The Ca2+-activated cation channel TRPM4 is a positive regulator of pressure overload-induced cardiac hypertrophy
Figures
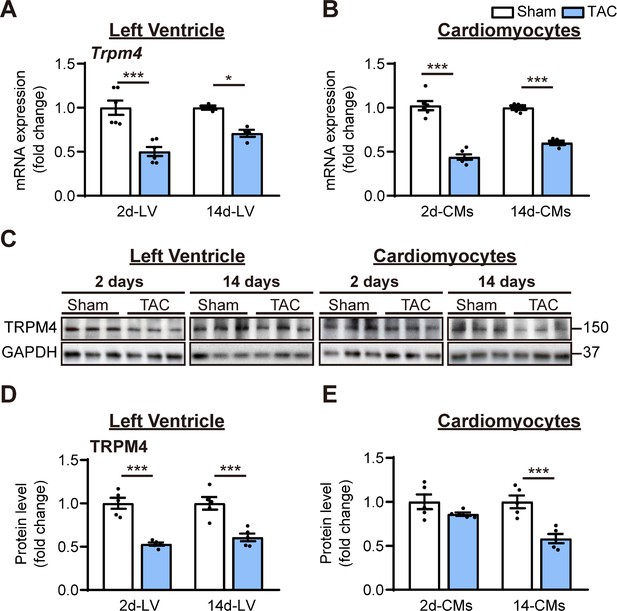
TRPM4 expression was downregulated in response to left ventricular (LV) pressure overload.
(A) Relative mRNA expression of Trpm4 in LV tissue and (B) in LV cardiomyocytes (CMs) after 2 days and 14 days of sham and TAC. (C) Representative western blots of TRPM4 protein expression in LV tissue (left panel) and in LV cardiomyocytes (right panel). (D) Western blots from LV tissue and (E) LV cardiomyocytes after 2 days and 14 days of TAC were quantified for TRPM4 protein expression. Relative TRPM4 mRNA and protein expression in the LV tissue and cardiomyocytes were normalised by GAPDH and calculated as fold change relative to sham in 2 days and 14 days groups, respectively. Results are presented as means ± SEM. *p<0.05, ***p<0.001 vs. sham-operated groups.
-
Figure 1—source data 1
Source data file (Excel) for Figure 1A,B,D and E.
- https://cdn.elifesciences.org/articles/66582/elife-66582-fig1-data1-v1.xlsx

Enlarged heart and cardiac fibrosis were detected 14 days after TAC.
(A) Representative photos of hearts from WT mice 2 days or 14 days after sham or TAC. (B) Representative photos of cardiac fibrosis, evaluated by Masson’s trichrome staining of LV tissue from WT mice subjected to TAC versus sham-operated controls. Scale bar = 100 µm.
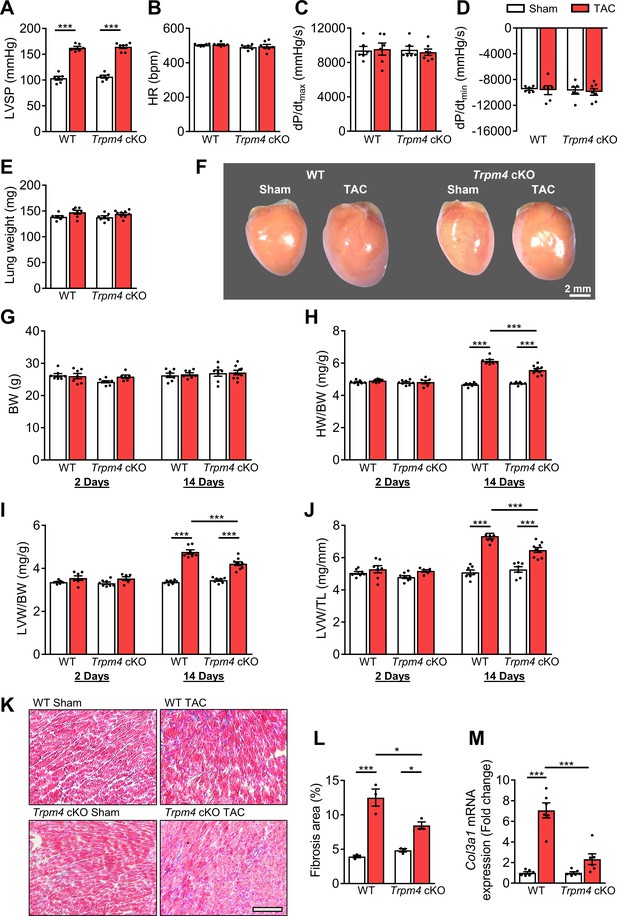
The hypertrophic response of WT and Trpm4 cKO mice to TAC-induced LV pressure overload.
