Electrical synaptic transmission requires a postsynaptic scaffolding protein
Figures
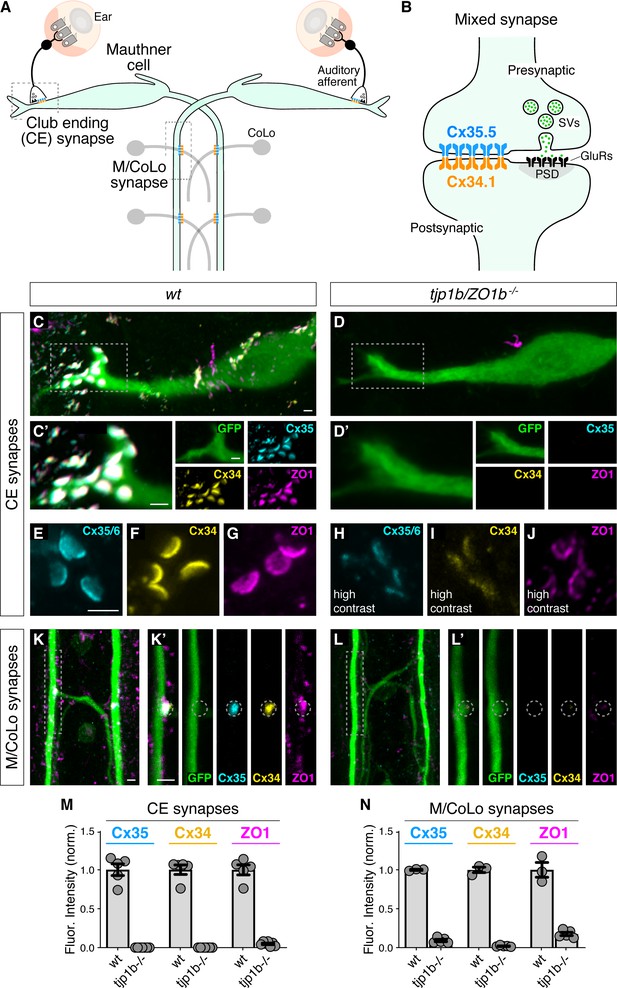
Localizing Connexin to electrical synapse contacts requires the intracellular scaffold protein ZO1b.
(A) Simplified diagram of the Mauthner cell circuit illustrating the electrical synapses of interest. The image represents a dorsal view with anterior on the top. Boxed regions indicate regions stereotypical synaptic contacts used for analysis. Presynaptic auditory afferents contact the postsynaptic Mauthner cell lateral dendrite in the hindbrain forming mixed electrical/glutamatergic Club Ending (CE) synapses. In the spinal cord, the presynaptic Mauthner axons form en passant electrical synapses with the postsynaptic CoLo interneurons (M/CoLo synapses) in each spinal cord hemisegment (2 of 30 repeating spinal segments are depicted). Electrical synapses are denoted as rectangles depicting the two Connexin (Cx) hemichannels (presynaptic Cx35.5 [cyan] and postsynaptic Cx34.1 [yellow]) that form the neuronal gap junction channels of this circuit. (B) Diagram of a mixed electrical/glutamatergic synapse as found at CEs. In the electrical component, molecularly asymmetric Connexin hemichannels (Cx35.5 [cyan], Cx34.1 (yellow)) directly couple cells. In the chemical component, presynaptic synaptic vesicles (SVs) release neurotransmitter (green circles) which align with postsynaptic glutamate receptors (GluRs). The formation and function of chemical synapses are regulated by scaffolds of the postsynaptic density (PSD, gray). (C–L) Confocal images of Mauthner circuit neurons and stereotypical electrical synapse contacts in 5-day-post-fertilization, zf206Et, transgenic zebrafish from wildtype (wt, C,E–G,K) and tjp1b/ZO1b-/- mutant animals (D,H–J,L). In panels (C,D,K,L) animals are stained with anti-GFP (green), anti-zebrafish-Cx35.5 (cyan), anti-zebrafish-Cx34.1 (yellow), and anti-human-ZO1 (magenta). In panels (E–J), animals are stained individually with the indicated antibody. Scale bar = 2 µm in all images. (C,D) Images of the Mauthner cell body and lateral dendrite in the hindbrain. Images are maximum intensity projections of ~15 µm. Boxes denote location of CE contact sites and this region is enlarged in C’ and D’. In C’ and D’ images are maximum-intensity projections of ~5 µm and neighboring panels show individual channels. (E–J) Images of the Mauthner CEs stained for individual electrical synapse components. Images are maximum-intensity projections of ~3.5 µm. In the tjp1b/ZO1b-/- mutant panels (H–J), the contrast for each channel was increased in order to visualize the staining that remained at the synapses. (K,L) Images of the Mauthner/CoLo processes and sites of contact in the spinal cord. Images are maximum-intensity projections of ~5 µm. Boxes denote regions enlarged in K’ and L’. In K’ and L’ images are individual Z-sections and neighboring panels show individual channels. (M,N) Quantification of Cx35.5 (cyan), Cx34.1 (yellow), and ZO1 (magenta) fluorescence intensities at CE (M) and M/CoLo (N) synapses for the noted genotypes. The height of the bar represents the mean of the sampled data normalized to the wt average, and circles represent the normalized value of each individual animal (CE synapses: wt n = 5, tjp1b/ZO1b-/- n = 7; M/CoLo synapses: wt n = 3, tjp1b/ZO1b-/- n = 5). Error bars are ± SEM. For each comparison, wt and tjp1b/ZO1b-/- values are significantly different (Welch's t-test, p<0.01). Associated experimental statistics can be found in Figure 1—source data 1.
-
Figure 1—source data 1
Source data for Figure 1.
- https://cdn.elifesciences.org/articles/66898/elife-66898-fig1-data1-v1.xlsx
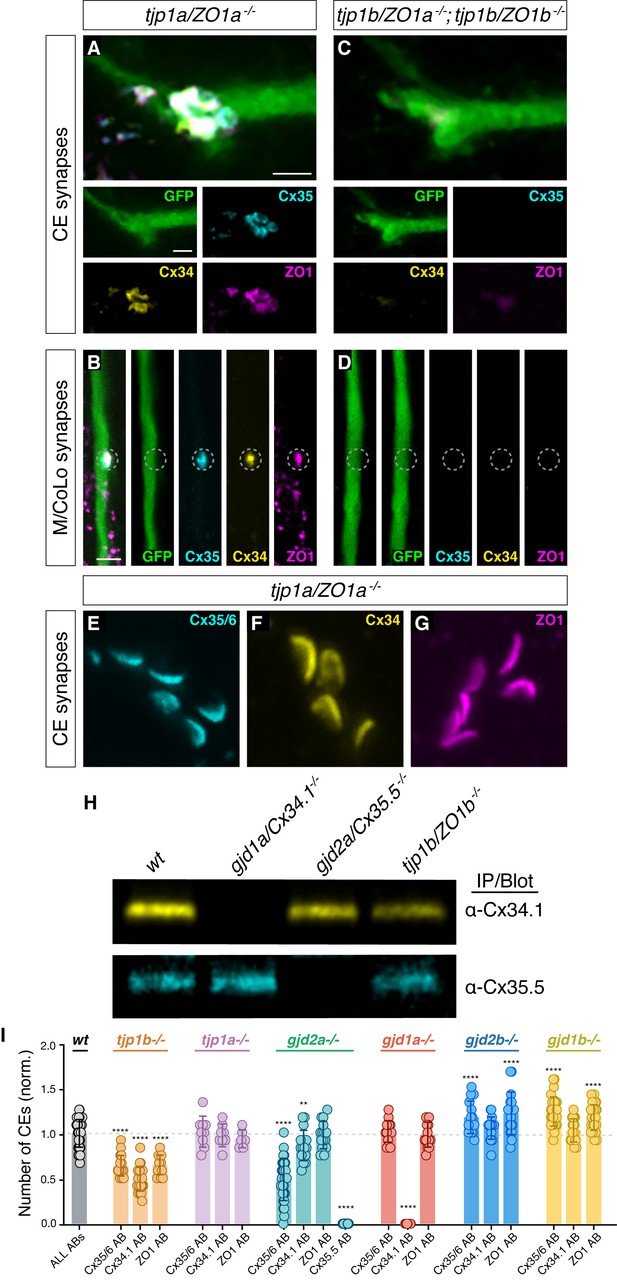
Characterization of ZO1 and Connexin mutants.
