Ciliary and extraciliary Gpr161 pools repress hedgehog signaling in a tissue-specific manner
Figures
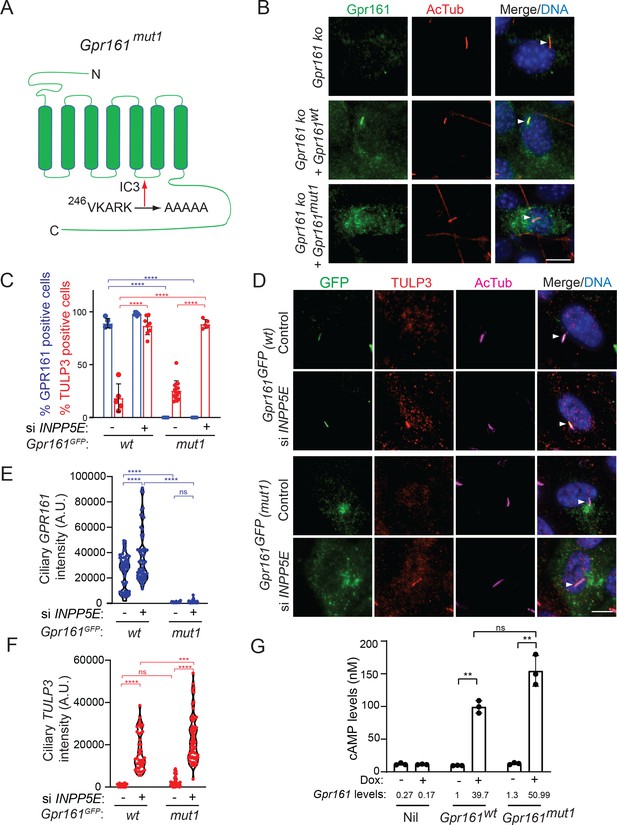
A ciliary localization defective Gpr161 mutant is competent for cAMP signaling.
(A) Cartoon representing the VKARK>AAAAA mut1 mutation in the third intracellular loop of mouse Gpr161. (B) Confluent NIH 3T3 Flp-In CRISPR-based Gpr161 knockout (ko) cells stably expressing untagged wildtype (wt) or Gpr161mut1 were starved for 24 hr, fixed, and immunostained with anti-Gpr161 (green), anti-acetylated tubulin (AcTub; red) antibodies and counterstained for DNA (blue). Arrowheads indicate cilia. (C – D) RPE hTERT cells stably expressing C-terminal LAP-tagged Gpr161wt or Gpr161mut1 constructs were sequentially transfected with control or INPP5E siRNA (100 nM) twice and cultured for a total of 72 hr. The cells were serum starved for the last 24 hr before fixation and immunostained with anti-GFP (green), anti-TULP3 (red), anti-acetylated tubulin (AcTub; magenta) antibodies and counterstained for DNA (blue). GPR161 and TULP3 positive cells were quantified. Arrowheads in (D) indicate cilia. Total 6–12 different images quantified from two experiments, and total 600–2000 cells counted/condition. Data shown as mean ± SD. ****, p<0.0001. Other pairwise comparisons are not significantly different. (E – F) Quantification of LAP-tagged Gpr161 (E) and TULP3 (F) pixel intensities from C shown as violin plots. Total counted cells were >60/condition. A.U., arbitrary units. ****, p<0.0001; ***, p p<0.001. (G) Doxycycline-inducible NIH 3T3 Tet-on 3G cells expressing untagged Gpr161wt or Gpr161mut1 were induced for 24 hr with 2 μg/ml doxycycline. The cells were subjected to TR-FRET cell-based assays for assaying cAMP. cAMP levels (nM) were calculated as interpolated values from a standard curve. Data from triplicate wells (mean ± SD) and is representative of 3 independent experiments. Mean Gpr161 transcript levels are shown below. **, p<0.01. ns, not significant. Scale: (B and D), 10 µm.
-
Figure 1—source data 1
Table showing data from experiments plotted in Figure 1C.
- https://cdn.elifesciences.org/articles/67121/elife-67121-fig1-data1-v2.xlsx
-
Figure 1—source data 2
Table showing cilia intensity data from plotted in Figure 1E and F.
- https://cdn.elifesciences.org/articles/67121/elife-67121-fig1-data2-v2.xlsx
-
Figure 1—source data 3
Table showing cAMP data plotted in Figure 1G.
- https://cdn.elifesciences.org/articles/67121/elife-67121-fig1-data3-v2.xlsx

Gpr161mut1 was not accumulated in cilia of β−arrestin 1/2 double knockout MEFs.
Wild type or β−arrestin 1/2 double knockout (Arrb1/Arrb2 dko) MEFs stably expressing LAP-tagged Gpr161wt or Gpr161mut1 were starved for 24 hr followed by SAG (500 nM) treatment for further 24 hr. Cells were fixed and immunostained with anti-GFP (green), anti-Gpr161 (red), anti-acetylated tubulin (AcTub; magenta) antibodies and counterstained for DNA (blue). Quantification below (mean ± SD) shows that Gpr161mut1 was not accumulated in cilia of Arrb1/Arrb2 dko MEFs irrespective of Hh pathway activation by SAG, despite accumulation of endogenous Gpr161 in the same cells. Arrows depict cilia. Scale, 10 μm.
