Biosynthesis of a sulfated exopolysaccharide, synechan, and bloom formation in the model cyanobacterium Synechocystis sp. strain PCC 6803
Figures
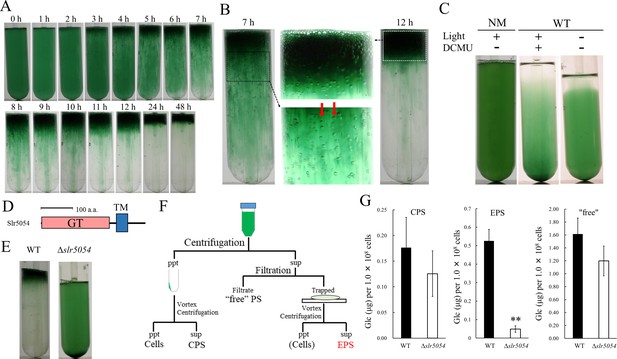
Bloom formation and exopolysaccharides (EPS) isolation.
(A) Time course of bloom formation by wild-type (WT) Synechocystis 6803 during the second step of culture. Extracellular gas bubbles are formed and trapped in viscous EPS (~1 hr). Green vertical columns with bubbles become apparent at 4 hr. Those trapped gas bubbles slowly rise together with the viscous columns. (B) Enlarged images showing gas bubbles trapped in EPS. Vertically aligned bubbles are indicated by red arrows. (C) Lack of bloom formation in the non-motile substrain (NM) or WT with or without light and the photosynthesis inhibitor DCMU at 48 hr of the second step of culture. (D) Domain architecture of Slr5054. GT, glycosyltransferase domain; TM, transmembrane region. (E) Lack of bloom formation in Δslr5054 after standing culture for 48 hr. (F) Isolation of EPS from the first step of culture. Cells and capsular polysaccharides (CPS) were removed from the culture by centrifugation, and EPS in the supernatant was separated from ‘free’ polysaccharide (PS) by membrane filtration followed by a second centrifugation to remove residual cells. CPS was collected from the cell pellet after vortexing and centrifugation. (G) Sugar content of fractions from WT and Δslr5054. Error bars represent SD (CPS, n = 6; others, n = 3; **p<0.005).
-
Figure 1—source data 1
Raw data of sugar quantification in wild type (WT) and Δslr5054.
- https://cdn.elifesciences.org/articles/66538/elife-66538-fig1-data1-v1.xlsx
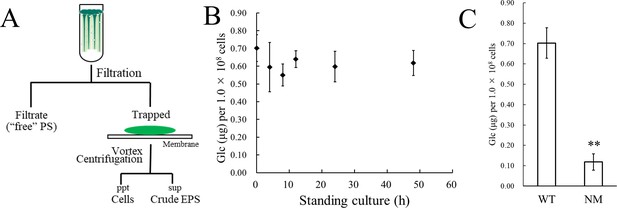
Isolation of crude exopolysaccharides (EPS) and sugar analysis.
(A) Protocol for isolating crude EPS from the bloomed culture. The bloom, including EPS and cells at the end of the second (standing) step of culture, was trapped by membrane filtration, and crude EPS was recovered from the bloom by vortexing and centrifugation. PS, polysaccharide; ppt, precipitate; sup, supernatant. (B) Time course of sugar accumulation in the crude EPS during the standing culture. (C) Sugar content of the crude EPS from WT, and a non-motile glucose-tolerant substrain (NM). Error bars represent SD (n = 3, **p<0.005).
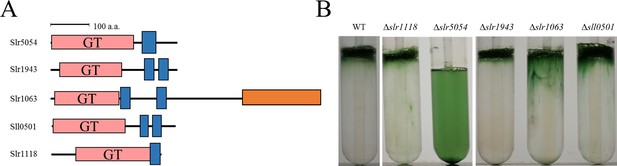
Bloom formation by several glycosyltransferase mutants.
(A) Domain architecture of membrane-bound glycosyltransferases. Red box, glycosyltransferase domain (GT); blue box, transmembrane region; orange box, glycogen phosphorylase domain. (B) Bloom formation by mutants after the second step of culture.
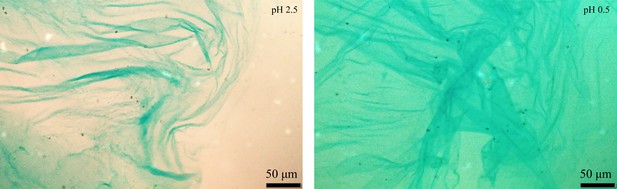
Alcian blue staining of isolated exopolysaccharides (EPS) from wild type (WT).