(A–G) Confocal images of Mauthner circuit neurons and stereotypical electrical synaptic contacts in 5-day-post-fertilization, zf206Et zebrafish larvae from tjp1a/ZO1a-/- mutant (A,B) and tjp1a/ZO1a-/-; tjp1b/ZO1b-/- double mutant animals (C,D). Animals are stained with anti-GFP (green), anti-zebrafish-Cx35.5 (cyan), anti-zebrafish-Cx34.1 (yellow), and anti-human-ZO1 (magenta). In panels (E–G) animals are stained individually with the indicated antibody. Scale bar = 2 µm in all images. (A,C) Images of the stereotypical location of CE contact sites on the Mauthner lateral dendrite. Images are maximum-intensity projections of ~5 µm and neighboring panels show individual channels. (B,D) Images of the sites of contact of Mauthner/CoLo processes in the spinal cord. Images are individual Z-sections and neighboring panels show individual channels. (E–G) Images of the Mauthner CEs stained for individual electrical synapse components. Images are maximum-intensity projections of E ~ 3.80 µm, F ~ 3.04 µm, G ~ 6.08 µm. (H) Zebrafish brain extracts from animals with indicated genotypes were immunoprecipitated and immunoblotted with anti-Cx34.1 antibody (top) or anti-Cx35.5 antibody (bottom). Results are representative of three independent experiments. (I) Quantification of CE counts based on individual stains in high-contrast images of the indicated genotypes. In this graph, wt data for all individual antibodies has been combined from all experiments depicted; individual data for each experiment can be found in the associated data table. The height of the bar represents the mean of the sampled data normalized to the wt average, and circles represent the normalized value of each individual animal (wt ALL ABs, n = 47; tjp1b Cx35/36 AB, n = 21; tjp1b Cx34.1 AB, n = 26; tjp1b ZO1 AB, n = 15; tjp1a Cx35/36 AB, n = 9; tjp1a Cx34.1 AB, n = 8; tjp1a ZO1 AB, n = 6; gjd2a Cx35/36 AB, n = 29; gjd2a Cx34.1 AB, n = 17; gjd2a ZO1 AB, n = 15; gjd2a Cx35.5 AB, n = 18; gjd1a Cx35/36 AB, n = 16; gjd1a Cx34.1 AB, n = 12; gjd1a ZO1 AB, n = 14; gjd2b Cx35/36 AB, n = 18; gjd2b Cx34.1 AB, n = 19; gjd2b ZO1 AB, n = 17; gjd1b Cx35/36 AB, n = 29; gjd1b Cx34.1 AB, n = 18; gjd1b ZO1 AB, n = 31). Error bars are ± SEM. Asterisks (*) indicate significant differences between wt and mutant counts (****=p<0.0001, **=p<0.01). Associated experimental statistics can be found in Figure 1—figure supplement 1—source data 1.
-
Figure 1—figure supplement 1—source data 1
Source data for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/66898/elife-66898-fig1-figsupp1-data1-v1.xlsx
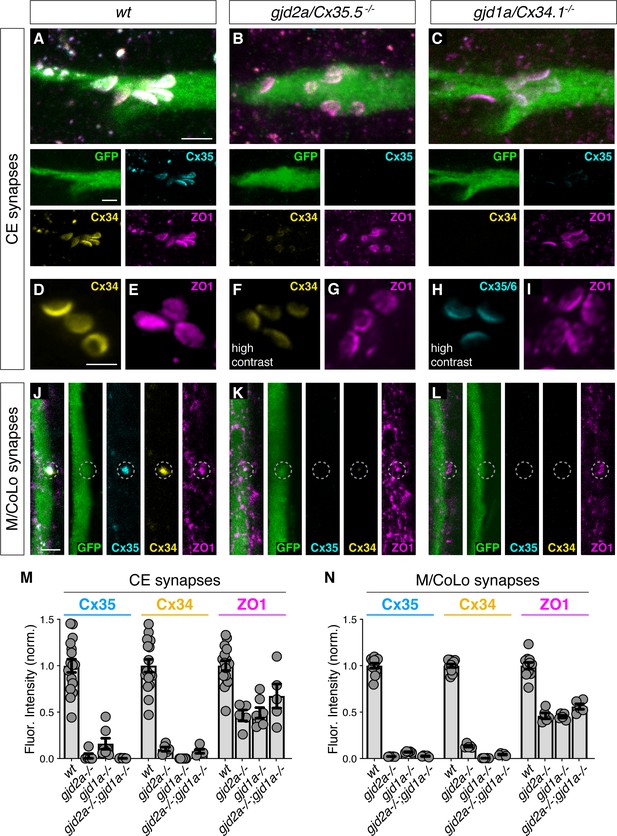
Localizing ZO1b to electrical synapses occurs independent of Connexins.
(A–L) Confocal images of Mauthner circuit neurons and stereotypical electrical synaptic contacts in 5-day-post-fertilization, zf206Et zebrafish larvae from wt (A,D,E,J), gjd2a/Cx35.5-/- mutant (B,F,G,K), and gjd1a/Cx34.1/- mutant animals (C,H,I,L). In panels (A–C,J–L) animals are stained with anti-GFP (green), anti-zebrafish-Cx35.5 (cyan), anti-zebrafish-Cx34.1 (yellow), and anti-human-ZO1 (magenta). In panels (D–I) animals are stained individually with the indicated antibody and in (F,H) the contrast is increased. Scale bar = 2 µm in all images. (A–C) Images of the stereotypical location of CE contact sites on the Mauthner lateral dendrite. Images are maximum-intensity projections of ~5 µm and neighboring panels show individual channels. (D–I) Images of the Mauthner CEs stained for individual electrical synapse components. Images are maximum-intensity projections of ~D ~ 2.66 µm, E ~ 1.90 µm, F ~ 1.90 µm, G ~ 0.72 µm, H ~ 2.28 µm, I ~ 2.16 µm. (F,H) Increased contrast for the Connexin channel reveals the residual staining at the synapses. (J–L) Images of the sites of contact of Mauthner/CoLo processes in the spinal cord. Images are individual Z-sections. Neighboring panels show individual channels. (M,N) Quantification of Cx35.5 (cyan), Cx34.1 (yellow), and ZO1 (magenta) fluorescence intensities at CE (M) and M/CoLo (N) synapses for the noted genotypes. wt data has been combined from all experiments. Individual data can be found in the Figure 2—source data 1. The height of the bar represents the mean of the sampled data normalized to the wt average. Circles represent the normalized value of each individual animal (CE synapse wt, mut paired experiments: wt n = 5, gjd2a-/- n = 5, wt n = 4, gjd1a-/- n = 7, wt n = 7, gjd2a-/-; gjd1a-/- n = 5; M/CoLo synapse wt, mut paired experiments: wt n = 5, gjd2a-/- n = 5, wt n = 3, gjd1a-/- n = 5, wt n = 3, gjd2a-/-; gjd1a-/- n = 5). Error bars are ± SEM. For each comparison, wt and mutant values are significantly different (Welch's t-test, p<0.01), except for the wt to gjd2a-/-; gjd1a-/- (Cx35.5/Cx34.1) double mutant comparison for ZO1 staining at CEs (p=0.842). Associated experimental statistics can be found in Figure 2—source data 1.
-
Figure 2—source data 1
Source data for Figure 2.
- https://cdn.elifesciences.org/articles/66898/elife-66898-fig2-data1-v1.xlsx
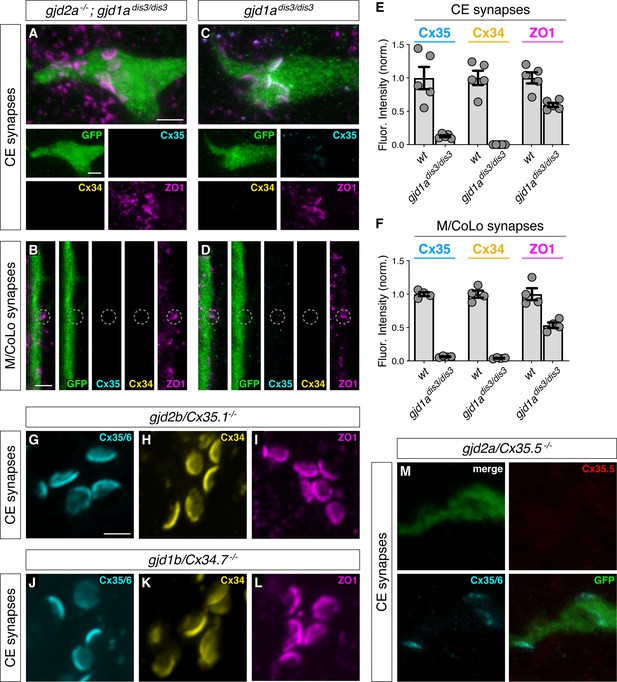
Characterization of Connexin mutants.