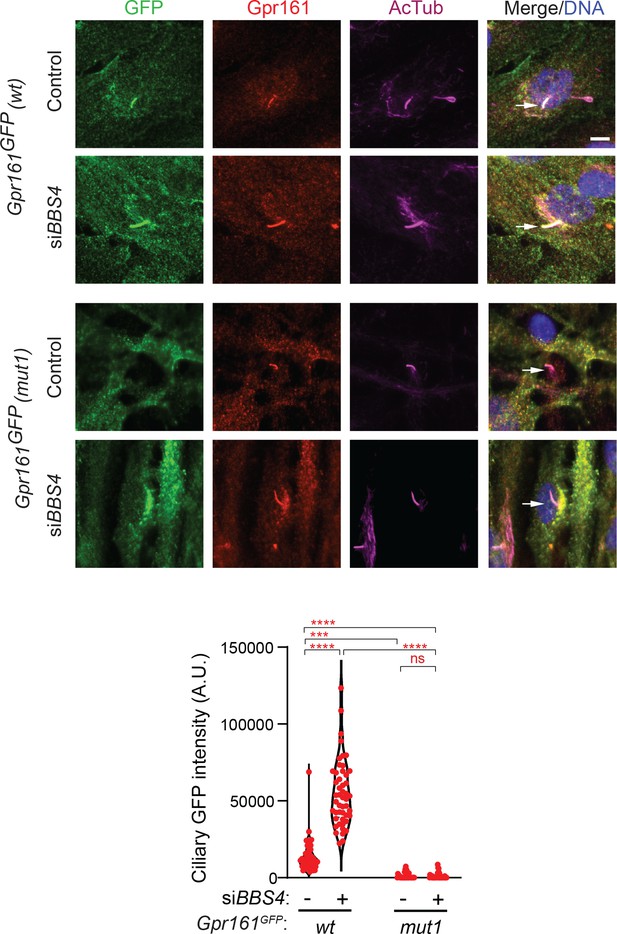
Gpr161mut1 was not accumulated in cilia upon BBS4 knockdown in RPE-hTERT cells.
RPE-hTERT cells stably expressing LAP-tagged Gpr161wt or Gpr161mut1 were treated with 100 nM siRNA against BBS4 for 48 hr before starving for 24 hr. Fixed cells were immunostained with anti-GFP (green), anti-Gpr161 (red), anti-acetylated tubulin (AcTub; magenta) antibodies and counterstained for DNA (blue). Violin plot of ciliary intensities shows that LAP-tagged Gpr161wt accumulated in cilia upon BBS4 knockdown in RPE-hTERT cells, but LAP-tagged Gpr161mut1 did not accumulate despite accumulation of endogenous Gpr161 in the same cells. Arrows depict cilia. ****, p<0.0001; ***, p<0.001; ns, not significant. Scale bar, 10 µm.
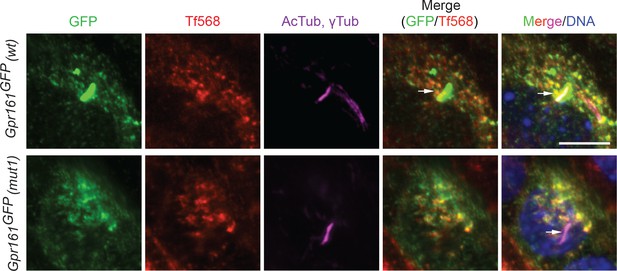
Gpr161mut1 containing vesicles co-label with endocytosed transferrin similar to Gpr161wt.
Wild-type MEFs stably expressing LAP-tagged Gpr161wt or Gpr161mut1 were starved for 48 hr followed by treatment with Transferrin conjugated with Alexa Fluor 568 (Tf568) or 30 min at 37°C. Fixed cells were immunostained with anti-GFP (green), anti-acetylated tubulin and γ-tubulin (AcTub; γTub in magenta) antibodies and counterstained for DNA. We previously demonstrated that Gpr161 containing vesicles to be recycling endosomes that co-label with endocytosed transferrin (Mukhopadhyay et al., 2013). Arrows point to cilia. Similar to LAP-tagged Gpr161wt, LAP-tagged Gpr161mut1 containing vesicles co-labeled with endocytosed transferrin. Scale bar, 10 µm.
