Myoglobin primary structure reveals multiple convergent transitions to semi-aquatic life in the world's smallest mammalian divers
Figures

Museum specimen photos illustrating the four major ecomorphotypes within the order Eulipotyphla.
Representative terrestrial (shrew-like mole, Uropsilus soricipes; bottom right), semi-aquatic (Russian desman, Desmana moschata; right centre), strictly fossorial (Eastern mole, Scalopus aquaticus; left), and semi-fossorial (Chinese long-tailed mole, Scaptonyx fusicaudus; top centre) talpid mole species are given. Photo by Kai He.
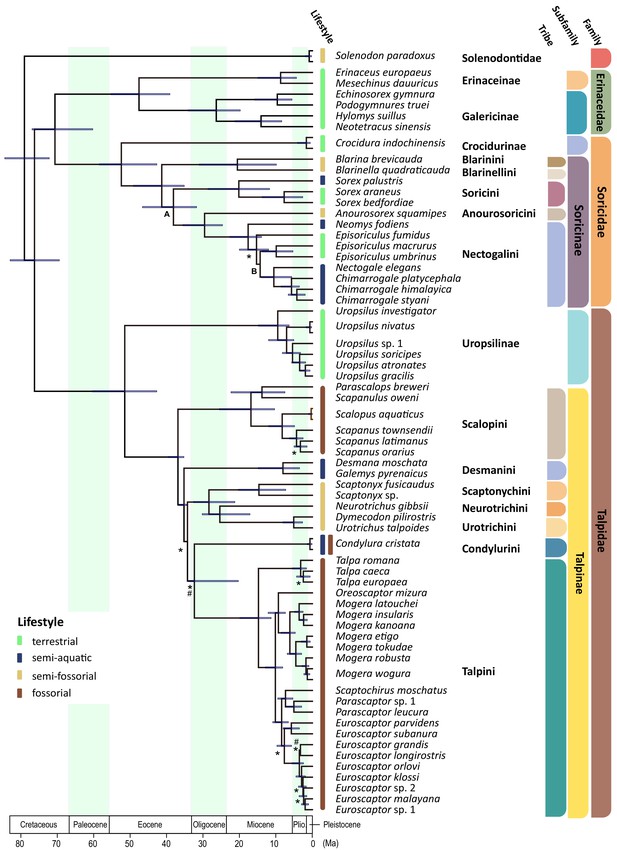
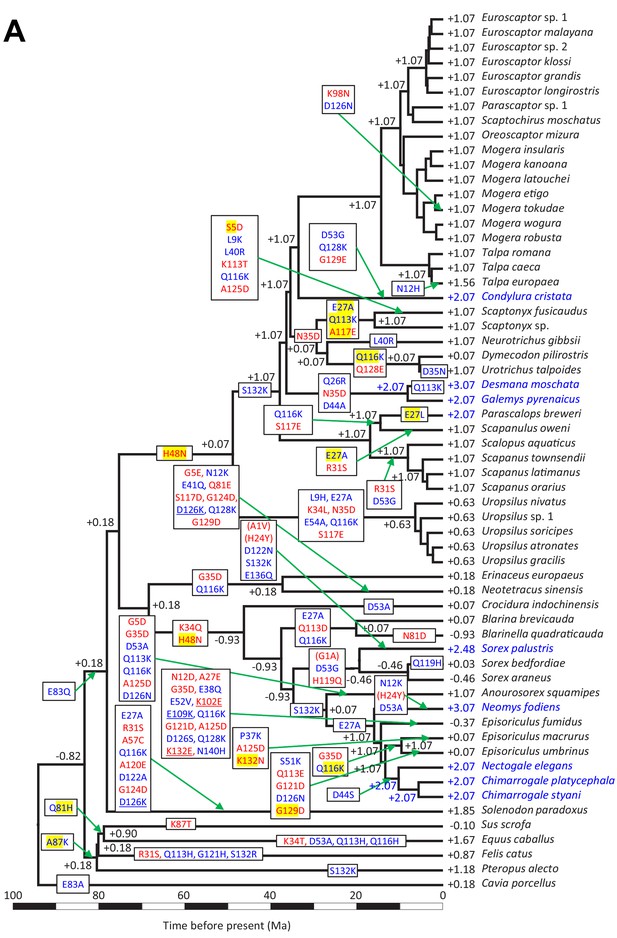
Time calibrated Bayesian phylogenetic tree of Eulipotyphla based on a concatenated alignment of 23 nuclear genes (outgroups not shown).