(A–D) Confocal images of Mauthner circuit neurons and stereotypical electrical synaptic contacts in 5-day-post-fertilization, zf206Et zebrafish larvae from gjd2a-/-; gjd1adis3/dis3 (Cx35.5/Cx34.1, respectively) mutant (A,B) and gjd1a/Cx34.1dis3/dis3 mutant larvae (C,D). Animals are stained with anti-GFP (green), anti-zebrafish-Cx35.5 (cyan), anti-zebrafish-Cx34.1 (yellow), and anti-human-ZO1 (magenta). The gjd1a/Cx34.1dis3/dis3 mutant was isolated from an ENU-based forward genetic screen for mutants affecting Mauthner electrical synapse formation (Miller et al., 2017). The effects observed from this mutation alone (C,D) are similar to the TALEN-induced frameshift mutation in gjd1a used in Figure 2. The TALEN-induced frameshift mutation is used throughout this paper, except here in this figure supplement, in the gjd2a-/-; gjd1adis3/dis3 mutant images in Figure 3—figure supplement 2, and in the gjd2a-/-; gjd1adis3/dis3 mutant quantitation presented in Figure 2 (M,N). Scale bar = 2 µm in all images. (A,C) Images of the stereotypical of CE contact sites on the Mauthner lateral dendrite. Images are maximum-intensity projections of ~5 µm and neighboring panels show individual channels. (B,D) Images of the sites of contact of Mauthner/CoLo processes in the spinal cord. Images are individual Z-sections and neighboring panels show individual channels. (E,F) Quantification of Cx35.5 (cyan), Cx34.1 (yellow), and ZO1 (magenta) fluorescence intensities at CE (E) and M/CoLo (F) synapses for the noted genotypes. The height of the bar represents the mean of the sampled data normalized to the wt average, and circles represent the normalized value of each individual animal (CE synapses: wt n = 5, gjd1adis3/dis3n = 5; M/CoLo synapses: wt n = 4, gjd1adis3/dis3n = 4). Error bars are ± SEM. For each comparison, wt and mutant values are significantly different (Welch's t-test, p<0.01). Associated experimental statistics can be found in Figure 2—figure supplement 1—source data 1. (G–L) Confocal images of Mauthner CEs synapses in 5-day-post-fertilization, zf206Et zebrafish larvae from gjd2b/Cx35.1-/- (G–I) and gjd1b/Cx34.7-/- (J–L) mutant animals. Images are maximum-intensity projections of G ~ 1.44 µm, H ~ 1.80 µm, I ~ 2.52 µm, J ~ 4.86 µm, K ~ 1.98 µm, L ~ 1.80. In each panel, animals are stained individually with the indicated antibody. Scale bar = 2 µm. (M) Confocal images of Mauthner CEs at 5-day-post-fertilization, zf206Et zebrafish larvae from a gjd2a/Cx35.5-/- mutant. Animal was stained with anti-GFP (green), anti-zebrafish-Cx35.5 (red), and anti-Cx35/6 (cyan). Images are maximum-intensity projections of ~3.80 µm. Scale bar = 2 µm. gjd2a/Cx35.5-/- mutants showed residual staining with the Cx35/6 antibody, but Cx35.5 staining at the synapses was not detected.
-
Figure 2—figure supplement 1—source data 1
Source data for Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/66898/elife-66898-fig2-figsupp1-data1-v1.xlsx
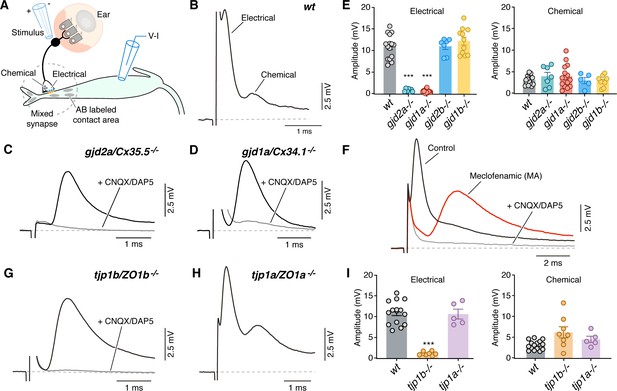
Electrical synaptic transmission at CEs requires ZO1b.
(A) Diagram illustrates the experimental paradigm to examine synaptic transmission. (B) The ‘mixed’ synaptic response in the Mauthner cell evoked by extracellular stimulation of auditory afferents known as club endings (CEs) is composed in wt zebrafish larvae of an early electrical and a delayed chemically mediated response (membrane potential = −79 mv). Traces here and elsewhere represent the average of at least 10 single synaptic responses. (C,D) gjd2a/Cx35.5-/- and gjd1a/Cx34.1-/- mutant zebrafish had no detectable electrical component (black traces). The remaining synaptic response was blocked by bath application of CNQX and DAP5 (20 µM each) that block AMPA and NMDA glutamate receptors, respectively (membrane potential = −83.2 and −81 mv, respectively). (E) Bar graphs summarize the maximal amplitude (mean ± SEM), at a stimulation strength at which all CEs are activated, for the electrical and chemical components in wt and Connexin mutant zebrafish. Left, electrical: wt: 10.9 ± 0.7 mV (n = 15); gjd2a/Cx35.5-/-: 0.8 ± 0.1 mV (p<0.0001, n = 7); gjd1a/Cx34.1-/-: 0.6 ± 0.1 mV (p<0.00001, n = 15); gjd2b/Cx35.1-/-: 11.0 ± 0.7 (n = 7); gjd1b/Cx34.7-/-: 12.2 ± 0.9 mV (n = 11). The values in mutants lacking electrical transmission represent the membrane potential measured at the delay, which show the expected electrical component. Right, chemical: wt: 3.1 ± 0.3 mV (n = 15); gjd2a/Cx35.5-/-: 3.9 ± 0.9 mV (n = 7); gjd1a/Cx34.1-/-: 3.9 ± 0.7 mV (n = 15); gjd2b/Cx35.1-/-: 3.0 ± 0.7 mV (n = 5); gjd1b/Cx34.7-/-:2.9 ± 0.4 mV (n = 9). (F) Blocking electrical transmission recapitulates gjd2a/Cx35.5-/- and gjd1a/Cx34.1-/- synaptic phenotypes. Synaptic responses are superimposed and obtained before (black trace) and after (red trace) adding Meclofenamic acid (MA, 200 µM) to the perfusion solution. The remaining synaptic response was blocked (gray trace) after adding CNQX/DAP5 (20 µM each) to the perfusion solution (membrane potential = −81 mv). (G) tjp1b/ZO1b-/- zebrafish lack electrical transmission (black trace). The remaining synaptic potential was blocked by CNQX/DAP5 (20 µM each; gray trace) (membrane potential = −82 mv). (H) Synaptic responses in tjp1a/ZO1a-/- zebrafish show both electrical and chemical components (membrane potential = −87 mv). (I) Bar graphs illustrate the maximal amplitude (mean ± SEM) for the electrical and chemical components of the synaptic response in wt and ZO1 mutant zebrafish. Left, Electrical: tjp1b/ZO1b-/-: 1.1 ± 0.2 mV (p-value<0.0005, n = 8); tjp1a/ZO1a-/-: 10.6 ± 1.2 mV (n = 5). Right, chemical: tjp1b/ZO1b-/-: 6.2 ± 1.3 mV (n = 8); tjp1a/ZO1a-/-: 4.5 ± 0.8 mV (n = 5). Associated experimental statistics can be found in Figure 3—source data 1.
-
Figure 3—source data 1
Source data for Figure 3.
- https://cdn.elifesciences.org/articles/66898/elife-66898-fig3-data1-v1.xlsx
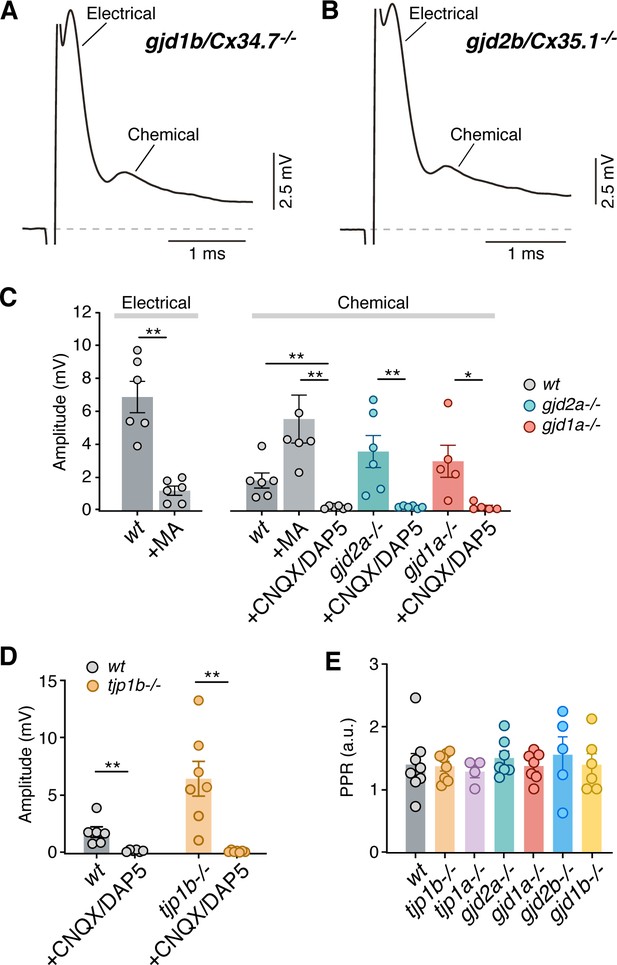
Electrophysiological characterization of Connexin and ZO mutants.
