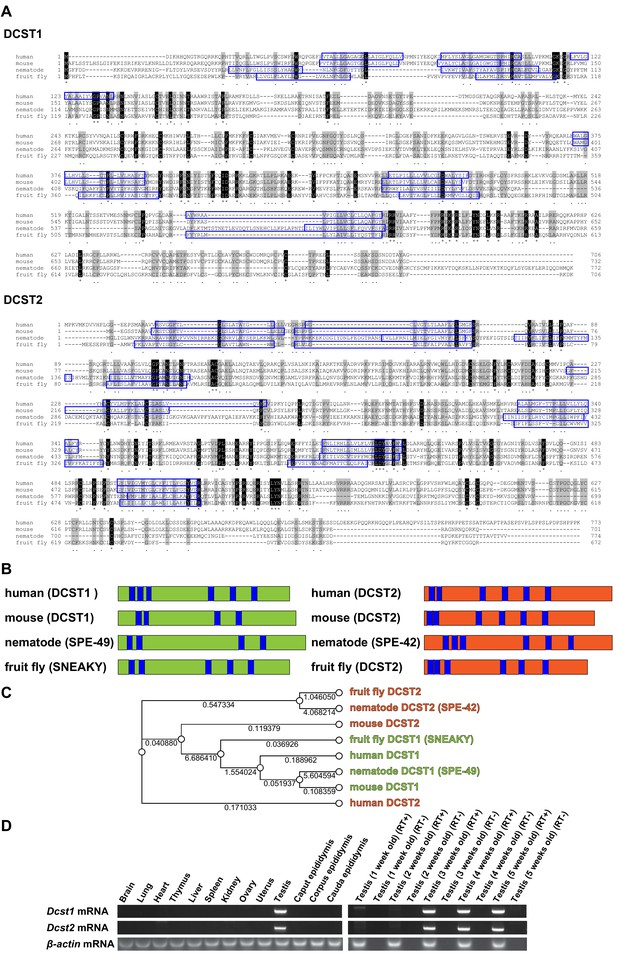
Alignments of dendrocyte expressed seven transmembrane protein domain-containing 1 (DCST1) and dendrocyte expressed seven transmembrane protein domain-containing 2 (DCST2) amino acid sequences, and mRNA expression profiles.
(A) Sequence alignments of DCST1 in Homo sapiens (human; GenBank: NP_689707.2), Mus musculus (mouse; GenBank: NP_084250.1), Caenorhabditis elegans (nematode; GenBank: NP_001343858.1), and Drosophila guanche (fruit fly; GenBank: XP_034122086.1) are shown. Sequence alignments of DCST2 in Homo sapiens (human; GenBank: NP_653223.2), Mus musculus (mouse; GenBank: NP_001357782.1), Caenorhabditis elegans (nematode; GenBank: NP_872213.4), and Drosophila mauritiana (fruit fly; GenBank: XP_033160419.1) are shown. Identical and similar amino acid residues are represented as asterisks and dots, respectively. The residues of putative transmembrane domains are indicated by blue boxes. (B) Schematic diagram of DCST1 and DCST2 in various species. The blue area indicates the putative transmembrane domain. (C) Phylogeny analysis of DCST1 and DCST2. Maximum likelihood phylogenetic tree was constructed using RAxML with 1000 bootstraps through Genetyx software. The distance is shown as a numerical value. (D) Tissue and age-dependent expression profile of Dcst1 and Dcst2 mRNA in mice. β-actin mRNA was used as an internal control.
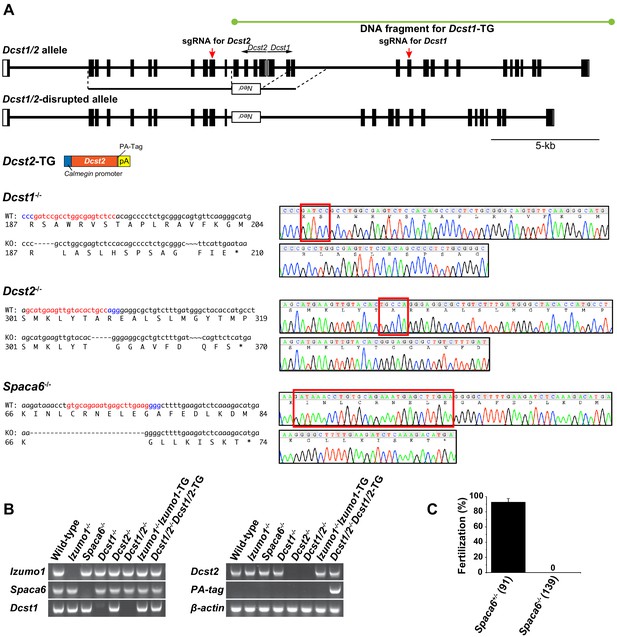
Strategy for Dcst1 and Dcst2 genes disruption and its validation.