The units of time are in millions of years (Ma). Branch lengths represent median ages. Node bars indicate the 95% confidence interval [CI] for each clade age. Unless specified, all relationships are highly supported. Relationships weakly supported in concatenation Bayesian and maximum likelihood (PP <0.97 and/or BS <80: #) as well as *BEAST and ASTRAL coalescent analyses (C-PP <0.97 and/or C-BS <80: *) are indicated. Note that an alternative position was recovered for Soricini in the ASTRAL tree (Node A; Figure 2—figure supplement 4), while both coalescent analyses (Node B; ASTRAL, *BEAST) favored Episoriculus monophyly (Figure 2—figure supplements 2–4). Colored bars at the tips of the tree denote terrestrial (green), semi-aquatic (blue), semi-fossorial (beige), and fossorial (brown) lifestyles in extant species.
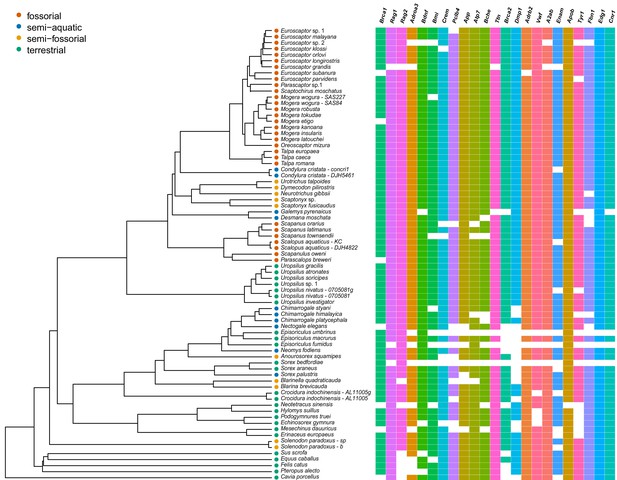
A heatmap produced using ggtree, showing tree-of-life genes included for each of the 76 samples used for phylogenetic analyses.
The ultrametric tree is the BEAST concatenation gene tree and a blank block indicates the gene is missing in the final dataset.
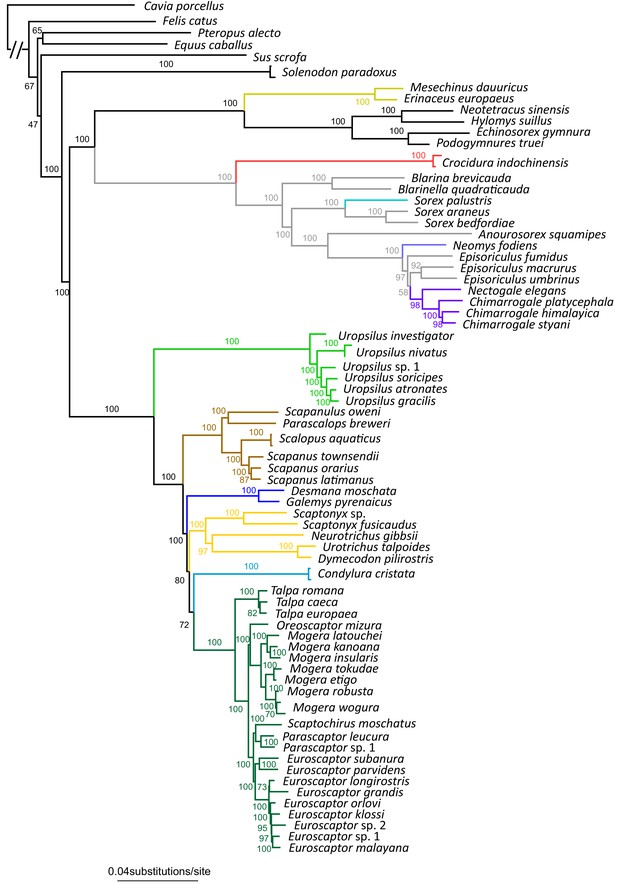
The full RAxML species tree constructed from 71 eulipotyphlan specimens.
Bootstrap supports are given next to internal nodes.