(A–B) Synaptic responses in gjd2b/Cx34.7-/- (A) and gjd1b/Cx35.1-/- (B) mutant zebrafish show electrical and chemical components of normal amplitudes (membrane potential = −80 and −82 mv, respectively). (C) Graph summarizes the maximal amplitude (mean ± SEM) of the electrical and chemical components of the synaptic response in control conditions and in the presence of MA (200 µM), or CNQX/DAP5 (20 µM) for wt, gjd2a/Cx35.5-/-, and gjd1a/Cx34.1-/- zebrafish. Electrical: amplitude in wt averaged 6.9 ± 0.9 mV (n = 6) and it was dramatically reduced after MA application to 1.2 ± 0.3 mV (n = 6; p<0.005). Chemical: amplitude in wt averaged 1.8 ± 0.5 mV (n = 6) and averaged 5.5 ± 1.5 mV after MA (n = 6; p<0.01) and was dramatically reduced after CNQX/DAP5 application to 0.2 ± 0.03 mV (n = 6, p<0.005). CNQX/DAP5 reduced amplitude in mutants from 3.6 ± 1.0 to 0.2 ± 0.03 mV in gjd2a/Cx35.5-/- (n = 6, p<0.005) and from 3.0 ± 1.0 to 0.2 ± 0.1 mV in gjd1a/Cx34.1-/- (n = 5, p<0.05). (D) Graph summarizes the amplitude of the chemical synaptic components in control conditions and in presence of CNQX/DAP5 (20 µM) for wt and the tjp1b/ZO1b-/- mutant. CNQX/DAP5 reduced amplitude from 1.8 ± 0.5 to 0.2 ± 0.03 mV (n = 6, p<0.005) in wt and from 6.4 ± 1.5 to 0.2 ± 0.03 mV (n = 7, p<0.005) in tjp1b/ZO1b-/-. (E) Neurotransmitter release properties of chemical transmission at CEs of wt, Connexin and ZO1 mutant zebrafish. Paired-pulse ratio (mean ± SEM; see methods) of the chemical responses in wt (1.4 ± 0.2 mV, n = 8), tjp1b/ZO1b-/- (1.4 ± 0.1 mV, n = 8), tjp1a/ZO1a-/- (1.3 ± 0.1 mV, n = 4), gjd2a/Cx35.5-/- (1.5 ± 0.1 mV, n = 7), gjd1a/Cx34.1-/- (1.4 ± 0.1 mV, n = 7), gjd2b/Cx34.7-/- (1.4 ± 0.2 mV, n = 6), and gjd1b/Cx35.1-/- (1.5 ± 0.3 mV, n = 5) zebrafish. Paired-pulse ratio values were not significantly different (Kruskal-Wallis ANOVA). Associated experimental statistics can be found in Figure 3—figure supplement 1—source data 1.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/66898/elife-66898-fig3-figsupp1-data1-v1.xlsx
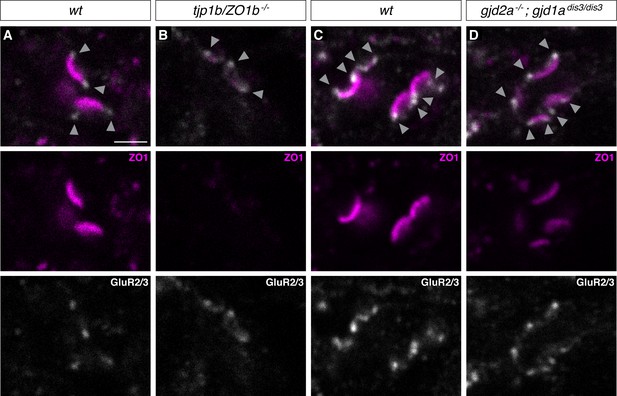
GluR2/3 localization is unaffected in ZO and Connexin mutants.
(A–D) Confocal images of Mauthner CEs in 5-day-post-fertilization, zf206Et zebrafish larvae from wt (A,C), tjp1b/ZO1b-/- (B), and gjd2a-/-; gjd1adis3/dis3 (Cx35.5/Cx34.1, respectively) mutant animals (D). Animals are stained with anti-human-ZO1 (magenta) and anti-rabbit-GluR2/3 (white), and images depict a single z-slice. Arrowheads highlight location of chemical component of mixed electrical/glutamatergic synapse. Scale bar = 2 µm.
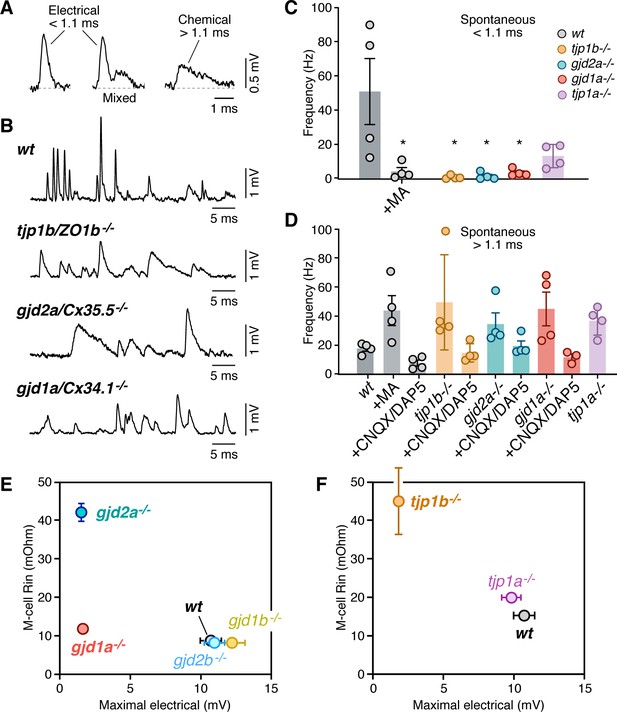
Lack of electrical transmission in tjp1b/ZO1b-/- is widespread and alters M-cell excitability.
(A) Spontaneous responses in wildtype (wt) zebrafish can be electrical, chemical, or mixed. Spontaneous electrical and chemical responses were identified for automated detection by their duration: electrical responses were brief (<1.1 ms), whereas chemical responses were longer lasting (>1.1 ms). Mixed responses combined both. (B) Representative single traces of spontaneous synaptic activity obtained from the Mauthner cells of wt, tjp1b/ZO1b-/-, gjd2a/Cx35.5-/-, and gjd1a/Cx34.1-/-. Note the lack of short-lasting spontaneous responses in mutant zebrafish (membrane potential = −89, –87, and −89 mV, respectively). (C) Bar graph summarize the frequency in Hz (mean ± SEM; each n represents a fish) of the spontaneous short-lasting (<1.1 ms) electrical responses in wt, tjp1b/ZO1b-/-, gjd2a/Cx35.5-/-, gjd1a/Cx34.1-/- and tjp1a/ZO1a-/- zebrafish. The frequency of events in wt zebrafish was 50.9 ± 19.2 Hz (n = 4) and was reduced by MA (200 µM) to 4.2 ± 2.3 Hz (p<0.05). The variability between WT fish reflects different states of the network. The frequency was dramatically reduced in mutant zebrafish lacking electrical transmission: tjp1b/ZO1b-/-: 0.85 ± 0.5 Hz (n = 4; p<0.05); gjd2a/Cx35.5-/-: 1.5 ± 1.0 Hz (n = 4; p<0.05); gjd1a/Cx34.1-/-: 3.4 ± 1.1 Hz (n = 4, p<0.05). Although reduced, the change was not significant in tjp1a/ZO1a-/-: 13.2 ± 3.4 Hz (n = 4). (D) Long-lasting (>1.1 ms) chemical responses. The frequency of events in wt zebrafish was 17.8 ± 1.8 Hz (n = 4) and increased after MA to 43.8 ± 10.2 Hz (n = 4; p<0.05). The frequency was also increased in mutant zebrafish: tjp1b/ZO1b-/-: 49.5 ± 16.3 Hz (n = 4, p<0.05); gjd2a/Cx35.5-/-: 34.6 ± 7.7 Hz (n = 4, p<0.05); gjd1a/Cx34.1-/-: 45.0 ± 11.6 Hz (n = 4, p<0.05); tjp1a/ZO1a-/-: 36.8 ± 4.9 Hz (n = 4, p<0.05). Spontaneous events > 1.1 ms were greatly reduced by glutamate receptor antagonists (20 µM, CNQX/DAP5). The remaining responses likely represent depolarizing inhibitory responses prominent in the Mauthner cell. (E–F) Changes in the excitability of the Mauthner cell in Connexin and ZO1 zebrafish mutants. The graphs plot the input resistance of the Mauthner cell (R–in) vs the maximal amplitude of the electrical component of the synaptic response for wt and Connexin (E) and ZO1 (F) mutants. See Table 1 for values of Rn and Figure 3 for those of maximal electrical amplitude. Bars represent standard deviation. Associated experimental statistics can be found in for Figure 4—source data 1.
-
Figure 4—source data 1
Source data for Figure 4.
- https://cdn.elifesciences.org/articles/66898/elife-66898-fig4-data1-v1.xlsx
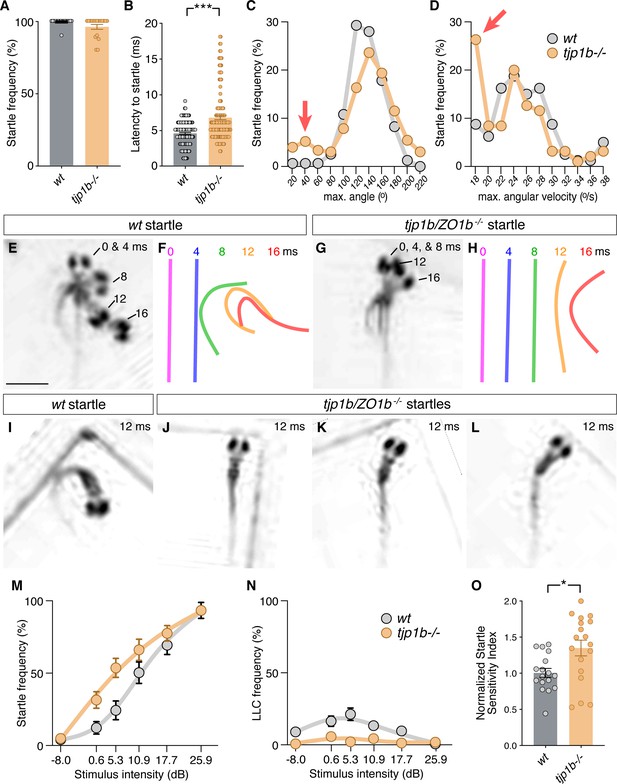
Mauthner-cell-initiated escape response parameters require ZO1b.