(A) Systolic pressure, (B) heart rate, (C, D) dP/dt after 14 days of sham or TAC in WT and Trpm4 cKO mice. (n = 6–7/group). (E) Lung weight after 14 days of sham or TAC in WT and Trpm4 cKO mice. (n = 7–9/group). (F) Representative photos indicate heart size differences after 14 days of sham or TAC in WT and Trpm4 cKO mice. (G) Body weight, (H) Heart weight, and (I, J) LV weight normalised to body weight and tibia length, in WT and Trpm4 cKO mice after 2 days and 14 days of sham or TAC. (n = 7–9/group). (K) Representative micrographs and (L) quantitation of Masson’s trichrome staining of LV tissue from WT mice and Trpm4 cKO mice after 14 days of sham or TAC (n = 3/group), scale bar = 200 µm in (K). (M) Relative collagen III (Col3a1) mRNA expression after 14 days of sham or TAC. (n = 6/group). The mRNA relative expression was normalised by comparison to GAPDH and calculated as fold change relative to sham in WT and Trpm4 cKO groups, respectively. Results are presented as means ± SEM. *p<0.05, ***p<0.001.
-
Figure 2—source data 1
Source data file (Excel) for Figure 2A,B,C,D,E,G,H,I,J,L, and M.
- https://cdn.elifesciences.org/articles/66582/elife-66582-fig2-data1-v1.xlsx
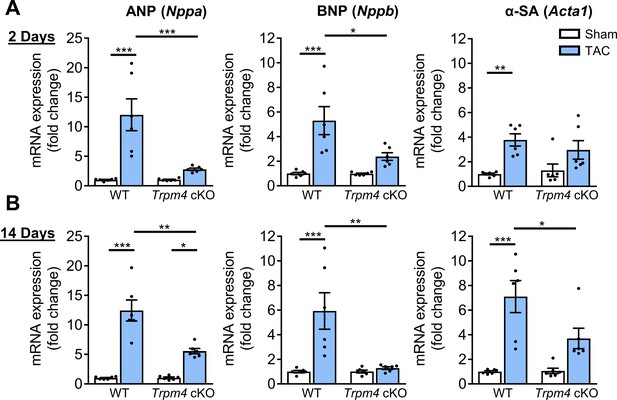
Comparison of gene expression of LVH markers in response to TAC-induced pressure overload in WT and Trpm4 cKO mice.
(A) Relative mRNA expression of ANP (Nppa), BNP (Nppb), and α-SA (Acta1) after 2 days of TAC compared to sham-operated mice. (n = 6/group). (B) Relative mRNA expression of ANP (Nppa), BNP (Nppb), and α-SA (Acta1) after 14 days of sham and TAC. (n = 6/group). The mRNA relative expression was normalised by GAPDH and calculated as fold change relative to WT sham in 2 days and 14 days groups, respectively. Results are presented as means ± SEM, *p<0.05, **p<0.01, ***p<0.001.
-
Figure 3—source data 1
Source data file (Excel) for Figure 3A,B.
- https://cdn.elifesciences.org/articles/66582/elife-66582-fig3-data1-v1.xlsx
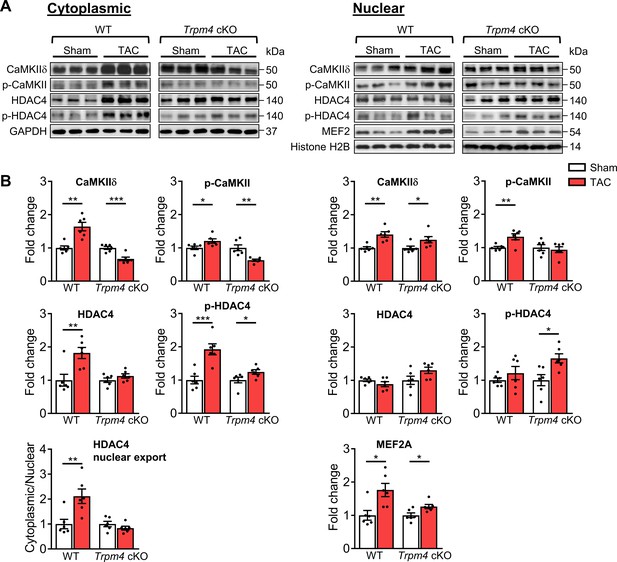
CaMKII-HDAC4-MEF2 signalling pathway in response to TAC after 2 days in WT and Trpm4 cKO mouse hearts.