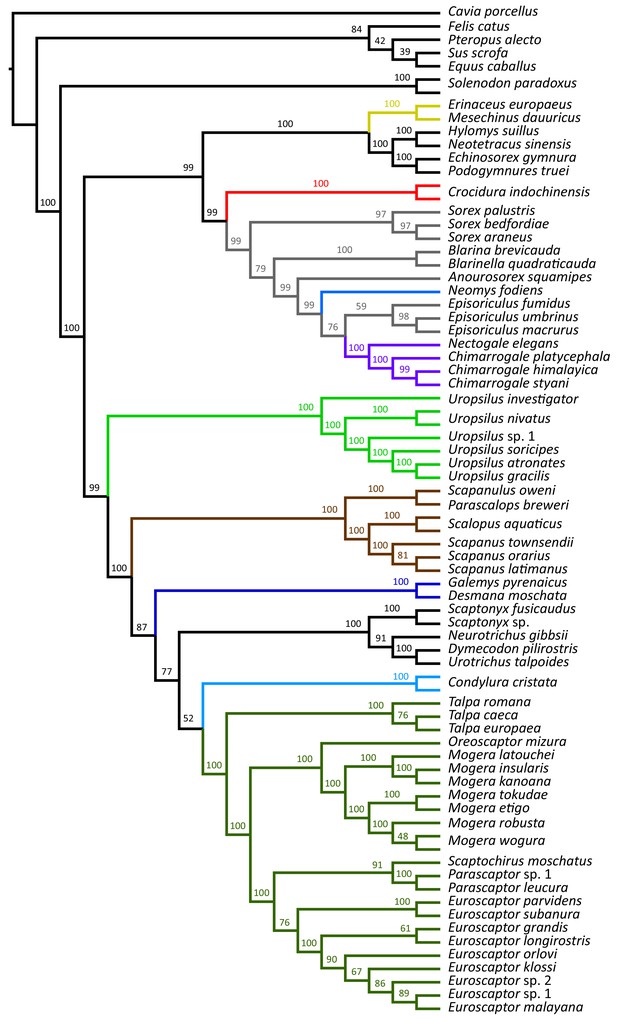
The full ASTRAL-III coalescent species tree constructed from 71 eulipotyphlan specimens.
Bootstrap supports are given next to internal nodes.
The full *BEAST coalescent species tree constructed from 71 eulipotyphlan specimens.
Posterior probabilities are given next to internal nodes.
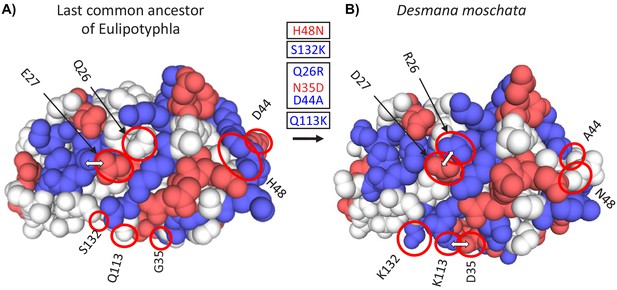
Three-dimensional structural models of myoglobin in (A) the last common ancestor of Eulipotyphla and (B) the semi-aquatic Russian desman (Desmana moschata) obtained by homology modelling using the SWISS-MODEL server (Waterhouse et al., 2018).
The structure of the last common ancestor of the group was modelled based on results of an amino acid sequence reconstruction (see text for details). Ancestral (left) and derived (right) states of charge-changing amino acid replacements are circled and indicated with positional number and one-letter amino acid code. Blue and red color indicate amino acids with positively (H, His; K, Lys; R, Arg) and negatively charged amino acid side chains (D, Asp; E, Glu), respectively. White double arrows indicate surface amino acid side chains involved in salt bridges that are affected by charge-changing substitutions. Text boxes indicate the reconstructed temporal order (top to bottom) of charge decreasing and charge increasing amino acid substitutions (red and blue font, respectively) in the Desmana lineage in one letter code from ancestral (left) to derived (right) separated by positional number. Note that charge neutral substitutions (e.g. G35N), are not given in the text boxes.
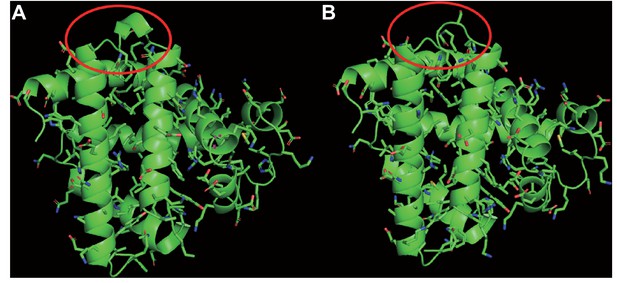
Structural model of myoglobin in (A) the last common ancestor of Eulipotyphla and (B) the semi-aquatic Russian desman (Desmana moschata).