(A) Frequency of responses classified as Mauthner-initiated startles in wt and tjp1b/ZO1b-/- mutants. Bar graphs show mean ± SEM. Each circle represents an individual animal’s average frequency of responses to 10 independent trials (wt n = 17, tjp1b/ZO1b-/- n = 18; Mann-Whitney test p=0.0947). (B) Latency of initiating startles in all individual trials. Bar graphs represent data as mean ± SEM with each circle representing individual latencies (wt n = 157, tjp1b/ZO1b-/- n = 165; Mann-Whitney test p<0.0001). (C,D) Kinematic analysis of the maximum turn angle (C) and the maximum angular velocity (D) of the startles plotted as frequency of events within the indicated bin. Red arrows indicate abnormal shallow angle and low velocity turns exhibited by tjp1b/ZO1b-/- mutants. (E–H) Time-lapse of wt (E,F) and tjp1b/ZO1b-/- mutant (G,H) startles. Scale bar = 1 mm. Individual snapshots taken at the indicated times (ms = milliseconds) are overlaid on an individual image (E,G). A line representing the midline body axis at each time was drawn to indicate the movement (F,H). (I–L) A wt startle bend at its maximum angle (I) compared to abnormally shaped bends executed by tjp1b/ZO1b-/- mutant larvae (J–L). (M–N) Mauthner-induced startle frequency (M) and long-latency C-bend (LLC) frequency (N) for 10 trials at six intensities with fit curves for wt and tjp1b/ZO1b-/- mutants. Each symbol represents data as mean ± SEM (wt n = 17, tjp1b/ZO1b-/- n = 18). (O) The startle sensitivity index is determined as the area under the curves for each individual animal in (M). Bar graphs represent data as mean ± SEM with each circle representing individual sensitivity indices (wt n = 17, tjp1b/ZO1b-/- n = 18; Mann-Whitney test p=0.03). Associated experimental statistics can be found in Figure 5—source data 1.
-
Figure 5—source data 1
Source data for Figure 5.
- https://cdn.elifesciences.org/articles/66898/elife-66898-fig5-data1-v1.xlsx
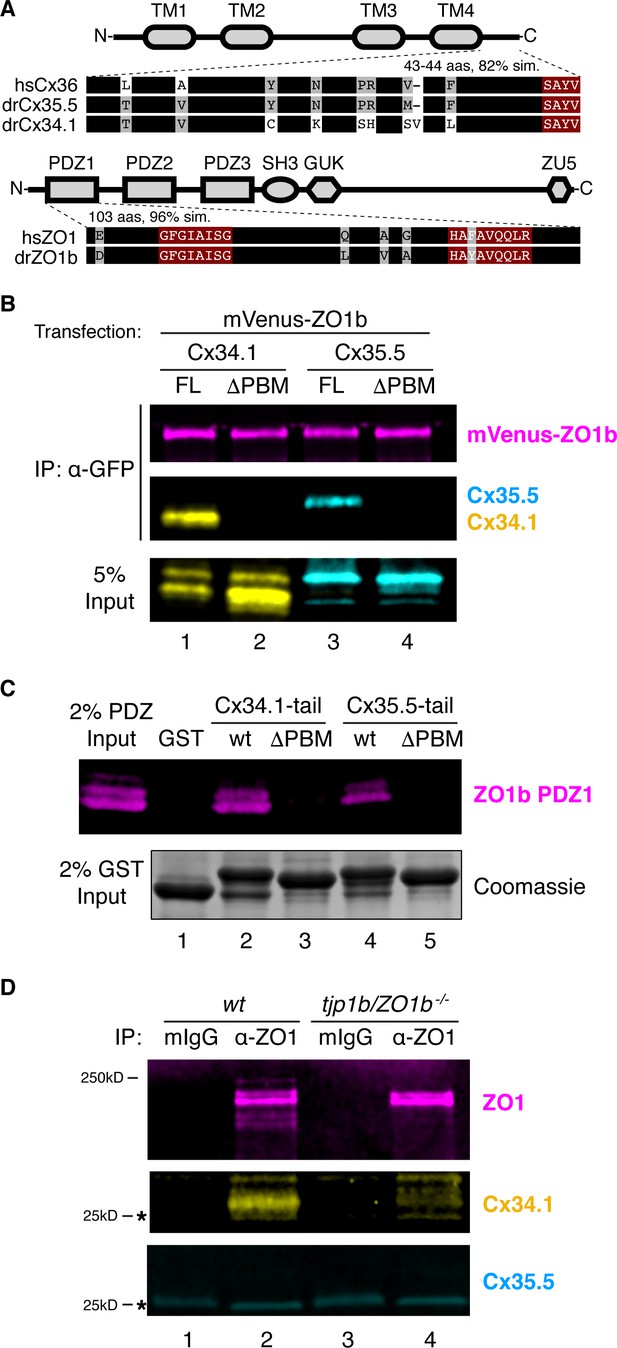
ZO1b scaffolds postsynaptic Cx34.1 in vivo.
(A) Schematic, linear diagrams of Cx36 and ZO1 homologues. Domains are depicted as gray shapes; TM = transmembrane, PDZ, SH3, GUK, and ZU5 = protein-protein interaction modules; hs = Homo sapiens, dr = Danio rerio. Amino acid alignments are shown for the indicated expanded regions. Black bars represent conserved amino acids; non-conserved amino acids are indicated. Maroon boxed amino acids represent the conserved PDZ-binding motif (PBM) of Cx36-family proteins (top) or the predicted PDZ1 residues of the conserved ligand-binding cleft of ZO1-family proteins (bottom). (B) HEK293T/17 cells were transfected with plasmids to express mVenus-ZO1b and either full-length Cx34.1 (lane 1), Cx34.1-∆PBM (lane 2), full-length Cx35.5 (lane 3), or Cx35.5-∆PBM (lane 4). Lysates were immunoprecipitated with anti-GFP antibody and analyzed by immunoblot for the presence of mVenus-ZO1b using anti-GFP antibody (upper, magenta), Cx34.1 protein using Cx34.1-specific antibody (middle, yellow), or Cx35.5 protein using Cx35.5-specific antibody (middle, cyan). Total extracts (bottom, 5% input) were blotted for Connexin proteins to demonstrate equivalent expression and uniform antibody recognition of expressed proteins. Results are representative of three independent experiments. (C) Bacterially purified GST (lane 1), GST-Cx34.1-tail (lane 2), GST-Cx34.1-tail-∆PBM (lane 3), GST-Cx35.5-tail (lane 4), or GST-Cx35.5-tail-∆PBM (lane 5) was immobilized on glutathione beads and incubated with purified ZO1b PDZ1 domain. The tail regions used are depicted in the expanded regions in (A). Bound proteins were analyzed by immunoblot for the presence of ZO1b PDZ1 using anti-TEV cleavage site antibody (top, magenta). Equal loading of GST proteins is indicated by Coomassie staining (bottom, 2% input). Results are representative of three independent experiments. (D) Zebrafish brain extract from wt (lanes 1,2) or tjp1b/ZO1b-/- mutant (lanes 3,4) animals was immunoprecipitated with control whole mouse IgG (lanes 1,3) or anti-ZO1 antibody (lanes 2,4). Immunoprecipitates were analyzed by immunoblot for the presence of ZO1 using anti-ZO1 antibody (top, magenta), Cx34.1 using Cx34.1-specific antibody (middle, yellow), and Cx35.5 using Cx35.5-specific antibody (bottom, cyan). Asterisks (*) indicate antibody light chain. Results are representative of three independent experiments.
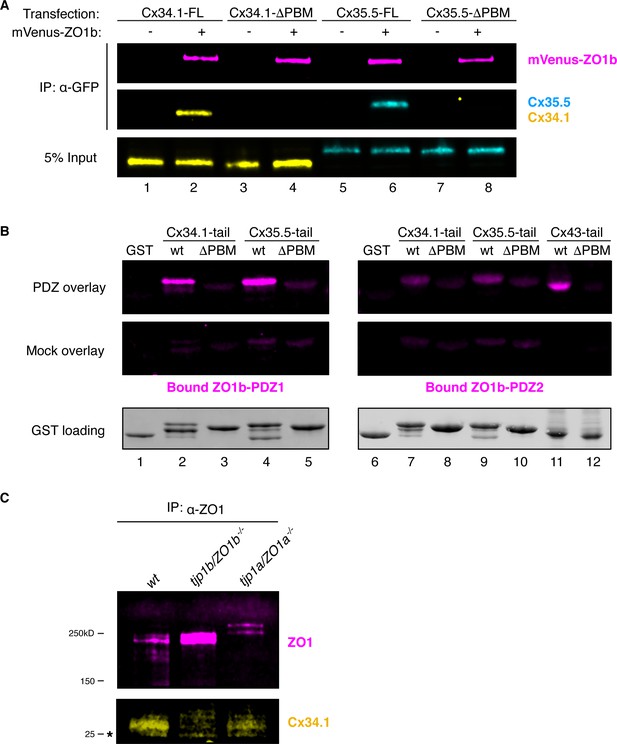
Biochemical characterization of ZO and Connexin interactions.