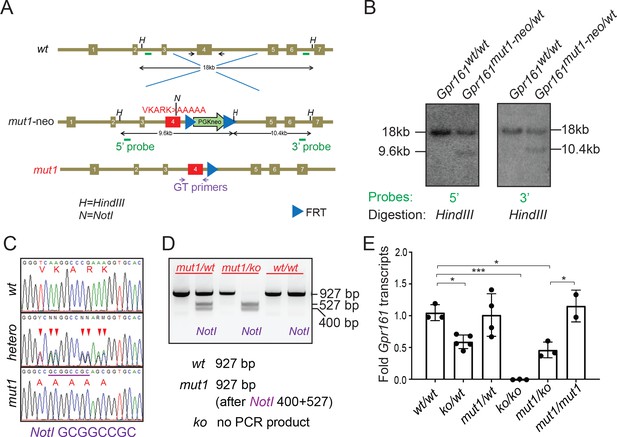
Generating ciliary localization defective endogenous knock-in Gpr161mut1 mouse model.
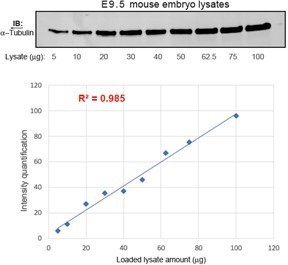
(A) The gene targeting strategy used to engineer the Gpr161mut1 allele. Exons are numbered based on NM_001081126.2. PGKneo and FRT cassettes and genotyping (GT) primer sequences are indicated. The mut1 sequence is located on Exon 4. (B) Southern blot analysis of representative ES cell clones using the 5’ and 3’ probes in A. (C) Sanger sequencing of Gpr161wt and Gpr161mut1 alleles in adult mouse-tail DNA. Double peaks in Gpr161wt/mut1 heterozygote indicated by arrowheads. The engineered NotI restriction site (GCGGCCGC) is indicated by a purple bar. (D) Genotyping for wild type, Gpr161mut1 and knockout (ko) alleles by PCR using designated primers shown in A and digesting with NotI. (E) qRT-PCR of Gpr161 transcripts normalized to Hprt in whole embryo extracts at E9.5 indicate diminished mRNA expression in the Gpr161 knockout (ko/ko) embryos compared to wild type (wt/wt), but unchanged in mut1/mut1 embryos. Data shown as mean ± SD. n=3 (wt/wt), 5 (ko/wt), 4 (mut1/wt), 3 (ko/ko), 3 (ko/mut1), 2 (mut1/mut1) embryos. *, p<0.05; ***, p<0.001. Other pairwise comparisons are not significantly different.
-
Figure 2—source data 1
Table showing transcript data plotted in Figure 2E.
- https://cdn.elifesciences.org/articles/67121/elife-67121-fig2-data1-v2.xlsx
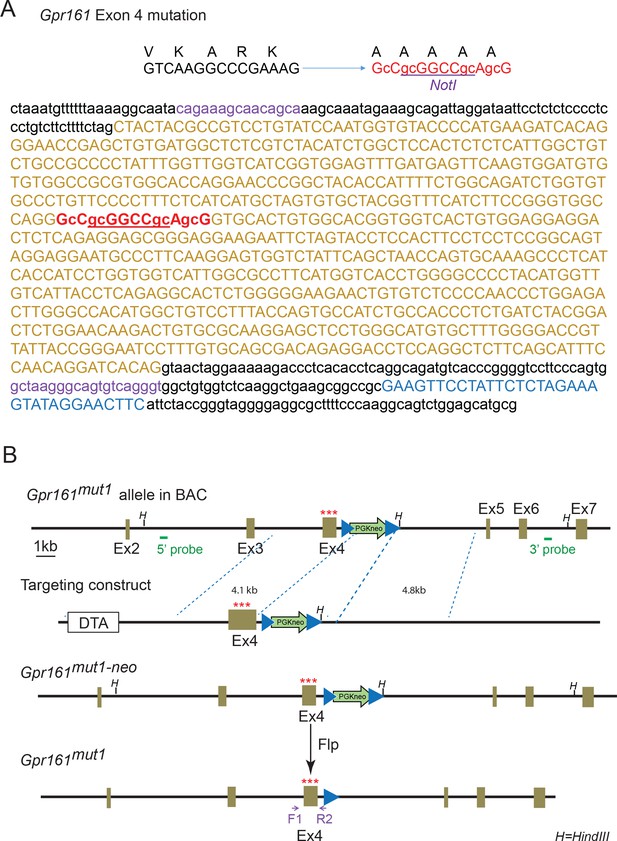
Genomic DNA sequence and scheme of Gpr161mut1 allele.
(A) Genomic DNA sequence of Gpr161mut1 allele, after deletion of the FRT-PGKneo-FRT cassette by crossing with Flp-O mice. (B) Scheme for generating mut1 allele. Details in Materials and methods.