Structures were obtained by homology modelling using the SWISS-MODEL server (Waterhouse et al., 2018) and the primary structures of myoglobin obtained by conceptual translation of the here determined nucleotide sequence (Russian desman) or by ancestral amino acid sequence reconstruction (see text for details). Structures were visualised in PyMOL version 2.1.1. Note that the gap at position 121 in the GH-loop of the tertiary structure (circled in red) of the Russian desman in (B) appears to exert negligible effect on the tertiary structure of the protein.
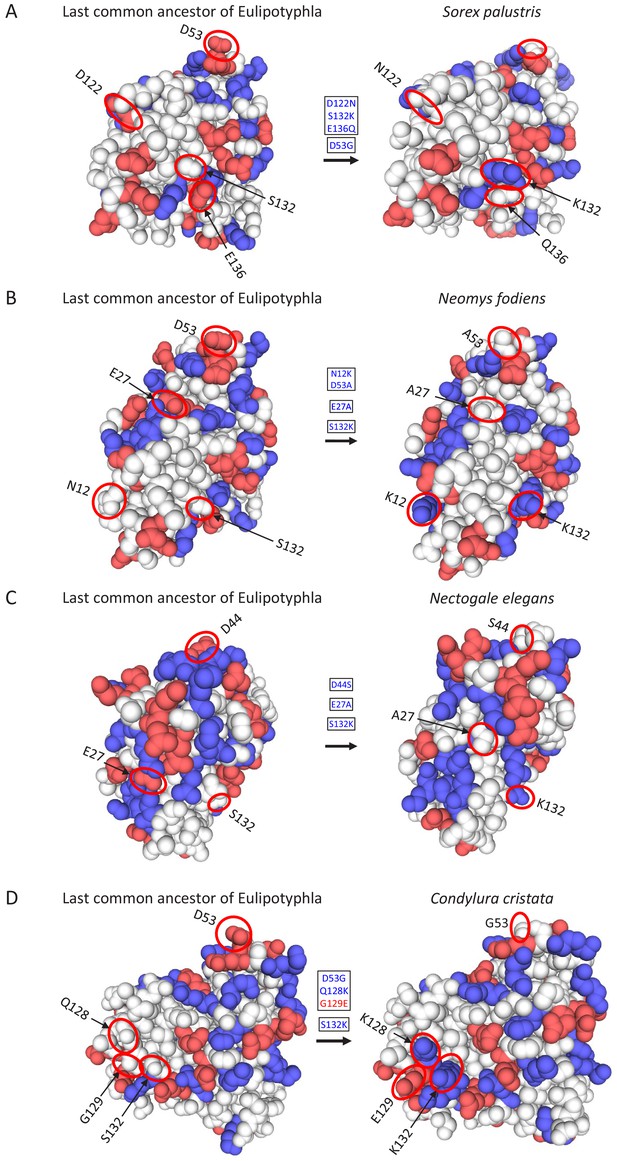
Location of charge-changing amino acid substitutions in the oxygen-storing protein myoglobin of four semi-aquatic species of moles and shrews in the mammalian insectivore order Eulipotyphla.
Three-dimensional structures were obtained by homology modelling using the SWISS-MODEL server (Waterhouse et al., 2018) and the amino acid sequences of (A) Sorex palustris, (B) Neomys fodiens, (C) Nectogale elegans, and (D) Condylura cristata. The three-dimensional myoglobin structure of the last common ancestor of the group was also modelled based on results of an amino acid sequence reconstruction (see text for details). Ancestral (left) and derived (right) states of charge-changing amino acid replacements are circled and indicated with positional number and one-letter amino acid code. Blue and red color indicate amino acids with positively (H, His; K, Lys; R, Arg) and negatively charged amino acid side chains (D, Asp; E, Glu). White double arrows indicate salt bridges that are affected by charge-changing substitutions. Image views between panels A-D have been rotated to maximally visualise lineage-specific replacements.
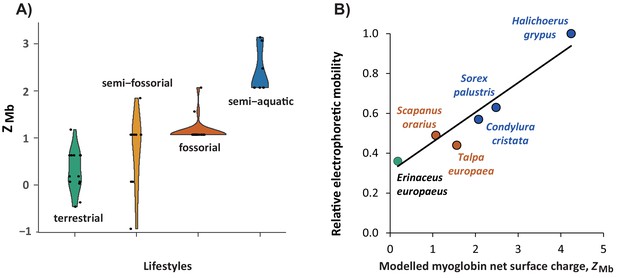
Relationship of modelled myoglobin net surface charge of eulipotyphlan mammals to lifestyle and relative electrophoretic mobility of native myoglobin proteins.