(A) Targeted disruption of the Dcst1/2, Dcst1, Dcst2, and Spaca6 genes, and constructs for Dcst1 and Dcst2 transgenic mice. Complete structures of the wild-type mouse Dcst1/2 alleles are shown. Exons and introns are represented by boxes and horizontal lines, respectively. An open reading frame for Dcst1 and Dcst2 genes is shown in black. For the targeted disruption of mouse Dcst1/2, the neomycin-resistance gene (Neor) was inserted between Dcst1 exon 3 and Dcst2 exon 4. A herpes simplex virus thymidine kinase gene was introduced into the targeting construct for negative selection. For the gene complementation experiments, transgenic mouse lines expressing dendrocyte expressed seven transmembrane protein domain-containing 1 (DCST1), in which full-length genome DNA fragment including all promoter region, exons, and introns was incorporated, and dendrocyte expressed seven transmembrane protein domain-containing 2 (DCST2), in which full-length cDNA with a PA-tag sequence was inserted between a testis-specific (pachytene spermatocyte to spermatid stage) Calmegin promoter and rabbit β-globin polyadenylation signal, were produced. For genome-editing of Dcst1, Dcst2, and Spaca6 using the CRISPR/Cas9 system, the red and blue letters show the target and protospacer adjacent motif (PAM) sequence, respectively (left panels). A DNA sequence chromatogram shows that Dcst1-/-, Dcst2-/-, and Spaca6-/- had 5-base, 5-base, and 28-base deletions, respectively. The deleted sequences are boxed in red (right panels). (B) Validation of gene disruption by reverse transcription polymerase chain reaction (RT-PCR). All primer sets employed wild-type mRNA-specific oligonucleotide sequences except for PA-tag. As a result, all gene disruptions and transgenes were confirmed appropriately. β-actin mRNA was used as an internal control. (C) In vitro fertilization analysis using Spaca6+/- and Spaca6-/- spermatozoa (n = 4 and 4, respectively). The fertilization rate was evaluated at the two-cell stage embryo. The numbers in parentheses indicate the numbers of oocytes used. The error bars represent standard error of the mean (s.e.m.).
-
Figure 2—source data 1
IVF in SPACA6 heterozygous and homozygous KO spermatozoa.
- https://cdn.elifesciences.org/articles/66313/elife-66313-fig2-data1-v1.xlsx
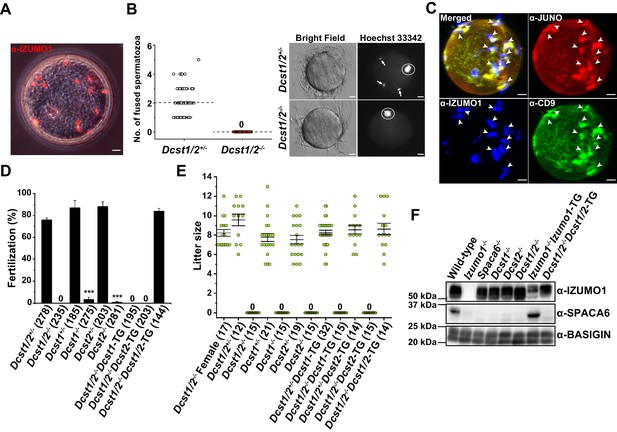
Identification of new gamete fusion-related factors dendrocyte expressed seven transmembrane protein domain-containing 1 (DCST1) and dendrocyte expressed seven transmembrane protein domain-containing 2 (DCST2).