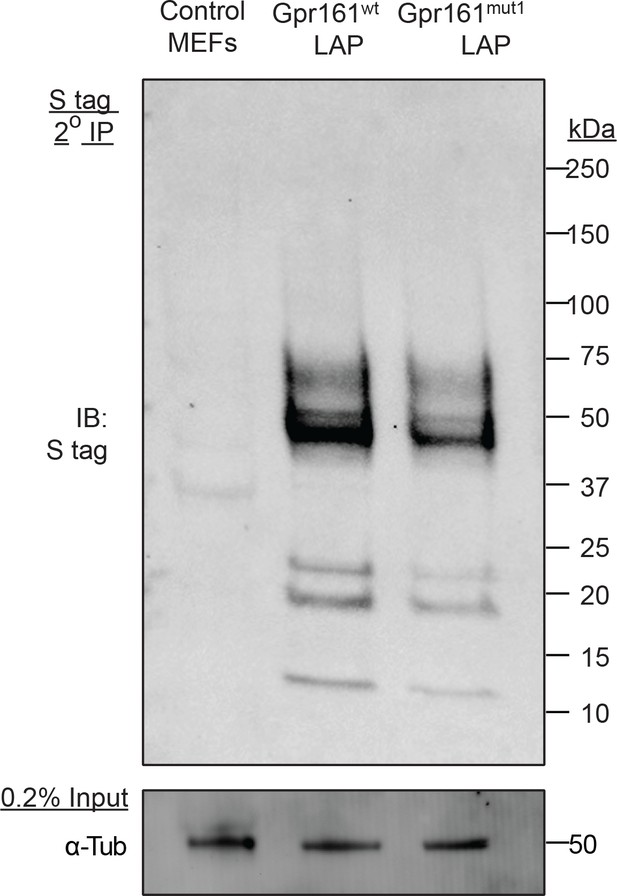
Tandem affinity purification of LAP-tagged Gpr161wt or Gpr161mut1 (see Materials and methods) stably expressed in wild type MEFs was followed by immunoblotting for S-tag.
Inputs show lysates before tandem affinity purification that were immunoblotted for α-tubulin.
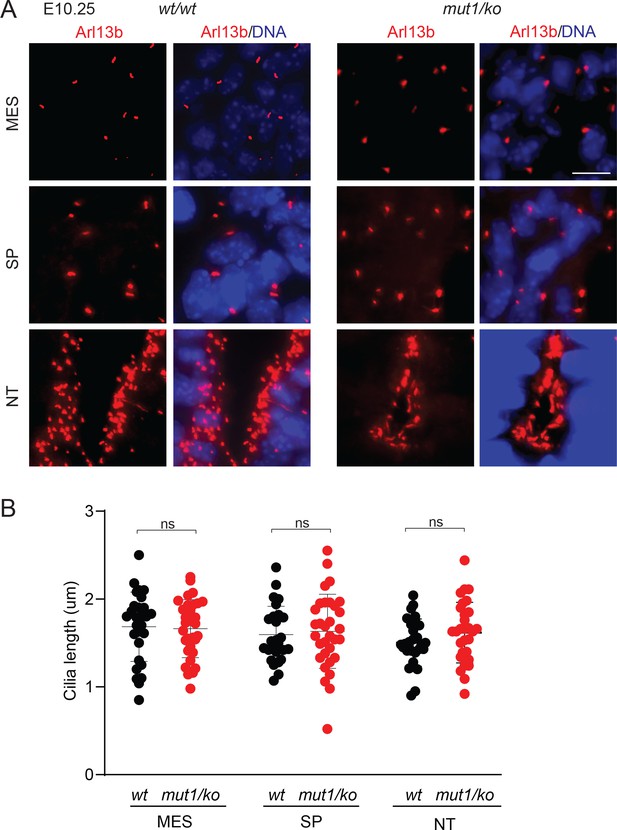
The ciliary morphologies (A), including ciliary lengths (B) in Gpr161ko/mut1 embryos at ~E10.25 in mesenchyme (MES), somatopleuric mesoderm (SP) or in the neural tube (NT) were unaffected compared to control wild type (wt) embryos.
Thorcaic level sections were counterstained with Arl13b (red) and DAPI (blue). Scale, 10 μm. Data shown as mean ± SD. Total counted cilia were >30.
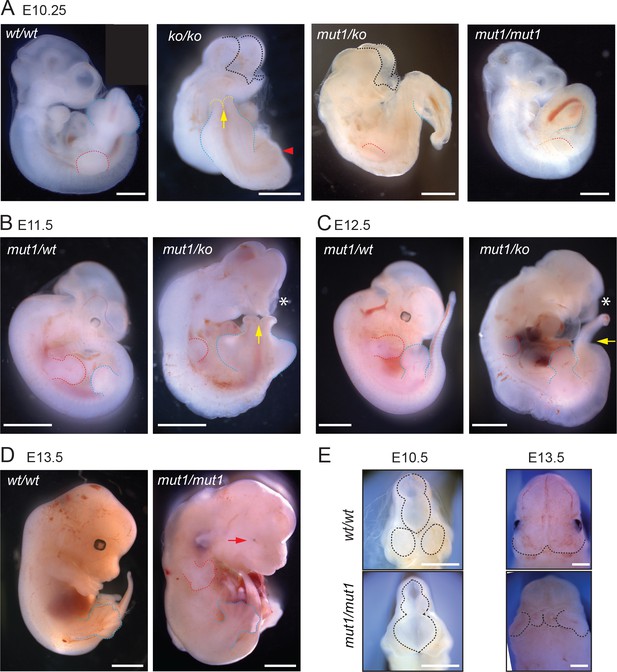
Gpr161mut1 allele is hypomorphic to knockout.