The microscopic images of isolated EPS from WT culture (Figure 1g) stained with alcian blue at pH 2.5 (left) and pH 0.5 (right).
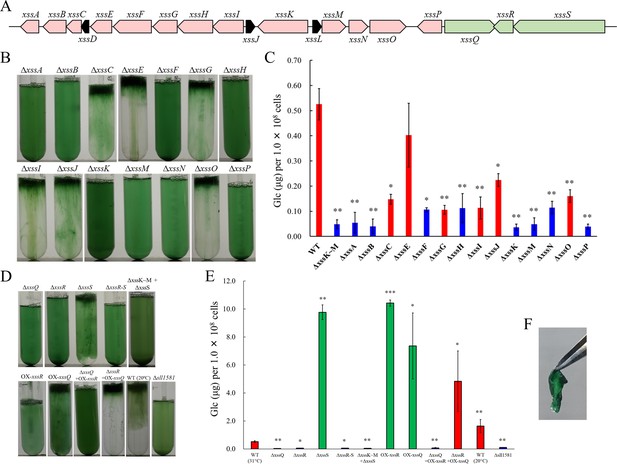
The extracellular sulfated polysaccharide biosynthesis (xss) gene cluster and phenotype of xss mutants.
(A) The xss gene cluster. Red, polysaccharide biosynthesis genes; green, regulatory genes; black, genes of unknown function. (B) Bloom formation by the mutants carrying disruptions in the polysaccharide biosynthesis xss genes. (C) Total sugar content (µg glucose per 1 × 108 cells) of the exopolysaccharides (EPS) fraction from mutants in b. Red bars, bloom-forming mutants; blue bars, non-bloom-forming mutants. Error bars represent SD (wild type [WT] grown at 20°C, n = 6; others, n = 3). Statistical significance was determined using Welch’s t test (*p<0.05, **p<0.005, ***p<0.0005). (D) Bloom formation by regulatory mutants, WT grown at 20°C, and outer-membrane polysaccharide export protein (OPX) mutant (∆sll1581). (E) Total sugar content of the EPS fraction from mutants in d. Red bars, bloom-forming mutants; green bars, excess-bloom-forming mutants. (F) A sheet of OX-xssR cells was stripped off from the agar plate by tweezers. The culture temperature was 31°C unless otherwise stated.
-
Figure 2—source data 1
Raw data of sugar quantification in wild type (WT) and mutants.
- https://cdn.elifesciences.org/articles/66538/elife-66538-fig2-data1-v1.xlsx
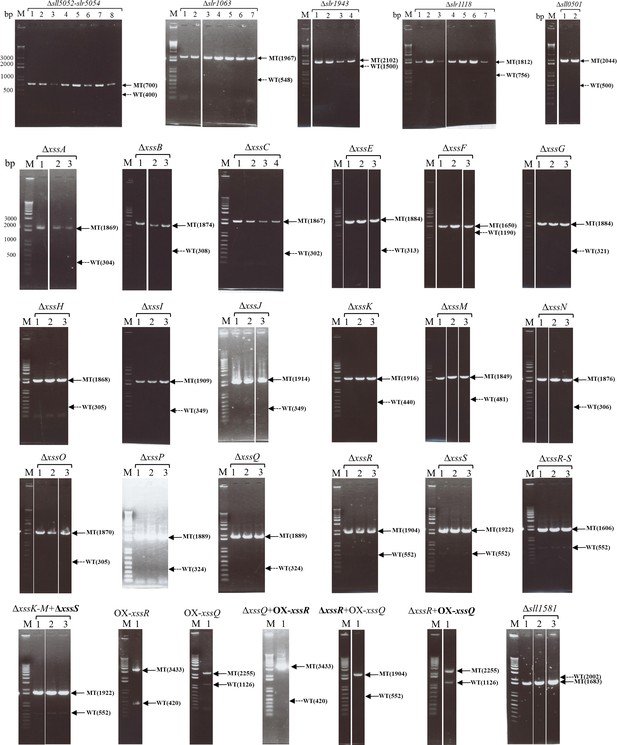
Agarose gel electrophoresis to assess segregation of mutants based on PCR data.
M indicates marker lane, and the numbers above each lane indicate the different clones. The band positions of wild type (WT) and mutants (MT) are shown at the right, with theoretical lengths in nt. A solid arrow indicates the existence of the band; a dotted arrow indicates the absence of the band. The bold text in the strain names indicates the region assessed by PCR.