(A) Accumulation of many acrosome-reacted (AR) IZUMO1 positive spermatozoa (red) in the perivitelline space of eggs recovered from the oviducts of females 8 hr after coitus with a Dcst1/2-/- male. Scale bar 10 µm. (B) Gamete fusion assay of Dcst1/2-disrupted spermatozoa. Gamete fusion was visualized with Hoechst 33342 dye transfer. The numbers of fused spermatozoa per oocyte from four independent experiments are shown. The total numbers of oocytes examined in Dcst1/2+/- and Dcst1/2-/- were 77 and 80, respectively (left graph). The broken lines indicate the average. The arrows and the circles in the representative photo show fused spermatozoa and meiosis metaphase II chromosomes, respectively (right picture). Scale bar 10 µm. (C) Localization of oocyte CD9 and JUNO upon Dcst1/2-/- sperm attachment. Sperm–oocyte interaction was visualized by staining with 0.25 µg ml−1 TH6–Alexa647 (JUNO: red), 0.5 µg ml−1 MZ3–FITC (CD9: green) and 0.5 µg ml−1 Mab125–Alexa546 (IZUMO1: blue). The eggs were fixed 30 min after insemination of Dcst1/2-/- spermatozoa. The arrowheads indicate the sites where JUNO and cluster of differentiation 9 (CD9) were concentrated in response to the AR spermatozoa (IZUMO1 positive: blue) bound to the oolemma. Scale bar 10 µm. (D) In vitro fertilization assay using Dcst1/2+/-, Dcst1/2-/-, Dcst1+/-, Dcst1-/-, Dcst2+/-, Dcst2-/-, Dcst1/2-/-Dcst1-TG, Dcst1/2-/-Dcst2-TG, and Dcst1/2-/-Dcst1/2-TG spermatozoa (n = 5, 5, 6, 7, 8, 8, 6, 6, and 6, respectively). The fertilization rate was evaluated at the two-cell stage embryo. The numbers in parentheses indicate the numbers of oocytes used. The error bars represent s.e.m. ***p<0.001 (paired two-tailed Student’s t-test, compared with each heterozygous mutants). (E) Fecundity of DCST1/2-related deficient male and female mice. The numbers in parentheses indicate the numbers of mating pairs. Regarding Dcst1/2-/-, Dcst1-/-, Dcst2-/-, Dcst1/2-/-Dcst1-TG, and Dcst1/2-/-Dcst2-TG males, the numbers of vaginal plug formations were counted. The error bars represent s.e.m. (F) Western blot analysis of various types of mouse lines with 1 µg ml−1 Mab18 (IZUMO1) and 1:500 SPACA6 anti-serum. Each lane was equally applied with 20 µg sperm lysates. BASIGIN was used as a loading control.
-
Figure 3—source data 1
Gamete fusion assay, IVF and litter size in DCST1/2-related mutants.
- https://cdn.elifesciences.org/articles/66313/elife-66313-fig3-data1-v1.xlsx
Videos
6 hr after insemination of Dcst1/2-disrupted spermatozoa.
The first and second half of the video show Dcst1/2 heterozygous and homozygous KO spermatozoa insemination, respectively.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | Dcst1/2 KO | This article | Deposited in RIKEN BRC (ID RBRC05733) | |
| Strain, strain background (Mus musculus) | Dcst1 KO | This article | Deposited in RIKEN BRC (ID RBRC10275) | |
| Strain, strain background (Mus musculus) | Dcst2 KO | This article | Deposited in RIKEN BRC (ID RBRC10366) | |
| Strain, strain background (Mus musculus) | Spaca6 KO | This article | Deposited in RIKEN BRC (ID RBRC10367) | |
| Strain, strain background (Mus musculus) | Izumo1 KO | Inoue et al., 2005 | RRID MGI_3576518 | |
| Strain, strain background (Mus musculus) | Dcst1 TG | This article | Deposited in RIKEN BRC (ID RBRC05856) | |
| Strain, strain background (Mus musculus) | Dcst2 TG | This article | ||
| Strain, strain background (Mus musculus) | Izumo1 TG | Inoue et al., 2005 | ||
| Antibody | Anti-mouse SPACA6 (rabbit polyclonal) | This article | (WB: 1:500) | |
| Antibody | Anti-mouse IZUMO1 (rat monoclonal) | Inoue et al., 2013 | Mab125 | Gift from Dr. Ikawa (IF: 0.5 µg ml−1) |
| Antibody | Anti-mouse IZUMO1 (rat monoclonal) | Inoue et al., 2015 | Mab18 | (WB: 1 µg ml−1) |
| Antibody | Anti-mouse FR4 (JUNO) (rat monoclonal) | BioLegend | TH6, RRID AB_1027724 | (IF: 0.25 µg ml−1) |
| Antibody | Anti-mouse CD9 (rat monoclonal) | BioLegend | MZ3, RRID AB_1279321 | (IF: 0.5 µg ml−1) |
| Antibody | Anti-mouse EMMPRIN (BASIGIN) (goat polyclonal) | Santa Cruz Biotechnology | G-19, RRID AB_2066959 | (WB: 0.2 µg ml−1) |
| Recombinant DNA reagent | Plasmid pX330 vector | Addgene | RRID Addgene_42230 | |
| Sequence-based reagent | Primers | This article | Detailed in 'Materials and methods' | |
| Peptide, recombinant protein | SPACA646-139 | This article | Detailed in 'Materials and methods' |