(A) HEK293T/17 cells were transfected with plasmids to express full-length Cx34.1 (lanes 1,2), Cx34.1-∆PBM (lanes 3,4), full-length Cx35.5 (lanes 5,6), or Cx35.5-∆PBM (lanes 7,8) together with empty vector (-) or mVenus-ZO1b (+). Lysates were immunoprecipitated with anti-GFP antibody and analyzed by immunoblot for the presence of mVenus-ZO1b using anti-GFP antibody (upper, magenta), Cx34.1 protein using Cx34.1-specific antibody (middle, yellow), or Cx35.5 protein using Cx35.5 specific antibody (middle, cyan). Total extracts (bottom, 5% input) were blotted for Connexin proteins to demonstrate equivalent expression and uniform antibody recognition of expressed proteins. Results are representative of three independent experiments. (B) Direct interaction of Connexin PBMs and the ZO1b PDZ1 domain by overlay assay. Bacterially purified GST (lanes 1,6), GST-Cx34.1-tail (lanes 2,7), GST-Cx34.1-tail-∆PBM (lanes 3,8), GST-Cx35.5-tail (lanes 4,9), GST-Cx35.5-tail-∆PBM (lanes 5,10), GST-Cx43-tail (lane 11), and GST-Cx43-tail-∆PBM were resolved by SDS-PAGE and transferred to nitrocellulose membrane. Membranes were incubated with 1 µM ZO1b PDZ1 domain (top left), 1 µM ZO1b PDZ2 domain (top right) or buffer alone (middle, mock overlay). Bound proteins were analyzed by immunoblot for the presence of ZO1b PDZ using anti-TEV cleavage site antibody (top and middle). Equal loading of GST proteins is indicated by Stain-Free technology (bottom). Results are representative of three independent experiments. (C) Zebrafish brain extract from wt (lane 1), tjp1b/ZO1b-/- (lane 2), or tjp1a/ZO1a-/- (lane 3) animals were immunoprecipitated with anti-ZO1 antibody. Immunoprecipitates were analyzed by immunoblot for the presence of ZO1 using anti-ZO1 antibody (top, magenta) or Cx34.1 using Cx34.1-specific antibody (bottom, yellow). Asterisk (*) indicates antibody light chain. Results are from one independent experiment.
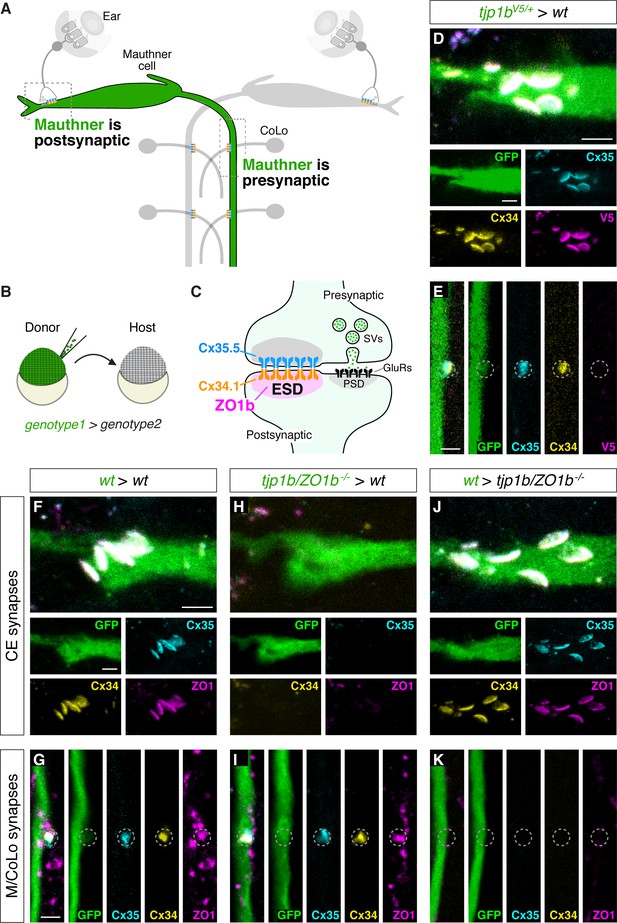
ZO1b localizes and functions postsynaptically.
(A) Schematic of the Mauthner circuit in chimeric animals. One Mauthner cell is derived from the GFP-expressing donor (green), while other neurons derive from the non-transgenic host (gray). The image represents a dorsal view with anterior to the top. Electrical synapses denoted as yellow (Cx34.1) and cyan (Cx35.5) rectangles. Boxed regions indicate regions imaged for analysis. (B) Diagram of experiment in which GFP-expressing donor cells are transplanted into a non-transgenic host to create chimeric embryos. GFP-expressing cells are of genotype1 while the rest of the cells in the chimeric embryo are derived from genotype2. (C) Diagram of a mixed electrical/chemical (glutamatergic) synapse summarizing data for ZO1b. ESD = electrical synapse density, see Discussion. (D–K) Confocal images of Mauthner circuit neurons and stereotypical electrical synaptic contacts in 5-day-post-fertilization, chimeric zebrafish larvae. Animals are stained with anti-GFP (green), anti-zebrafish-Cx35.5 (cyan), and anti-zebrafish-Cx34.1 (yellow). In panels (D–E), animals are stained with anti-V5 (magenta), and in (F–K) animals are stained with anti-human-ZO1 (magenta). The genotype of the donor cell (green, genotype1) and host (genotype2) varies and is noted above each set of images (genotype1 > genotype2). Images of CEs (D,F,H,J) are maximum-intensity projections of ~5 µm. Images of M/CoLo synapses (E,G,I,K) are single Z-slices. Neighboring panels show individual channels. Scale bar = 2 µm in all images.
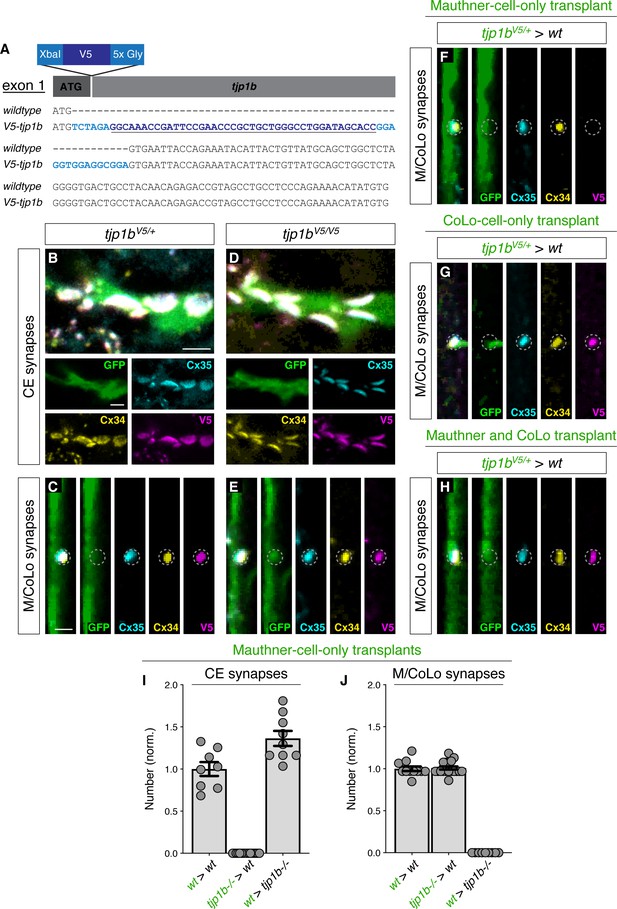
Characterization of ZO1b localization and function.
(A) Schematic and verified exon 1 sequence of V5 epitope tag insertion into the N-terminus of the endogenous tjp1b locus. Bases in dark blue indicate V5 sequence. Light blue bases indicate the XbaI restriction enzyme cut site and the 5x glycine linker sequence. (B–H) Confocal images of Mauthner circuit neurons and stereotypical electrical synaptic sites of formation in 5-day-post-fertilization, V5-tjp1b heterozygous (B,C) and V5-tjp1b homozygous (D,E), and chimeric (F–H) zebrafish larvae. For chimeric larvae, the genotype of the donor cell (green, genotype1) and host (black, genotype2) varies and is noted above each set of images (genotype1 > genotype2). Animals are stained with anti-GFP (green), anti-zebrafish-Cx35.5 (cyan), anti-zebrafish-Cx34.1 (yellow), and anti-V5 (magenta). Images have been passed through a 3 × 3 median filter. Scale bar = 2 µm in all images. (F–H) Confocal images of M/CoLo sites of contact in 5-day-post-fertilization chimeric animals. In each set of images, GFP-expressing cells (Mauthner (F), CoLo (G), or both Mauthner and CoLo (H)) are derived from a V5-tjp1b+/-; zf206Et donor. Animals are stained with anti-GFP (green), anti-zebrafish-Cx35.5 (cyan), anti-zebrafish-Cx34.1 (yellow), and anti-V5 (magenta). (I–J) Quantification of the number of stereotypical electrical synaptic structures labeled with both anti-Cx34.1 and anti-Cx35.5 in chimeric animals of the indicated genotypes with a GFP-positive Mauthner cell. The height of the bar represents the mean of the sampled data normalized to the wt>wt average, and circles represent the normalized value of each individual animal (CE synapses: wt>wt n = 8, tjp1b/ZO1b-/->wt n = 16, wt>tjp1b/ZO1b-/- n = 9; M/CoLo synapses: wt>wt n = 11, tjp1b/ZO1b-/->wt n = 18, wt>tjp1b/ZO1b-/- n = 9). Error bars are ± SEM. Associated experimental statistics can be found in Figure 7—figure supplement 1—source data 1.