(A) Violin plot showing the distribution (y-axis) and probability density (x-axis) of modelled myoglobin net surface charge, ZMb, among living species (black dots) of the four prevalent eulipotyphlan ecomorphotypes. (B) Correlation between ZMb and electrophoretic mobility of native myoglobin from five eulipotyphlan insectivores; data from the grey seal (Halichoerus grypus) is added for comparison. ZMb was calculated as the sum of the charge of all ionisable groups at pH 6.5 by modelling myoglobin primary structures onto the tertiary structure and using published, conserved, site-specific ionisation constants (McLellan, 1984; Mirceta et al., 2013). Electrophoretic mobility was assessed relative to the mobility of grey seal myoglobin using native polyacrylamide gel electrophoresis of heart or skeletal muscle protein extracts of the indicated species. Green, orange, brown, and blue areas (A) or symbols and fonts (B) indicate terrestrial, semi-fossorial, fossorial, and semi-aquatic/aquatic species, respectively. Phylogenetic Generalised Least Squares analysis in panel (B) revealed a highly significant positive correlation (R2 = 0.897, p<0.005) between the two parameters (solid line, y = 0.1488 x+0.3075).
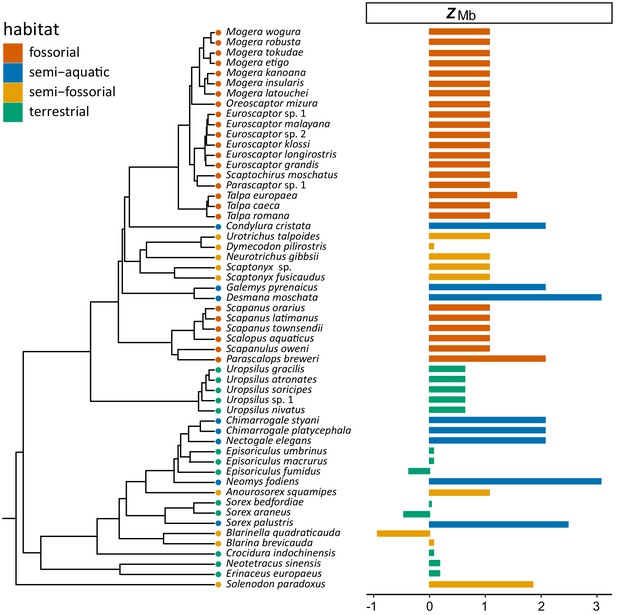
Time-calibrated tree of 55 eulipotyphlan species for which complete myoglobin coding sequences were determined (left).
Horizontal bars on the right indicate the calculated ZMb for each species, which are color coded according to species lifestyle.
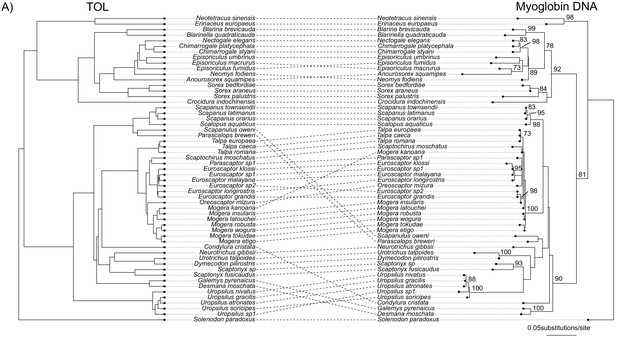
Comparisons of the Bayesian concatenation species tree estimated using the tree-of-life genes with myoglobin RAxML gene trees estimated using nucleotide sequences.
Bootstrap supports are given next to internal nodes on the myoglobin gene trees. Only Bootstrap supports higher than 70 are shown.
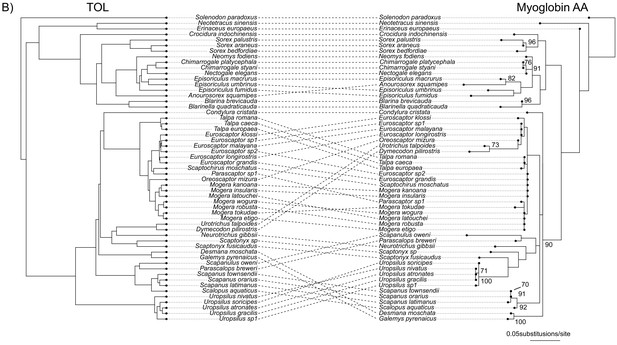
Comparisons of the Bayesian concatenation species tree estimated using the tree-of-life genes with myoglobin RAxML gene trees estimated using amino-acid sequences.