(A) Representative western blots showing the expression of key proteins in the CaMKII-HDAC4-MEF2 signalling pathway in the cytoplasm (left) and nucleus (right). (B) Cytoplasmic (left) and nuclear (right) quantitative data were normalised by GAPDH and Histone H2B, respectively. Fold changes and cytoplasmic/nuclear ratios were calculated relative to sham groups, in each genotype. Results are presented as means ± SEM, n = 6/group, *p<0.05, **p<0.01, ***p<0.001.
-
Figure 4—source data 1
Source data file (Excel) for Figure 4B.
- https://cdn.elifesciences.org/articles/66582/elife-66582-fig4-data1-v1.xlsx
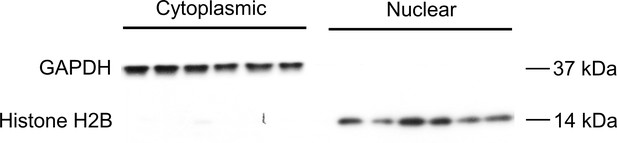
Demonstration of successful fractionation of compartments.
The purity of the fractions extracted from the LV tissue was assessed by western blot using specific marker proteins: GAPDH for cytoplasmic fraction and histone H2B for nuclear fraction. Each fraction (n = 6/group) was run side-by-side on the same blot and then probed separately against each of two primary antibodies: anti-GAPDH and anti-Histone H2B to validate purity of fraction.

Cytoplasmic/nuclear ratio of the key proteins in CaMKII-HDAC4-MEF2 signalling pathway.
Cytoplasmic/nuclear ratios are shown as fold changes relative to sham groups, in each genotype. Results are presented as means ± SEM, n = 6/group, *p<0.05, ***p<0.001.
-
Figure 4—figure supplement 2—source data 1
Source data file (Excel) for Figure 4—figure supplement 2.
- https://cdn.elifesciences.org/articles/66582/elife-66582-fig4-figsupp2-data1-v1.xlsx
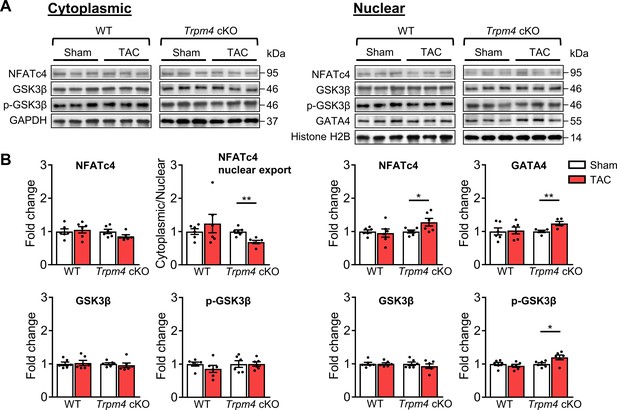
Calcineurin-NFAT signalling pathway in response to TAC after 2 days in WT and Trpm4 cKO mouse hearts.
(A) Representative western blots showing the expression of key proteins in the calcineurin-NFAT signalling pathway in cytoplasm (left) and nucleus (right). (B) Cytoplasmic (left) and nuclear (right) quantitative data were normalised by GAPDH and Histone H2B, respectively. Fold changes and cytoplasmic/nuclear ratios were calculated relative to sham groups, in each genotype. Results are presented as means ± SEM, n = 5–6/group, *p<0.05, **p<0.01.
-
Figure 5—source data 1
Source data file (Excel) for Figure 5B.