-
Figure 7—figure supplement 1—source data 1
Source data for Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/66898/elife-66898-fig7-figsupp1-data1-v1.xlsx
Tables
Mauthner cell electrophysiological properties from wildtype (wt), Connexin, and ZO-1 mutant zebrafish, Related to Figure 4.
Average measurements of resting potential (Vrest), firing threshold (Vthreshold), input resistance (Rin) and Rheobase obtained in Mauthner cells of wt, tjp1b/ZO1b-/-, tjp1a/ZO1a-/-, gjd2a/Cx35.5-/-, gjd1a/Cx34.1-/-, gjd2b/Cx34.7-/-, and gjd1b/Cx35.1-/- zebrafish. Each ‘n’ represents a fish (only one Mauthner cell was recorded in each fish). Associated experimental statistics can be found in Table 1—source data 1.
| Ephys. Prop. | wt (n = 10) | tjp1b-/- (n = 8) | tjp1a-/- (n = 5) | gjd2a-/- (n = 6) | gjd1a-/- (n = 9) | gjd2b-/- (n = 5) | gjd1b-/- (n = 6) |
|---|---|---|---|---|---|---|---|
| Vrest (mV) | −83.5 ± 1.6 | −73.7 ± 2.2 | −85.8 ± 1.8 | −89.3 ± 2.4 | −86.9 ± 1.3 | −83.9 ± 0.9 | −84.6 ± 1.5 |
| Vthreshold (mV) | −59.1 ± 2.3 | −46.5 ± 1.7 | −55.3 ± 3.3 | −61.8 ± 2.2 | −60.5 ± 1.3 | −56.5 ± 3.6 | −58.3 ± 2.9 |
| Rin (MOhm) | 6.1 ± 0.8 | 45.4 ± 8.6 | 11.7 ± 1.3 | 42.0 ± 2.3 | 11.8 ± 0.9 | 5.5 ± 0.6 | 5.5 ± 0.7 |
| Rheobase (nA) | 4.0 ± 0.4 | 1.5 ± 0.2 | 2.7 ± 0.4 | 1.2 ± 0.2 | 2.6 ± 0.2 | 4.4 ± 0.1 | 4.9 ± 0.6 |
-
Table 1—source data 1
Source data for Table 1.
- https://cdn.elifesciences.org/articles/66898/elife-66898-table1-data1-v1.xlsx
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Danio rerio) | gjd1a | ZFIN | ZDB-GENE-080723–77 | |
| Gene (Danio rerio) | gjd1b | ZFIN | ZDB-GENE-100921–89 | |
| Gene (Danio rerio) | gjd2a | ZFIN | ZDB-GENE-111020–17 | |
| Gene (Danio rerio) | gjd2b | ZFIN | ZDB-GENE-030911–1 | |
| Gene (Danio rerio) | tjp1a | ZFIN | ZDB-GENE-031001–2 | |
| Gene (Danio rerio) | tjp1b | ZFIN | ZDB-GENE-070925–1 | |
| Strain, strain background (Danio rerio) | AB x Tübingen | University of Oregon fish facility | ZFIN: ZDB-GENO-010924–10; PubMed: PMC4667794 | |
| Strain, strain background (Danio rerio) | M/CoLo:GFP (zf206Et) | Satou et al., 2009 | ZFIN: ZDB-ALT-110217–6; PubMed: 19474306 | |
| Strain, strain background (Danio rerio) | tjp1bΔ16bp (fh448) | Shah et al., 2015 | ZFIN: ZDB-ALT-160825–6; PubMed: PMC4667794 | |
| Strain, strain background (Danio rerio) | tjp1aΔ2bp (fh463) | Marsh et al., 2017 | ZFIN: ZDB-ALT-180920–6; Pubmed: PMC5698123 | |
| Strain, strain background (Danio rerio) | gjd1adis3 (fh360) | Miller et al., 2017 | ZFIN: ZDB-ALT-160825–2: Pubmed: PMC5462537 | |
| Strain, strain background (Danio rerio) | gjd2a∆5bp (fh437) | Shah et al., 2015 | ZFIN: ZDB-TALEN-170822–4; Pubmed: PMC4667794 | |
| Strain, strain background (Danio rerio) | gjd1a ∆8bp (fh436) | Shah et al., 2015 | ZFIN: ZDB-TALEN-170822–3; Pubmed: PMC4667794 | |
| Strain, strain background (Danio rerio) | V5-tjp1b (b1406) | This paper | N/A | See Materials and methods |
| Strain, strain background (Homo sapiens) | HEK293T/17 cells | ATCC | CRL-11268 | |
| Strain, strain background (Escherichia coli) | BL21(DE3) | New England BioLabs | C2527I | |
| Strain, strain background (Escherichia coli) | DH5alpha | Zymo Research | T3009 | |
| Antibody | Chicken monoclonal anti-GFP IgY | Abcam | ab13970; RRID:AB_300798 | (1:500) |
| Antibody | Rabbit monoclonal anti-GFP | Abcam | Ab290 | (1:1000) |
| Antibody | Mouse monoclonal IgG1 anti-ZO1 | ThermoFisher | 33–9100; RRID: AB_2533147 | (1:350) |
| Antibody | Rabbit monoclonal anti-Cx35.5 | Miller et al., 2015 | clone 12H5 | (1:800) |
| Antibody | Rabbit monoclonal anti-Cx35.5-IRDye 680LT conjugated | Fred Hutch Antibody Technology Facility, Miller lab conjugated | clone 12H5 | (1:1000) |
| Antibody | Mouse monoclonal IgG1 anti-Cx35.5 | Miller et al., 2015 | clone 4B12a | (1:1000) |
| Antibody | Rabbit monoclonal anti-Cx34.1 | Miller et al., 2015 | clone 3A4 | (supernatant) |
| Antibody | Rabbit monoclonal anti-Cx34.1-680LT conjugated | Fred Hutch Antibody Technology Facility, Miller lab conjugated | clone 3A4 | (supernatant) |
| Antibody | Mouse monoclonal IgG2A anti-Cx34.1 | Miller et al., 2015 | clone 5C10A | (1:200) |
| Antibody | Rabbit monoclonal anti -GluR2/3 | Millipore Sigma | 07–598; RRID:AB_11213931 | (1:250) |
| Antibody | Rabbit monoclonal anti-TEV Cleavage Site | Invitrogen | PA1-119; RRID: AB_2539888 | (1:2000) |
| Antibody | Goat monoclonal anti-rabbit Alexa 405 | Invitrogen | A31556; RRID:AB_221605 | (1:500) |
| Antibody | Donkey monoclonal anti-chicken IgGY Alexa 488 | Jackson Immuno Research Laboratories | 703-545-155 | (1:500) |
| Antibody | Goat monoclonal anti-mouse IgG2a Alexa 555 | Invitrogen | A21137 | (1:500) |
| Antibody | Goat monoclonal anti-mouse IgG1 Alexa 633 | Invitrogen | A21126 | (1:500) |
| Antibody | Mouse monoclonal IgG2a anti-V5 peptide | Invitrogen | R960-25 | (1:1000) |
| Antibody | ChromPure mouse monoclonal IgG, whole molecule | Jackson ImmunoResearch Laboratories | 015-000-003; RRID: AB_2337188 | (1:1000) |
| Antibody | IRDye 680LT goat monoclonal anti-rabbit secondary | LI-COR | 925–68021; RRID: AB_2713919 | (1:10000) |
| Antibody | IRDye 800CW goat monoclonal anti-mouse secondary | LI-COR | 925–32210; RRID: AB_2687825 | (1:10000) |
| Antibody | Mouse monoclonal IgG kappa binding protein (m-IgGκ BP) conjugated to CruzFluor 790 (CFL 790) secondary | Santa Cruz Biotechnology | sc-516181 | (1:10000) |
| Recombinant DNA reagent | pCMV mammalian expression plasmid | J. O’Brien lab | N/A | |
| Recombinant DNA reagent | pGEX bacterial expression plasmid | K. Prehoda lab | N/A | |
| Recombinant DNA reagent | pBH bacterial expression plasmid | K. Prehoda lab | N/A | |
| Sequence-based reagent | tjp1bΔ16bp genotyping primers: Fwd, TCTCTTTCCTTCTTTCTGTGTGTTT; Rev, AAAAGTGAAATTCTCACCCTGTG | Marsh et al., 2017 | N/A | |
| Sequence-based reagent | gjd2a∆5bp genotyping primers: Fwd, GATGAGCAGCGATGGGAGAAT; Rev, CTTGAATTTCGGCGTCAGACAG | Miller et al., 2015 | N/A | |
| Sequence-based reagent | gjd1a ∆8bp genotyping primers: Fwd, CTCAGGCTGAAGGTCGGCAGGGAAG; Rev, GCTGTACCGCAGCCTCCAGCAAC | Miller et al., 2015 | N/A | |
| Sequence-based reagent | gjd1adis3 genotyping primers: Fwd, AGTGCGACCGCTACCCTTGC; Rev, AGCACCACGCAGATTCCGCT, | Miller et al., 2015 | N/A | |
| Sequence-based reagent | tjp1aΔ2bp genotyping primers: Fwd, GTACAACAATGGAGGAAACTGTCA; Rev, AAAGAAGCTATGTTCAACACTCACC | Marsh et al., 2017 | N/A | |