Bootstrap supports are given next to internal nodes on the myoglobin gene trees. Only Bootstrap supports higher than 70 are shown.
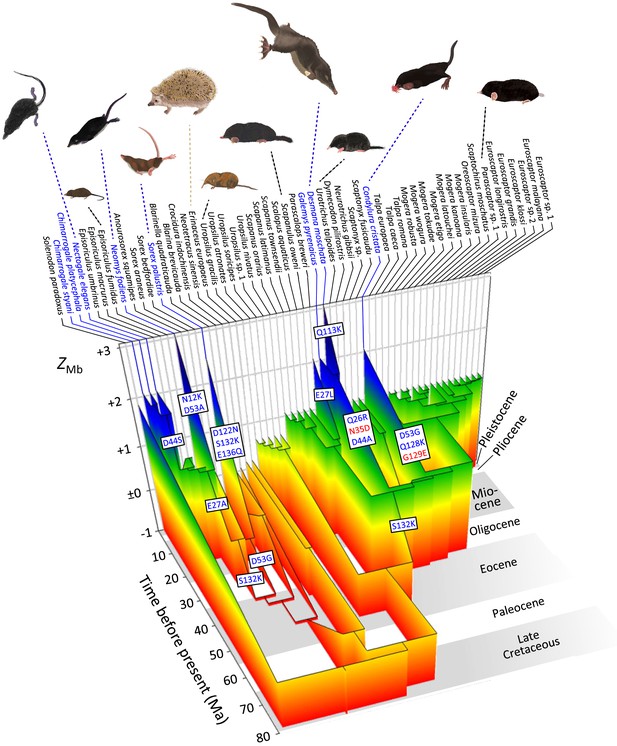
Evolutionary reconstruction of myoglobin net surface charge ZMb in 55 eulipotyphlan insectivores mapped onto the time calibrated phylogeny of Figure 2.
Ancestral ZMb was modelled from primary structures as in Figure 3 and after maximum likelihood ancestral sequence reconstruction. Major charge increasing (blue font) and charge decreasing (red font) amino acid substitutions, from ancestral to derived and separated by positional number, inferred for the immediate ancestry of semi-aquatic species (blue font) are indicated in textboxes alongside the respective branches. Grey and white background shading indicates geologic epochs. See Figure 5—figure supplement 1A for a complete account of charge-changing substitutions, reconstructed ZMb values, and outgroup information. Paintings of representative species by Umi Matsushita.
Evolutionary reconstructions of myoglobin net surface charge (ZMb) in eulipotyphlan mammals.
Maximum likelihood ancestral (A) amino-acid-based and (B) codon-based sequence reconstruction and net surface charge (ZMb) calculation of eulipotyphlan myoglobin mapped onto the time calibrated phylogeny of Figure 2. Results of a separate amino-acid based reconstruction based on the *BEAST species tree reconstruction (Figure 2—figure supplement 3) is provided in (C) with Episoriculus fumidus as the species with an alternative position compared to A and B in red font. Only charge-changing substitutions are shown, with blue and red font in text boxes indicating charge increasing and charge decreasing substitutions, respectively. Absolute charge-changing substitutions > 1.0 are underlined, whereas those <0.10 are shown in brackets. Yellow highlighting indicates amino acids reconstructed with p<0.95 for which less likely but differentially charged amino acids have been reconstructed with p>0.05 (see text for details). Reconstructed net charges are shown at nodes, though have been omitted in some cases if they were identical to values on the preceding node, for clarity. Terminal species considered semi-aquatic are indicated by blue font, with ZMbvalues >+ 2.0 also given in blue font.

Ancestral reconstruction of semi-aquatic lifestyles (blue filled circles) within Eulipotyphla based on both the (A) RAxML concatenation gene tree and (B) the *BEAST species tree.
For each tree, reconstructions were constructed using either a maximum parsimony (left) or a threshold (right) model. Note that Episoriculus fumidus (red font in B) was supported at different positions on the two trees.