- https://cdn.elifesciences.org/articles/66582/elife-66582-fig5-data1-v1.xlsx
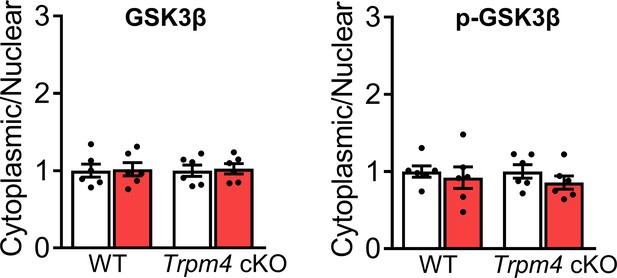
Cytoplasmic/nuclear ratio of the key proteins in calcineurin-NFAT signalling pathway.
Cytoplasmic/nuclear ratios are shown as fold changes relative to sham groups, in each genotype. Results are presented as means ± SEM, n = 6/group.
-
Figure 5—figure supplement 1—source data 1
Source data file (Excel) for Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/66582/elife-66582-fig5-figsupp1-data1-v1.xlsx

Schematic of the putative TAC-induced pathway that culminates in left ventricular hypertrophy.
A Ca2+-permeable MS channel (e.g. Piezo1, TRPV2, TRPV4) acts as the mechanotransducer providing local Ca2+ that in turn stimulates TRPM4. The Na+-permeable TRPM4 activity then could either stimulate voltage-gated Ca2+ channels through membrane depolarisation or induce reverse activity of the Na+/ Ca2+exchanger through local Na+ loading. Either of these outcomes would lead to a high-amplitude increase in local Ca2+. Calmodulin then responds to this high-amplitude Ca2+ stimulus through the lower affinity Ca2+ binding site at its N-lobe which subsequently activates CaMKIIδ and thus stimulates the CaMKII-HDAC4-MEF2 pathway as shown in Yu et al., 2021. Calcineurin activation is inhibited by the activated CaMKIIδ and is activated preferentially by low-amplitude Ca2+ signalling via Gq-coupled receptors and calmodulin (see Discussion). ECM: extracellular matrix, MS: mechanosensitive, Cav: voltage-gated Ca2+ channel, ΔVm: membrane depolarisation, CaM: calmodulin.
Tables
Haemodynamic parameters were measured in wild-type (WT) mice 14 days after subjected to TAC versus sham-operated controls (n = 7–11/group).
Post-mortem analysis of mice 2 days or 14 days after sham or TAC; LVH developed 14 days after TAC, indicated by the ratios of HW/BW, LVW/BW, and LVW/TL in WT mice subjected to TAC versus sham-operated controls. Cardiac fibrosis was evaluated by Masson’s trichrome staining of LV tissue from WT mice subjected to 2 days or 14 days of TAC versus sham-operated controls; cardiac fibrosis areas were graded (n = 5–6/group). Relative Collagen III (Col3a1) mRNA expression was normalised by GAPDH and calculated as fold change relative to sham in 2 days and 14 days groups, respectively (n = 4/group). LVSP: left ventricular systolic pressure; HR: heart rate; dP/dt: first derivative of pressure with respect to time. BW: body weight; HW: heart weight; LVW: left ventricular weight; LW: lung weight; TL: tibia length; HW/BW: heart weight to body weight ratio; LVW/BW: LV weight to body weight ratio; LVW/TL: LV weight to tibia length ratio; LW/BW: lung weight to body weight ratio. Results are presented as means ± SEM. **p<0.01, ***p<0.001, compared between sham- and TAC-operated groups.