(A – F) Bright-field images of wild type (wt/wt), Gpr161 knockout (ko/ko), mut1/ko heterozygote, and mut1 homozygote (mut1/mut1) at indicated time points. Red arrowhead indicates no limb bud in knockout embryo. En face view of E10.5 and E13.5 embryos in (E). Black dotted line or asterisk, rostral malformation; yellow arrow and dotted line, spina bifida; red dotted line, forelimb; blue dotted line, hindlimb. See also Tables 1 and 2. Scale: (A), 1 mm; (B–D), 2 mm; (D); (E), 1 mm.
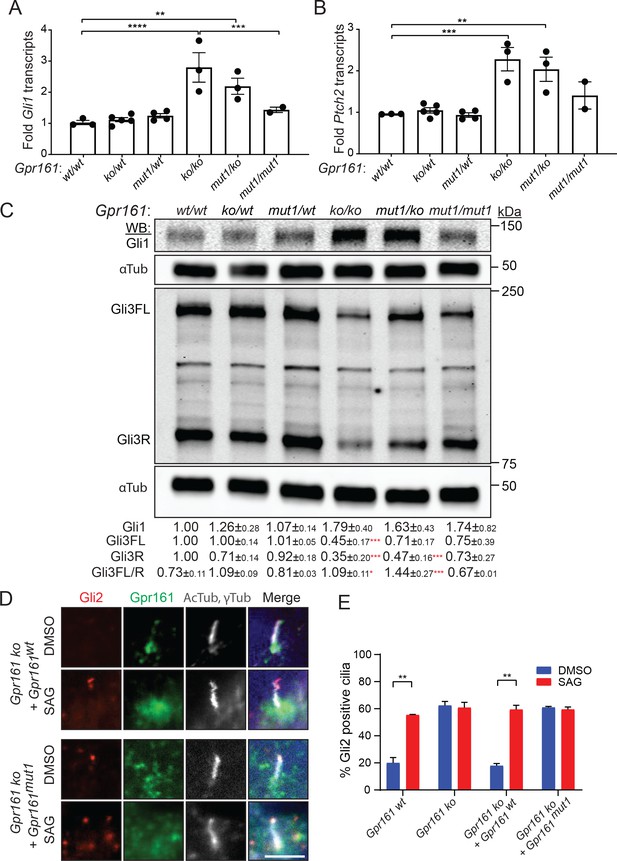
Gpr161mut1/ko embryos have reduced Hh pathway hyperactivation compared to knockouts.
(A – B) Gli1 (A) and Ptch2 (B) transcript levels in whole-embryo extracts at E9.5 by qRT-PCR, normalized to Gapdh, n = 3–4 embryos each, data shown as mean ± SEM. **, p<0.01, ***, p<0.001, ****, p<0.0001. Only significant differences are marked. (C) Immunoblotting for Gli1, Gli3 and α-tubulin in whole-embryo lysates at E9.5. n=2 or 3 independent experiments for Gli1 or Gli3 immunoblotting, respectively. Data shown as mean ± SD normalized to α-tubulin. *, p<0.05; **, p<0.01; ***, p<0.001; ****, p<0.0001. (D) NIH 3T3 Flp-In Gpr161 ko cells stably expressing untagged wildtype (wt) or Gpr161mut1 were starved for 24 hr upon confluence and were treated for further 24 hr ± SAG (500 nM). After fixation, cells were immunostained with anti-Gli2 (red), anti-Gpr161 (green), anti-acetylated, and γ-tubulin (AcTub, γTub in grey) antibodies. (E) Quantification of Gli2 positive cilia from (D). n=100 cells counted/condition from two coverslips each. Data shown as mean ± SD. **, p<0.01. Scale: (E), 5 μm.
-
Figure 4—source data 1
Table showing transcript data plotted in Figure 4A and B.
- https://cdn.elifesciences.org/articles/67121/elife-67121-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Table showing data from individual experiments in Figure 4C.
- https://cdn.elifesciences.org/articles/67121/elife-67121-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Table showing Gli2 positive cilia data plotted in Figure 4E.
- https://cdn.elifesciences.org/articles/67121/elife-67121-fig4-data3-v2.xlsx
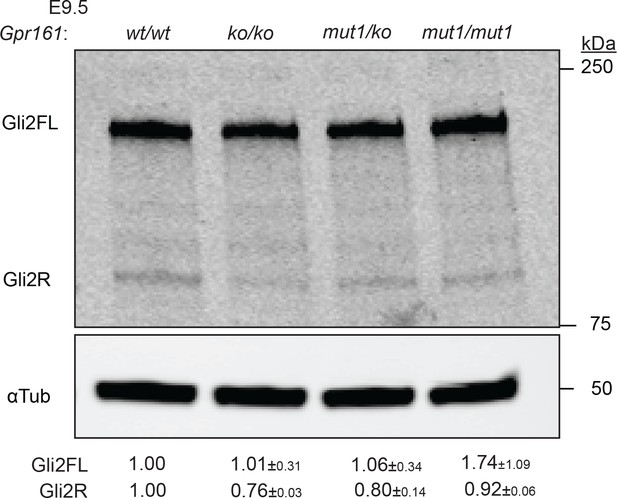
Immunoblotting for Gli2 and α-tubulin in E9.5 whole-embryo lysates.
n = 2. Data shown as mean ± SD.