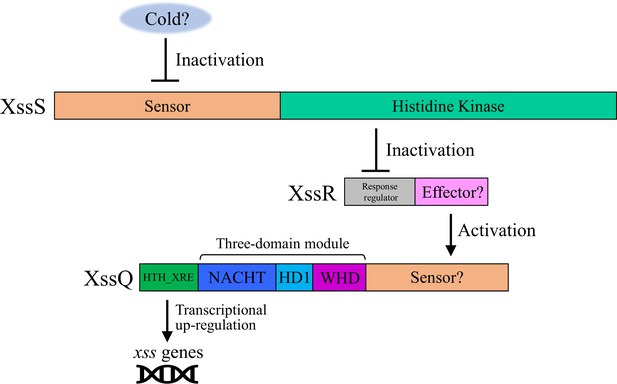
Domain architecture and proposed signal transduction pathway for XssS/XssR/XssQ.
Hybrid-type histidine kinase XssS presumably senses a cold signal and transduces it to the response regulator XssR, which in turn activates the transcription of extracellular sulfated polysaccharide biosynthesis (xss) genes via the signal transduction ATPase with numerous domains (STAND) protein XssQ. Generally, STAND proteins consist of the three-domain module, sensor region, and effector region13. In XssQ, the three-domain module consisting of the NACHT (NAIP [neuronal apoptosis inhibitor protein], CIIA [MHC class II transcription activator], HET-E, and TP1 [mammalian telomerase-associated proteins]) domain (PF05729), HD1 (helical domain 1), and WHD (winged helix domain) responds to a signal from XssR and oligomerizes, leading to the activation of the N-terminal effector domain (helix-turn-helix XRE family domain [SM00530]).
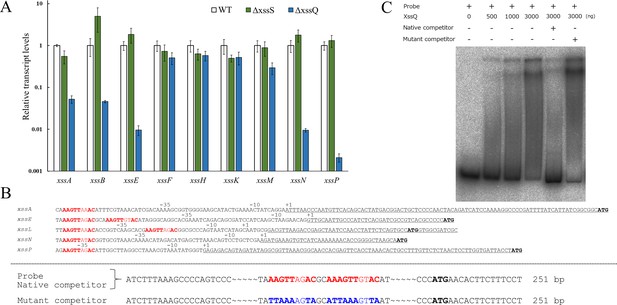
Transcriptional regulation of extracellular sulfated polysaccharide biosynthesis (xss) genes.
(A) Transcript levels for xss genes in wild type (WT), ΔxssS, and ΔxssQ measured by quantitative PCR (qPCR). The internal standard was rnpB. Relative expression levels were obtained by normalization to the transcript levels of each gene in WT. Error bars represent SD (n = 3, biological triplicates). (B) Upper: Sequence comparison of upstream regions of the five regulated genes. Putative consensus regions are shown in red and fully conserved nucleotides are shown in bold letter. Underlines represent transcribed regions based on the report (Kopf et al., 2014b) and initiation codons of regulated genes are shown in black/bold letters. (B) Lower: Sequences of DNA probe and competitors for xssE (native and mutant) used for electrophoretic mobility shift assay (EMSA) of C. Consensus regions are shown in red, and mutated regions are shown in blue. Total DNA size is 251 bp, where identical sequences are mostly not shown except 20 bp at both ends. (C) The autoradiogram image of EMSA of XssQ protein and the DNA probe of xssE with some competitors.
-
Figure 3—source data 1
Raw data of RT-qPCR analysis in wild type (WT), ΔxssS, and ΔxssQ.
- https://cdn.elifesciences.org/articles/66538/elife-66538-fig3-data1-v1.xlsx
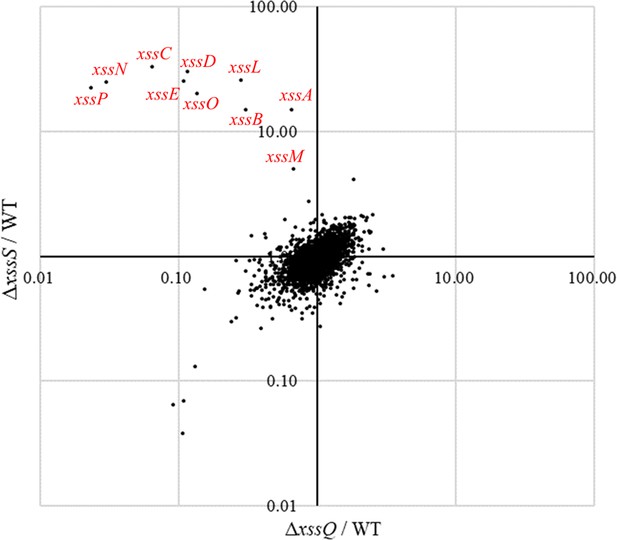
Scatter plot of transcriptome comparison among wild type (WT), ΔxssS, and ΔxssQ.