| Sequence-based reagent | tjp1b N-terminus Crispr target: GGATTTCTGGTAATTCACCA | This paper | N/A | See Materials and Methods |
| Sequence-based reagent | tjp1b N-terminus oligo: GAGCCAGCTGCATAACAGTAATGTATTTCTGGTAATTCACTCCGCCTCCACCTCCGGTGCTATCCAGGCCCAGCAGCGGGTTCGGAATCGGTTTGCCTCTAGACATGGTACTGTTCACCGCTTTTTTGAAACACAAAAATCCGCA | This paper | N/A | See Materials and Methods |
| Sequence-based reagent | V5-tjp1b screening primers: Fwd, GGGAGTAGGAGGAGAAGGA; Rev, GTTTTCTGGGAGGCAGGCTA | This paper | N/A | See Materials and Methods |
| Peptide, recombinant protein | GST-Cx34.1 (aa 256–299) | This paper | GST-Cx34.1-tail wt | See Materials and Methods |
| Peptide, recombinant protein | GST-Cx34.1 (aa 256–295) | This paper | GST- Cx34.1-tail ∆PBM | See Materials and Methods |
| Peptide, recombinant protein | GST-Cx35.5 (aa 267–309) | This paper | GST-Cx35.5-tail wt | See Materials and Methods |
| Peptide, recombinant protein | GST-Cx35.5 (aa 267–305) | This paper | GST- Cx35.5-tail ∆PBM | See Materials and Methods |
| Peptide, recombinant protein | 6xHIS-TEV cleavage site-ZO1b (aa105-207) | This paper | ZO1b PDZ1 | See Materials and Methods |
| Peptide, recombinant protein | 6xHIS-TEV cleavage site-ZO1b (aa 298–387) | This paper | ZO1b PDZ2 | See Materials and Methods |
| Peptide, recombinant protein | mVenus-ZO1b (aa 2–1778)−8xHIS | This paper | mVenus-ZO1b | See Materials and Methods |
| Peptide, recombinant protein | Cx34.1 (aa 1–299) | This paper | Cx34.1-FL | See Materials and Methods |
| Peptide, recombinant protein | Cx34.1 (aa 1–295) | This paper | Cx34.1-∆PBM | See Materials and Methods |
| Peptide, recombinant protein | Cx35.5 (aa 1–309) | This paper | Cx35.5-FL | See Materials and Methods |
| Peptide, recombinant protein | Cx35.5 (aa 1–305) | This paper | Cx35.5-∆PBM | See Materials and Methods |
| Commercial assay or kit | Taq 2X Master Mix | NEB | M0270L | |
| Commercial assay or kit | Universal Mycoplasma Detection Kit | ATCC | 30–1012K | |
| Chemical compound, drug | ProLong Gold antifade reagent | ThermoFisher | P36930 | |
| Chemical compound, drug | n-Octyl-β-D-Glucopyranoside, Anagrade | Anatrace | O311 | |
| Chemical compound, drug | Protease Inhibitor Mini Tablets, EDTA-free | Pierce | A32955 | |
| Chemical compound, drug | Lipofectamine 3000 | Invitrogen | L3000008 | |
| Chemical compound, drug | Dulbecco's Modified Eagle's Medium (DMEM) | ATCC | 30–2002 | |
| Chemical compound, drug | Opti-MEM | Gibco | 31-985-062 | |
| Chemical compound, drug | Glutathione resin | Pierce | PI16100 | |
| Chemical compound, drug | His60 Ni Superflow resin | TaKaRa | 635659 | |
| Chemical compound, drug | Protein A/G Agarose | Pierce | PI20421 | |
| Chemical compound, drug | 4–15% Criterion TGX Stain-Free Protein Gel | BioRad | 5678083 | |
| Chemical compound, drug | 4–20% Mini-PROTEAN TGX Stain-Free Protein Gels | BioRad | 4568095 | |
| Chemical compound, drug | (+)-Tubocurarine chloride pentahydrate | Sigma | 93750 | |
| Chemical compound, drug | Ethyl 3-aminobenzoate methanesulfonate salt | Sigma | A5040 | |
| Chemical compound, drug | Sodium chloride | Sigma | 567440 | |
| Chemical compound, drug | Potassium chloride | Sigma | P3911 | |
| Chemical compound, drug | Calcium chloride | Sigma | 21115 | |
| Chemical compound, drug | Magnesium chloride | Sigma | M1028 | |
| Chemical compound, drug | HEPES | Sigma | H3375 | |
| Chemical compound, drug | D-(+)-Glucose | Sigma | G8270 | |
| Chemical compound, drug | Sodium hydroxide | Sigma | S5881 | |
| Chemical compound, drug | Potassium methanesulfonate | Sigma | 83000 | |
| Chemical compound, drug | EGTA | Sigma | E3889 | |
| Chemical compound, drug | Adenosine 5′-phosphosulfate sodium salt | Sigma | A5508 | |
| Chemical compound, drug | Guanosine 5′-triphosphate tris salt | Sigma | G9002 | |
| Chemical compound, drug | Creatine Phosphate, Dipotassium Salt | Sigma | 237911 | |
| Chemical compound, drug | D-Mannitol | Sigma | M4125 | |
| Chemical compound, drug | Potassium Hydroxide | Sigma | P5958 | |
| Chemical compound, drug | Meclofenamic acid sodium salt | Sigma | M4531 | |
| Chemical compound, drug | Dimethyl sulfoxide | Sigma | D8418 | |
| Chemical compound, drug | CNQX | Tocris | 0190 | |
| Chemical compound, drug | DAP-5 | Tocris | 0106 | |
| Chemical compound, drug | Capillary Glass 1.5 mm OD, 1.12 mm ID | WPI | TW150F-3 | |
| Chemical compound, drug | SYLGARD 184 Silicone Elastomer Kit | DOW | https://www.dow.com/en-us/pdp.sylgard-184-silicone-elastomer-kit.01064291z.html | |
| Software, algorithm | GraphPad Prism | Graph Pad Software | https://www.graphpad.com/ | |
| Software, algorithm | Adobe Photoshop CC 2015 | Adobe | https://www.adobe.com/ | |
| Software, algorithm | Adobe Illustrator CC 2015 | Adobe | https://www.adobe.com/ | |
| Software, algorithm | scikit-image | van der Walt et al., 2014 | https://peerj.com/articles/453/ | |
| Software, algorithm | SciPy | Virtanen et al., 2020 | https://www.nature.com/articles/s41592-019-0686-2?luicode=10000011&lfid=1008082086c7dfebc09fc300733002ea997ba2_-_feed&u=https%3A%2F%2Fwww.nature.com%2Farticles%2Fs41592-019-0686-2 | |
| Software, algorithm | FiJi | Schindelin et al., 2012 | PubMed: 22743772; https://fiji.sc/ | |
| Software, algorithm | OriginPro | OriginLab Corp. | https://www.originlab.com/ | |
| Software, algorithm | Clampex | Molecular Devices | ||
| Other | Leica TCS SP8 Confocal | Leica | http://www.leica-microsystems.com/products/confocal-microscopes/details/product/leica-tcs-sp8/ | |
| Other | 40X/1.10 Water Objective | Leica | 11506357 | |
| Other | 63X/1.40 Oil Objective | Leica | 15506350 | |
| Other | Amicon Ultra-0.5 Centrifugal Filter Units, 10K MWCO | MilliporeSigma | UFC501008 | |
| Other | Amicon Ultra-4 Centrifugal Filter Units 10 kDa MWCO | MilliporeSigma | UFC801008 | |
| Other | Upright Axio Examiner Microscope | Carl Zeiss Microscopy, LLC | https://www.zeiss.com/corporate/int/home.html?vaURL=www.zeiss.de/en | |
| Other | 20X/0.5 W-N-Achroplan | Carl Zeiss Microscopy, LLC | 420957–9900 | |
| Other | 40X/1.0 VIS-IR W-Plan-Apochromatic | Carl Zeiss Microscopy | 421462–9900 | |
| Other | Multiclamp 700B amplifier | Molecular Devices | https://www.moleculardevices.com/ | |
| Other | Digidata 1440A | Molecular Devices | https://www.moleculardevices.com/ |