-
Figure 4—figure supplement 1—source data 1
Table showing data from individual experiments i Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/67121/elife-67121-fig4-figsupp1-data1-v2.xlsx
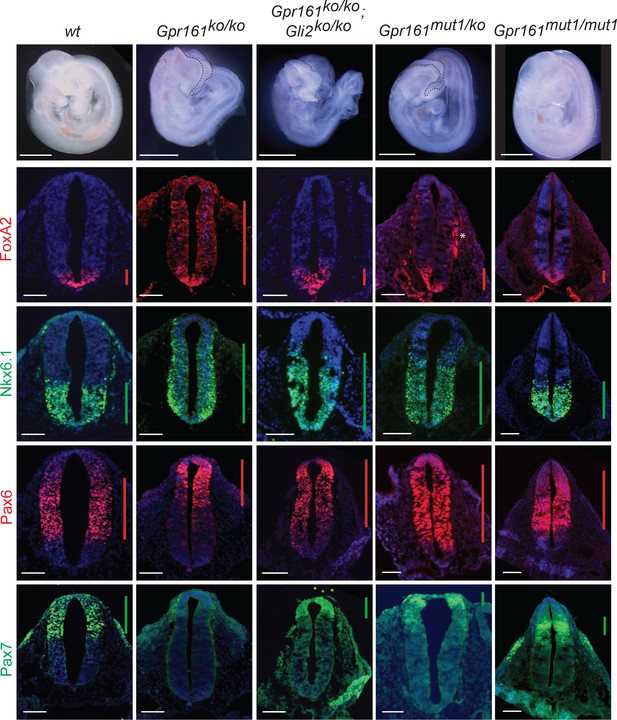
Gpr161mut1/ko embryos exhibit less ventralized neural tube compared to Gpr161 knockouts.
Topmost panels show bright-field images of wildtype (wt), Gpr161 ko, Gpr161 mut1/ko, Gpr161 mut1/mut1, Gpr161; Gli2 double ko, and Gli2 ko whole-mount embryos at E9.5. Bottom panels show rostral neural tube horizontal sections immunostained using designated markers. All images are counterstained with Hoechst. Black dotted line mark rostral malformations. Vertical bars show the extent of dorsoventral expression of markers. Asterix, nonspecific background staining outside neural tube. n=2–4 embryos each genotype and immunostaining. Scale: 50 µm.
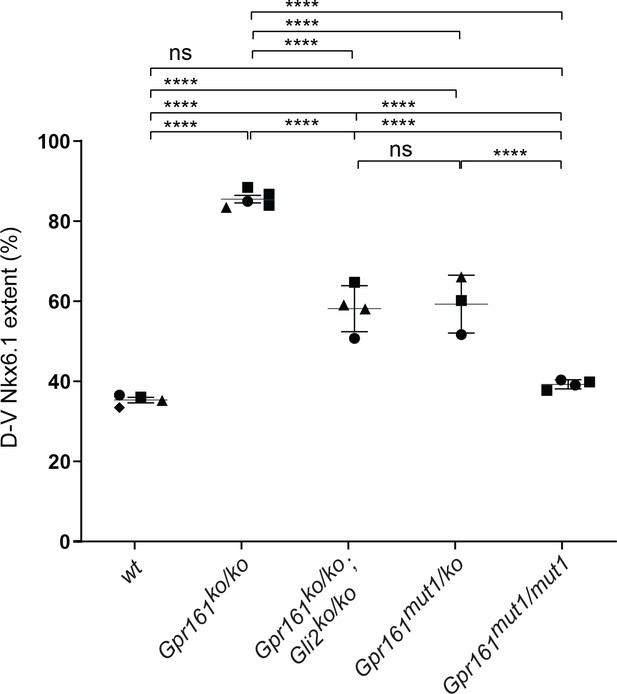
Quantification of dorsoventral (D–V) extent of Nkx6.1 expression in Gpr161 mutants.
D–V extent of Nkx6.1 expression to that of the neural tube (Figure 5) shows similar levels of partial ventralization in Gpr161 mut1/ko and Gpr161; Gli2 double ko compared to Gpr161 ko/ko and normal extent of expression in Gpr161 mut1/mut1. Quantification (mean ± SEM) at thoracic region from two to four embryos (each shown by a different shape). For each embryo, average from two or more sections were taken. ****, p<0.0001.