Transcript levels were based on RPKM (reads per kilobase of exon per million mapped reads) value shown in Figure 3—figure supplement 1—source data 1. Dots in the second quadrant mean down-regulation in ΔxssQ and up-regulation in ΔxssS. Most xss genes were in this quadrant.
-
Figure 3—figure supplement 1—source data 1
RPKM (reads per kilobase of exon per million mapped reads) values of RNA-seq analysis of Synechocystis sp.
PCC 6803 wild type (WT), ΔxssS, and ΔxssQ.
- https://cdn.elifesciences.org/articles/66538/elife-66538-fig3-figsupp1-data1-v1.xlsx
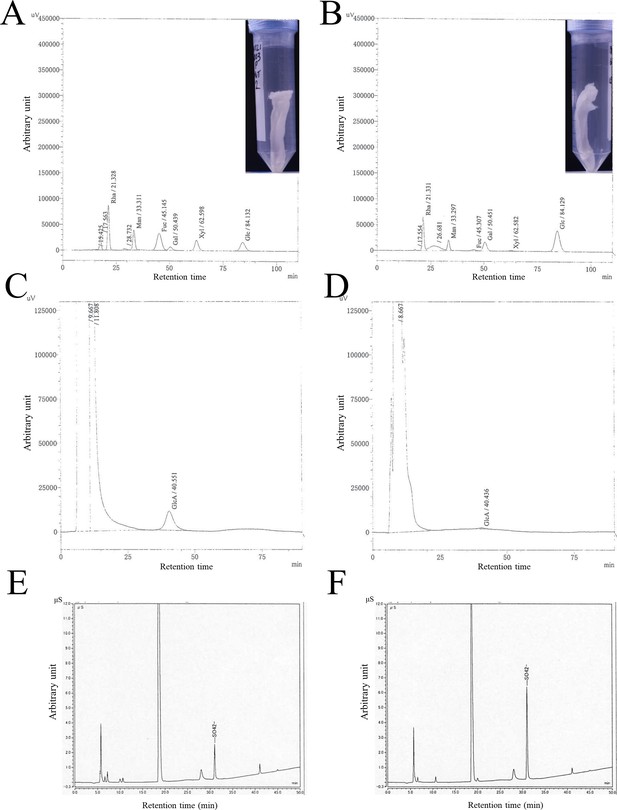
Chromatograms of HPLC and anion exchange column chromatography of exopolysaccharides (EPS).
(A and B) HPLC profiles for neutral sugars in wild-type EPS (A) and ΔxssS EPS (B). (C and D) HPLC profiles for uronic acids in wild-type EPS (C) and ΔxssS EPS (D). The corresponding monosaccharide and retention time are noted at each peak. (E and F) HPLC profiles for SO42– after hydrolysis of EPS samples of wild type (E) and ΔxssS (F).
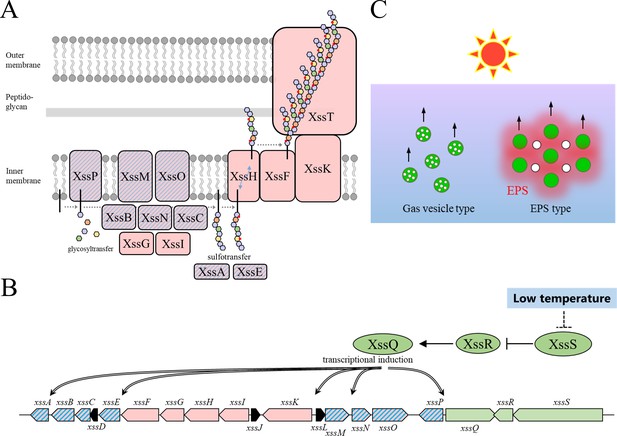
Proposed models for the synechan biosynthesis apparatus, transcriptional regulation, and bloom formation.