| 2 days | 14 days | |||
|---|---|---|---|---|
| Sham | TAC | Sham | TAC | |
| Haemodynamic parameter | ||||
| n | 7 | 7 | ||
| HR (bpm) | 506 ± 4 | 506 ± 3 | ||
| Aortic systolic pressure (mmHg) | 103 ± 1 | 164 ± 2*** | ||
| Aortic diastolic pressure (mmHg) | 76 ± 1 | 74 ± 1 | ||
| LV systolic Pressure (mmHg) | 105 ± 3 | 164 ± 8*** | ||
| dP/dtmax (mmHg/s) | 9438 ± 367 | 9838 ± 259 | ||
| dP/dtmin (mmHg/s) | −9666 ± 377 | −10108 ± 364 | ||
| Anatomical parameter | ||||
| n | 8 | 8 | 11 | 11 |
| BW (g) | 28.5 ± 0.3 | 27.7 ± 0.5 | 28.6 ± 0.3 | 27.2 ± 0.5 |
| HW (mg) | 136.7 ± 2.2 | 132.8 ± 1.3 | 133.1 ± 1.9 | 176.1 ± 3.6 *** |
| LVW (mg) | 98.0 ± 2.0 | 97.7 ± 1.4 | 96.4 ± 1.8 | 136.1 ± 1.4 *** |
| LW (mg) | 141.9 ± 0.9 | 143.6 ± 1.5 | 146.9 ± 1.8 | 147.0 ± 1.9 |
| TL (mm) | 17.4 ± 0.1 | 17.5 ± 0.2 | 17.5 ± 0.2 | 17.2 ± 0.1 |
| HW/BW (mg/g) | 4.8 ± 0.1 | 4.8 ± 0.1 | 4.6 ± 0.1 | 6.6 ± 0.1 *** |
| LVW/BW (mg/g) | 3.4 ± 0.1 | 3.5 ± 0.1 | 3.4 ± 0.1 | 5.1 ± 0.1 *** |
| LVW/TL (mg/mm) | 5.6 ± 0.1 | 5.6 ± 0.1 | 5.3 ± 0.1 | 7.9 ± 0.1 *** |
| LW/BW (mg/g) | 5.0 ± 0.1 | 5.2 ± 0.1 | 5.2 ± 0.1 | 5.4 ± 0.1 |
| Assessment of cardiac fibrosis | ||||
| n | 5 | 5 | 6 | 6 |
| Fibrosis areas (%) | 4.0 ± 0.2 | 3.6 ± 0.2 | 4.4 ± 0.1 | 12.4 ± 0.5*** |
| n | 4 | 4 | 4 | 4 |
| Collagen III mRNA expression (fold change) | 1.0 ± 0.1 | 5.7 ± 0.8** | 1.0 ± 0.1 | 5.1 ± 0.7** |
-
Table 1—source data 1
Haemodynamic and anatomical parameters.
- https://cdn.elifesciences.org/articles/66582/elife-66582-table1-data1-v1.xlsx
Early markers of LVH induction in response to left ventricular pressure overload in WT mice.
Relative mRNA expression of ANP (Nppa), BNP (Nppb), and a-SA (Acta1) after 2 days or 14 days of TAC compared to sham (n = 4–5/group). The relative mRNA expression was normalised by GAPDH and calculated as fold change relative to sham in 2 days and 14 days groups, respectively.Results are presented as means ± SEM. **p<0.01, ***p<0.001, compared between sham- and TAC-operated groups.
| 2 days | 14 days | |||
|---|---|---|---|---|
| Sham | TAC | Sham | TAC | |
| LVH markers (fold change) | ||||
| n | 4 | 4 | 5 | 5 |
| ANP | 1.0 ± 0.1 | 9.9 ± 1.1** | 1.0 ± 0.1 | 9.6 ± 0.7*** |
| BNP | 1.0 ± 0.1 | 8.1 ± 0.8** | 1.0 ± 0.2 | 7.5 ± 0.4*** |
| α-SA | 1.0 ± 0.1 | 4.5 ± 0.5** | 1.0 ± 0.1 | 4.2 ± 0.4*** |
-
Table 2—source data 1
Early gene markers.