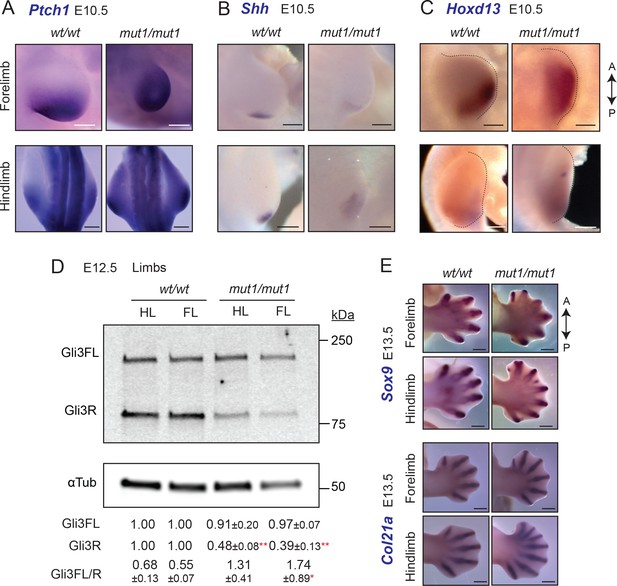
Gpr161mut1/mut1 embryos exhibit high Hh signaling and polydactyly in limb buds.
(A–C) RNA in situ hybridization for E10.5 Ptch1 (A), Shh (B) Hoxd13 (C) in limb buds. Ptch1 and Hoxd13 were expanded anteriorly in Gpr161mut1/mut1 limb buds. Shh was diffusely expressed posteriorly but was not ectopically expressed anteriorly in Gpr161mut1/mut1 limb buds. n=2–5 each. (D) Immunoblotting of forelimb (FL) and hindlimb (HL) buds for Gli3 and α-tubulin shows decreased Gli3R levels at E12.5 wildtype (wt) versus Gpr161mut1/mut1. Quantification shown is normalized to α-tubulin. n = three experiments. *, p<0.05; **, p<0.01. (E) RNA in situ hybridization for Sox9 and Col2a1 in E13.5 wildtype and Gpr161mut1/mut1 limb buds. Gpr161mut1/mut1 limb buds show polydactyly. n=four each. Scale: (A–C), 50 µm; (E), 500 µm.
-
Figure 6—source data 1
Table showing data from individual experiments in Figure 6D.
- https://cdn.elifesciences.org/articles/67121/elife-67121-fig6-data1-v2.xlsx
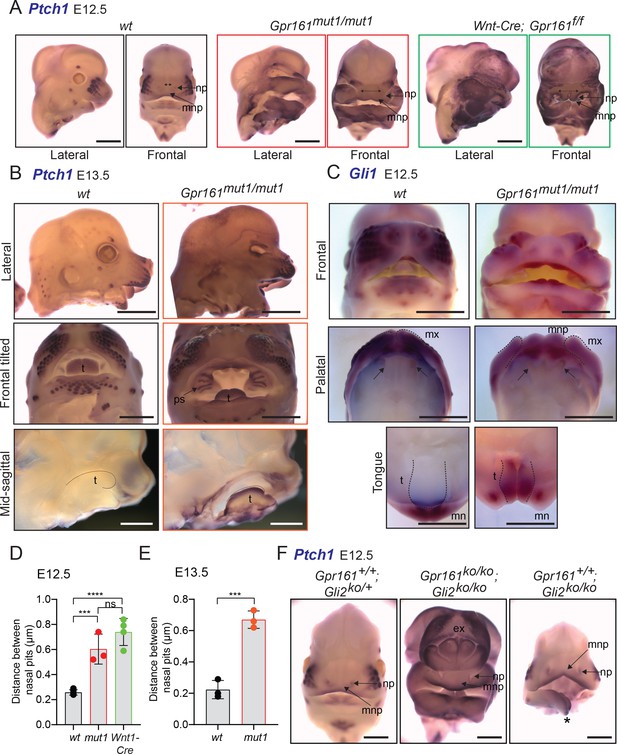
Gpr161mut1/mut1 embryos show high Hh signaling and mid face widening.