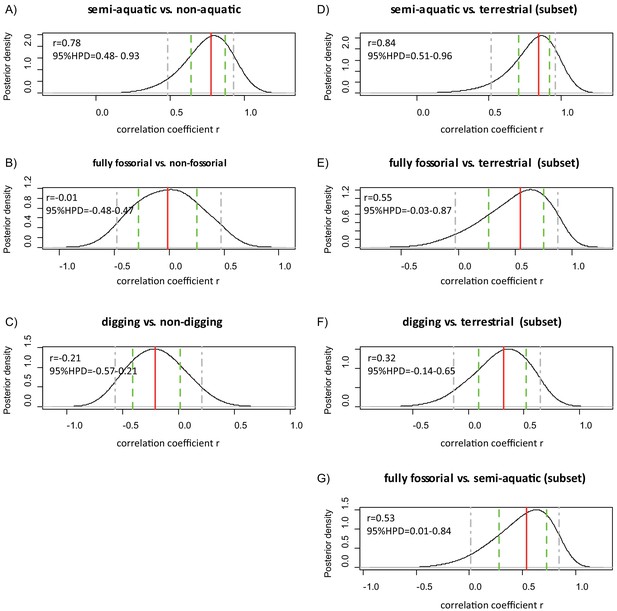
Threshold analyses of myoglobin net surface charge and lifestyle of 55 eulipotyphlan mammals.
Posterior density distribution of the correlation coefficient (r) between (A) semi-aquatic, (B) fully fossorial, and (C) digging (to include fossorial and semi-fossorial lifestyles) and ZMb as estimated using the threshBayes function (D-G) show the results based on the analyses of subsets of samples that included only: (D) terrestrial and semi-aquatic species, (E) terrestrial and fully fossorial species, (F) terrestrial and digging species, or (G) semi-aquatic and fully fossorial species. The solid red line indicates the grand mean, the green dashed lines indicate the mean of the 80% confidence intervals, and the grey dashed lines indicate the mean of the 95% confidence intervals.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Commercial assay or kit | Qiagen DNeasy Blood and Tissue Kit | Qiagen | 69504 | |
| Peptide, recombinant protein | NEBNext dsDNA Fragmentase | New England BioLabs | M0348 | |
| Commercial assay or kit | NEBNext Fast DNA Library Prep Set for Ion Torrent kit | New England BioLabs | E6270L | |
| Sequence-based reagent | NEXTflex DNA Barcodes for Ion Torrent | BIOO Scientific | NOVA-401004 | |
| Commercial assay or kit | E-Gel EX Gel, 2% | Invitrogen | G402002 | |
| Peptide, recombinant protein | NEBNext High-Fidelity 2X PCR Master Mix | New England BioLabs | M0541L | |
| chemical compound, drug | Sera-mag speedbeads | ThermoFisher | 09-981-123 | |
| Commercial assay or kit | myBaits custom target capture kit | Arbor Biosciences | personalized | |
| Commercial assay or kit | Ion 318 Chip Kit v2 BC | ThermoFisher | 4488146 | |
| Software, algorithm | Torrent Suite | ThermoFisher https://github.com/iontorrent/TS copy archived at swh:1:rev:7591590843c967435ee093a3ffe9a2c6dea45ed8 Bridenbecker et al., 2020 | v4.0.2 | |
| Software, algorithm | AlienTrimmer | https://research.pasteur.fr/en/software/alientrimmer/ | RRID:SCR_011835 | v0.3.2 |
| Software, algorithm | SolexaQA++ | http://solexaqa.sourceforge.net/ | RRID:SCR_005421 | v3.1 |
| Software, algorithm | ParDRe | https://sourceforge.net/projects/pardre/ | v2.25 | |
| Software, algorithm | Karect | https://github.com/aminallam/karect, copy archived at swh:1:rev:ba3ad54e5f8ccec5fa972333fcf441ac0c6c2be0 Allam, 2015 | ||
| Software, algorithm | Abyss | http://www.bcgsc.ca/platform/bioinfo/software/abyss | RRID:SCR_010709 | v2.0 |
| Software, algorithm | MIRA | http://sourceforge.net/p/mira-assembler/wiki/Home/ | RRID:SCR_010731 | v4.0 |
| Software, algorithm | SPAdes | http://bioinf.spbau.ru/spades/ | RRID:SCR_000131 | v3.10 |
| Software, algorithm | Geneious | http://www.geneious.com/ | RRID:SCR_010519 | R11 |
| Software, algorithm | FastQC | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | RRID:SCR_014583 | v0.11.5 |
| Software, algorithm | Trimmomatic | http://www.usadellab.org/cms/index.php?page=trimmomatic | RRID:SCR_011848 | v0.39 |
| Software, algorithm | PHYLUCE | https://github.com/faircloth-lab/phyluce, copy archived at swh:1:rev:66ff432f95cb8430d23f6c66a7981d57e8e06902Faircloth et al., 2021 | v1.6.0 | |
| Software, algorithm | MAFFT | http://mafft.cbrc.jp/alignment/server/ | RRID:SCR_011811 | v6.864 |
| Software, algorithm | FastTree | http://www.microbesonline.org/fasttree/ | RRID:SCR_015501 | v2.1.5 |
| Software, algorithm | ASTRAL III | https://github.com/smirarab/ASTRAL copy archived at swh:1:rev:05a85064da2ace5236dba94907bb3c45f45f9597 Mirarab et al., 2021 | v5.15.0 | |
| Software, algorithm | RDP | http://web.cbio.uct.ac.za/~darren/rdp.html | RRID:SCR_018537 | v5.5 |