- https://cdn.elifesciences.org/articles/66582/elife-66582-table2-data1-v1.xlsx
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-TRPM4 (rabbit polyclonal) | Alomone Labs | Cat# ACC-044, RRID:AB_2040250 | Western blot (1:200) |
| Antibody | Anti-CaMKII delta (rabbit monoclonal) | Abcam | Cat# ab181052, RRID:AB_2891241 | Western blot (1:1000) |
| Antibody | Anti-p-CaMKII (Thr287) (rabbit polyclonal) | Thermo Fisher Scientific | Cat# PA5-37833, RRID:AB_2554441 | Western blot (1:5000) |
| Antibody | Anti-HDAC4 (rabbit monoclonal) | Cell Signaling Technology | Cat# 7628 RRID:AB_10860255 | Western blot (1:1500) |
| Antibody | Anti-p-HDAC4 (Ser246) (rabbit monoclonal) | Cell Signaling Technology | Cat# 3443 RRID:AB_2118723 | Western blot (1:1500) |
| Antibody | Anti-MEF2A (rabbit polyclonal) | Cell Signaling Technology | Cat# 9736 RRID:AB_10691852 | Western blot (1:3000) |
| Antibody | Anti-NFATc4 (rabbit polyclonal) | Abcam | Cat# ab99431, RRID:AB_10675673 | Western blot (1:1500) |
| Antibody | Anti-GSK3β (rabbit monoclonal) | Cell Signaling Technology | Cat# 9315, RRID:AB_490890 | Western blot (1:500) |
| Antibody | Anti-p-GSK3β (Ser9) (rabbit polyclonal) | Cell Signaling Technology | Cat# 9336, RRID:AB_331405 | Western blot (1:1500) |
| Antibody | Anti-GATA4 (mouse monoclonal) | Santa Cruz Biotechnology | Cat# sc-25310, RRID:AB_627667 | Western blot (1:1000) |
| Antibody | Anti-GAPDH (rabbit monoclonal) | Cell Signaling Technology | Cat# 2118, RRID:AB_561053 | Western blot (1:10,000) |
| Antibody | Anti-Histone H2B (rabbit polyclonal) | Abcam | Cat# ab1790, RRID:AB_302612 | Western blot (1:5000) |
| Antibody | Goat anti-rabbit IgG (goat polyclonal) | Abcam | Cat# ab6721, RRID:AB_955447 | Western blot (1:10,000) |
| Antibody | Rabbit anti-mouse IgG (rabbit polyclonal) | Abcam | Cat# ab6728, RRID:AB_955440 | Western blot (1:5000) |
| Sequence-based reagent | ANP (Nppa)_F | Sigma-Aldrich | PCR primers | TGATAGATGAAGGCAGGAAGCCGC |
| Sequence-based reagent | ANP(Nppa)_R | Sigma-Aldrich | PCR primers | AGGATTGGAGCCCAGAGTGGACTAGG |
| Sequence-based reagent | BNP (Nppb)_F | Sigma-Aldrich | PCR primers | TCTCCAGAGCAATTCAAGAT |
| Sequence-based reagent | BNP (Nppb)_R | Sigma-Aldrich | PCR primers | AACAACTTCAGTGCGTTACA |
| Sequence-based reagent | α-SA (Acta1)_F | Sigma-Aldrich | PCR primers | GTGAGATTGTGCGCGACATC |
| Sequence-based reagent | α-SA (Acta1)_R | Sigma-Aldrich | PCR primers | GGCAACGGAAACGCTCATT |
| Sequence-based reagent | Collagen III (Col3A1)_F | Sigma-Aldrich | PCR primers | GACAGATTCTGGTGCAGAGA |
| Sequence-based reagent | Collagen III (Col3A1)_R | Sigma-Aldrich | PCR primers | CATCAACGACATCTTCAGGAAT |
| Sequence-based reagent | Trpm4_F | Sigma-Aldrich | PCR primers | GAGAAGCCCACAGATGCCTATG |
| Sequence-based reagent | Trpm4_R | Sigma-Aldrich | PCR primers | AGCACCGACACCACCAAGTTTG |
Additional files
-
Source data 1
Western blots.
- https://cdn.elifesciences.org/articles/66582/elife-66582-data1-v1.zip
-
Supplementary file 1
Haemodynamic and anatomical parameters after 2 days and 14 days of sham/TAC in WT and Trpm4 cKO mice.
Haemodynamic measurements include heart rate (HR), aortic systolic and diastolic pressure, LV systolic pressure, dP/dtmax and dP/dtmin; anatomical measurements include body weight (BW), heart weight (HW), LV weight (LVW), lung weight (LW), tibial length (TL), heart weight normalised by body weight (HW/BW), LV weight normalised by body weight and tibial length (LVW/BW; LVW/TL), lung weight normalised by body weight (LW/BW). Data are presented as means ± SEM. Comparison between sham and TAC in WT or Trpm4 cKO groups: **p<0.01, ***p<0.001; Comparison between WT and Trpm4 cKO TAC groups: #p<0.05, ###p<0.001.
- https://cdn.elifesciences.org/articles/66582/elife-66582-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66582/elife-66582-transrepform-v1.docx