(A) Model for the synechan biosynthesis apparatus with sugar polymerization and modification. Red boxes represent biosynthesis components, and red boxes with blue stripes represent transcriptionally regulated components. A putative lipid linker is shown as a black rod. Each monosaccharide is shown as a small hexagon, and each sulfate group is shown as a red spot. (B) Signaling and transcriptional regulation model. Green arrows and ellipses represent regulatory genes and proteins, respectively. Genes for synechan biosynthesis are shown in red, and transcriptionally regulated genes are depicted with blue stripes. Arrows with double lines represent transcriptional activation. (C) Two flotation models in cyanobacteria. Left, flotation of cells (green circles) with intracellular gas-filled vesicles (white circles). Right, flotation of exopolysaccharides (EPS) (red shading)-entrapped cells (green circles) and extracellular gas bubbles (white circles), which are generated by photosynthesis.
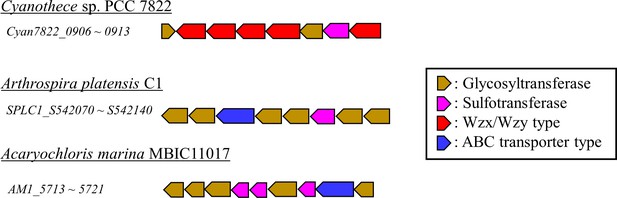
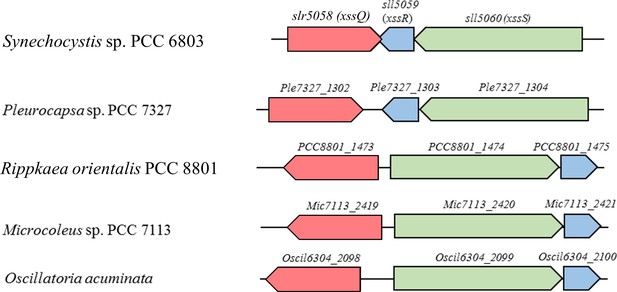
Typical examples of putative gene clusters for biosynthesis of sulfated polysaccharides in cyanobacteria.
A part of each cluster harboring genes for sulfotransferases, glycosyltransferases, and polysaccharide biosynthesis/export systems (Wzx/Wzy type and ABC transporter type) in cyanobacterial genomes.
Tables
Chemical composition of the exopolysaccharides (EPS) from wild-type (WT) Synechocystis 6803 and ΔxssS mutant.
| Sugars | Sulfate residues | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neutral sugars (mol/mol% ) | Uronic acids (mol/mol % ) | Substitution degree | ||||||||||||
| Rhamnose | Ribose | Mannose | Fucose | Galactose | Xylose | Glucose | Total | Galacturonic acid | Glucuronic acid | Total | (mol/mol%) | |||
| WT | 16.6 | N.D. | 25.7 | 16.2 | 4.7 | 10.6 | 23.1 | 96.9 | ND | 3.1 | 3.1 | 10.4 | ||
| ΔxssS | 13.1 | N.D. | 14.2 | 1.2 | 12.5 | 1.0 | 57.9 | 99.9 | ND | 0.1 | 0.1 | 26.6 | ||
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Synechocystis sp. PCC 6803) | slr1943 | GenBank | Gene ID:952818 | |
| Gene (Synechocystis sp. PCC 6803) | slr1043 | GenBank | Gene ID:953647 | |
| Gene (Synechocystis sp. PCC 6803) | sll0501 | GenBank | Gene ID:953286 | |
| Gene (Synechocystis sp. PCC 6803) | slr1118 | GenBank | Gene ID:952865 | |
| Gene (Synechocystis sp. PCC 6803) | sll5042 | GenBank | Gene ID:2655985 | |
| Gene (Synechocystis sp. PCC 6803) | sll5043 | GenBank | Gene ID:2655983 | |
| Gene (Synechocystis sp. PCC 6803) | sll5044 | GenBank | Gene ID:2655981 | |
| Gene (Synechocystis sp. PCC 6803) | ssl5045 | GenBank | Gene ID:2655897 | |
| Gene (Synechocystis sp. PCC 6803) | sll5046 | GenBank | Gene ID:2655982 | |
| Gene (Synechocystis sp. PCC 6803) | sll5047 | GenBank | Gene ID:2655980 | |
| Gene (Synechocystis sp. PCC 6803) | sll5048 | GenBank | Gene ID:2655974 | |
| Gene (Synechocystis sp. PCC 6803) | sll5049 | GenBank | Gene ID:2655975 | |
| Gene (Synechocystis sp. PCC 6803) | sll5050 | GenBank | Gene ID:2655972 | |
| Gene (Synechocystis sp. PCC 6803) | slr5051 | GenBank | Gene ID:2655936 | |
| Gene (Synechocystis sp. PCC 6803) | sll5052 | GenBank | Gene ID:2655973 | |
| Gene (Synechocystis sp. PCC 6803) | slr5053 | GenBank | Gene ID:2655990 | |