| Software, algorithm | RAXML | https://github.com/stamatak/standard-RAxML, copy archived at swh:1:rev:a33ff40640b4a76abd5ea3a9e2f57b7dd8d854f6 Stamatakis et al., 2018 | RRID:SCR_006086 | v8.2 |
| Software, algorithm | Newick utilities | http://cegg.unige.ch/newick_utils | v1.6 | |
| Software, algorithm | Tree Graph 2 | http://treegraph.bioinfweb.info/ | v2 | |
| Software, algorithm | BEAST | BEAST 2 https://www.beast2.org | RRID:SCR_010228 | v2.5 |
| Software, algorithm | MEGA-X | http://megasoftware.net/ | RRID:SCR_000667 | Version X |
| Software, algorithm | PAML | http://abacus.gene.ucl.ac.uk/software/paml.html | RRID:SCR_014932 | v4.8 |
| Software, algorithm | EasyCodeML | https://github.com/BioEasy/EasyCodeML, copy archived at swh:1:rev:744a2480e2071c85e044155d8699e87b46356eb9Chen, 2021 | v1.31 | |
| Software, algorithm | FastML | https://swissmodel.expasy.org/ | RRID:SCR_000305 | v3.11 |
| Software, algorithm | PyMol | Schrödinger, LLC (http://www.pymol.org) | RRID:SCR_000305 | v2.1.1 |
| Software, algorithm | SWISS-MODEL server | https://swissmodel.expasy.org/ | RRID:SCR_018123 | |
| Software, algorithm | R | https://www.r-project.org/ | v3.6 | |
| Software, algorithm | CAPER | https://cran.r-project.org/web/packages/caper/index.html | v1.0.1 | |
| Software, algorithm | phytools | https://cran.r-project.org/web/packages/phytools/index.html | RRID:SCR_015502 | v0.7 |
| Software, algorithm | castor | https://cran.r-project.org/web/packages/castor/index.html | v1.6.7 | |
| Software, algorithm | ggtree | https://bioconductor.org/packages/ggtree/ | RRID:SCR_018560 | v3.12 |
Additional files
-
Supplementary file 1
Supplementary information for myoglobin primary structure reveals multiple convergent transitions to semi-aquatic life in the world's smallest mammalian divers.
(a) Sample information of specimens used in this study. (b) Hybridisation capture results of tree-of-life gene segments from 61 eulipotyphlan DNA libraries. Numbers in each column represent total base pairs captured; NA: no data. (C) Result of likelihood-based Shimodaira–Hasegawa test to compare the best scoring RAxML concatenated gene tree and alternative evolutionary hypotheses. (d) Myoglobin amino acid alignment used for modeling myoglobin net surface charge (ZMb) and ancestral sequence reconstructions. Myoglobin helices A to H are highlighted in yellow, with amino acid positions and helical notations indicated above and below the graphic, respectively. Internal amino acid residue positions are shaded in light grey, while deleted residues are indicated by a dash mark. Strongly anionic residues (D [Asp] and E [Glu]) are shaded in red, with strongly (K [Lys] and R [Arg]) and weakly (H [His]) cationic residues shaded in dark and light green, respectively. (e) Charge increasing (blue font) and decreasing (red font) residue substitutions reconstructed for semi-aquatic eulipotyphlan branches. (f) Evolutionary models estimated using bModelTest in BEAST, and used for BEAST and *BEAST analyses. (g) RAxML best scoring gene trees used for ASTRAL-III coalescent analysis before and after collapsing 0% Shimodaira–Hasegawa (SH) scores in Newick format. (h) Calibrations used for estimating divergence times in the BEAST analyses. (i) The best scoring concatenation species trees estimated using BEAST and RAxML, and the best species coalescence trees estimated using ASTRAL-III and *BEAST, in Newick format. (j) Primers used to amplify and sequence the protein coding exons of myoglobin.
- https://cdn.elifesciences.org/articles/66797/elife-66797-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/66797/elife-66797-transrepform-v2.pdf