| Gene (Synechocystis sp. PCC 6803) | slr5054 | GenBank | Gene ID:2655991 | |
| Gene (Synechocystis sp. PCC 6803) | slr5055 | GenBank | Gene ID:2655867 | |
| Gene (Synechocystis sp. PCC 6803) | slr5056 | GenBank | Gene ID:2655868 | |
| Gene (Synechocystis sp. PCC 6803) | sll5057 | GenBank | Gene ID:2655970 | |
| Gene (Synechocystis sp. PCC 6803) | slr5058 | GenBank | Gene ID:2655931 | |
| Gene (Synechocystis sp. PCC 6803) | sll5059 | GenBank | Gene ID:2655971 | |
| Gene (Synechocystis sp. PCC 6803) | sll5060 | GenBank | Gene ID:2655968 | |
| Gene (Synechocystis sp. PCC 6803) | sll1581 | GenBank | Gene ID:953845 | |
| Strain, strain background (Synechocystis sp. PCC 6803) | Wild-type strain; WT | Doi.org/10.1093/dnares/dsr042 | PCC-P | Motile |
| Strain, strain background (Synechocystis sp. PCC 6803) | Non-motile strain; NM | Doi.org/10.1093/dnares/dsr042 | GT-I | Glucose-tolerant |
| Strain, strain background (Escherichia coli) | JM109 | Takara | Takara:9052 | |
| Recombinant DNA reagent | pPCR-Script (plasmid) | STRATAGENE | STRATAGENE:211186 | |
| Commercial assay or kit | In-Fusion HD Cloning | Clontech | Clontech:639635 | |
| Chemical compound, drug | Alcian Blue 8GX | MERCK | MERCK:05500 |
Additional files
-
Source data 1
Source data for Supplementary file 4.
- https://cdn.elifesciences.org/articles/66538/elife-66538-data1-v1.xlsx
-
Supplementary file 1
Summary of Synechocystis 6803 Xss proteins.
Function: GT, glycosyltransferase; ST, sulfotransferase; PCP, polysaccharide co-polymerase; OPX, outer-membrane polysaccharide export protein. Regulation: transcriptional regulation of xss genes by XssQ/R/S. n.d., not determined.
- https://cdn.elifesciences.org/articles/66538/elife-66538-supp1-v1.xlsx
-
Supplementary file 2
Exopolysaccharides (EPS) accumulation and bloom formation by wild-type (WT) Synechocystis 6803 and extracellular sulfated polysaccharide biosynthesis (xss) mutants.
Classification: ST, sulfotransferase; GT, glycosyltransferase; PCP, polysaccharide co-polymerase; OPX, outer-membrane polysaccharide export protein. Total sugar content of the EPS fraction is expressed as µg glucose/1 × 108 cells. The errors are based on SD (n = 3). *Close to the detection limit. EPS accumulation ratio, percent of WT grown at 31°C. Bloom formation is summarized from Figure 2.
- https://cdn.elifesciences.org/articles/66538/elife-66538-supp2-v1.xlsx
-
Supplementary file 3
Number of sulfotransferase genes (STs) in the genome of cyanobacteria collected from various habitats. * mentioned in the text.
- https://cdn.elifesciences.org/articles/66538/elife-66538-supp3-v1.xlsx
-
Supplementary file 4
Transcriptional levels of extracellular sulfated polysaccharide biosynthesis (xss) genes in the motile (WT) and non-motile (NM) substrains.
Transcriptional level is given as RPKM (reads per kilobase of exon per million mapped reads).
- https://cdn.elifesciences.org/articles/66538/elife-66538-supp4-v1.xlsx
-
Supplementary file 5
Primer sequences.
- https://cdn.elifesciences.org/articles/66538/elife-66538-supp5-v1.xlsx
-
Supplementary file 6
List of mutants.
*Almost complete segregation. #Partial segregation.
- https://cdn.elifesciences.org/articles/66538/elife-66538-supp6-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66538/elife-66538-transrepform-v1.docx