(A) RNA in situ hybridization for Ptch1 in wildtype (wt), Gpr161mut1/mut1, and Wnt1-Cre; Gpr161f/f E12.5 head. Distances between nasal pits are shown as bidirectional arrows. Lateral and frontal views are shown. n=3–4 each. (B) RNA in situ hybridization for Ptch1 in E13.5 wildtype (wt), Gpr161mut1/mut1 head. Top panels, lateral view; middle panels, frontal tilted view shows palate and tongue; bottom panels, sagittal section showing tongue. Black dotted lines in the bottom panels indicate tongue. Bidirectional arrows show increased distance between nasal pits. Arrow points to prominent palatal shelf in Gpr161mut1/mut1. n=3 each. (C) RNA in situ hybridization for Gli1 in E12.5 wildtype (wt), Gpr161mut1/mut1 head. Top panels, frontal view. Middle panels show palates imaged from below (palatal view) and bottom panels show lower jaw viewed from above (tongue view) after separating the jaws. Arrows in upper panel show secondary palatal shelves. Black dotted lines in the bottom panels indicate tongue. Note increased gap between maxillary processes by ingression of median nasal processes. n=3 each. (D–E) Quantification of distance between nasal pits as shown in (A). The colors are matched with each strain in A and B. Error bars represent SEM. ***, p<0.001; ****, p<0.0001, unpaired t-test. n=3–4 each. (F) RNA in situ hybridization for Ptch1 in E12.5 control (Gli2-/+), Gpr161; Gli2 double ko and Gli2 ko head. Note persistent exencephaly in Gpr161; Gli2 double ko and midfacial widening. Displaced lower jaw is an artifact (*). n=1–2 each. Scale: (A and F), 1 mm; (B and C) 2 mm. Abbreviations: ex, exencephaly; mnp, medial nasal process; mx, maxillary process; mn, mandibular process; np, nasal pit; ps, palatal shelf; t, tongue.
-
Figure 7—source data 1
Table showing data for distance between nasal pits plotted in Figure 7D and E.
- https://cdn.elifesciences.org/articles/67121/elife-67121-fig7-data1-v2.xlsx
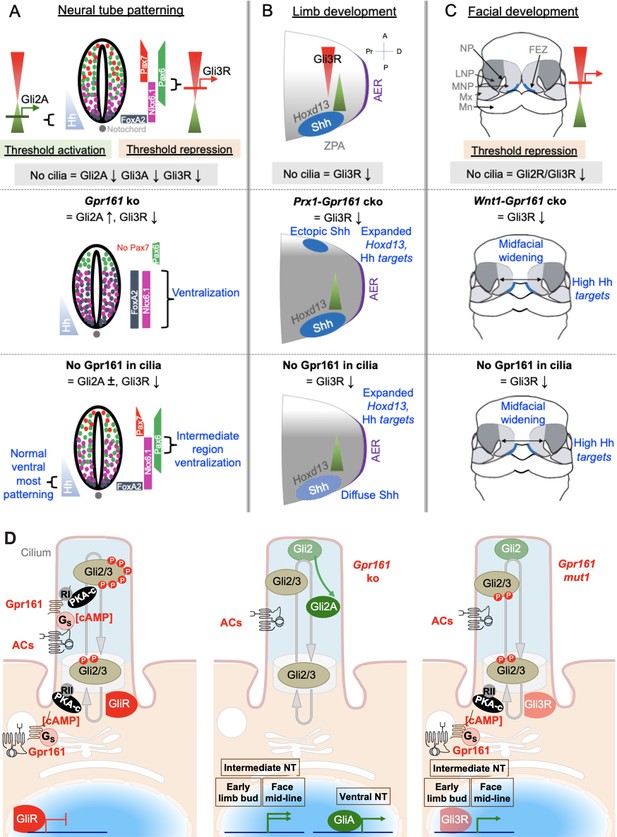
Gpr161 ciliary pools determine Hh pathway repression-regulated morpho-phenotypic spectrum.
(A) Neural tube development. Hh is expressed from the notochord. Gli2A-mediated threshold activation mediates floorplate and ventral-most progenitor patterning. Gli3R regulates intermediate level patterning. Ciliary disruption prevents patterning of all ventral progenitors. Complete loss of Gpr161 in ko causes ventralization of all ventral progenitors from excessive Gli2A generation and loss of Gli3R. Lack of Gpr161 ciliary pools in Gpr161mut1/ko reduces Gli3R formation sufficiently to cause intermediate level expansion of Nkx6.1 but does not generate excessive GliA to cause ventralization of floor plate markers. Caudal spina bifida and exencephaly persists in Gpr161mut1/ko similar to Gpr161 ko. (B) Limb development. Shh expression from ZPA starting from E9.75 establishes posterior gradient of pathway targets such as Ptch1/Gli1. Anterior Gli3R gradient also limits expression of genes such as Hoxd13 to the posterior mesenchyme. Ciliary disruption causes preaxial polydactyly from lack of Gli3R induced anterior expansion of 5’Hoxd gene expression. Gpr161 ko prevents forelimb formation. Conditional deletion of Gpr161 in limb mesenchyme using Prx1-Cre; Gpr161f/f (Prx1-Gpr161 cko) causes increased Hh pathway targets, reduced Gli3R, and expanded Hoxd13 in limb buds contributing to polydactyly. Loss of Gpr161 ciliary pools in Gpr161mut1/mut1 causes increased Hh pathway targets, reduced Gli3R, and expanded Hoxd13 in limb buds contributing to polydactyly. Shh expression in the posterior limb bud is diffuse, likely from lack of counter-antagonism between Gli3R and dHand. Abbreviations: AER, anterior ectodermal ridge; ZPA, zone of polarizing activity; A, anterior; P, posterior, Pr, proximal; D, distal. (C) Facial development. Shh is expressed from the frontonasal ectodermal zone in the medial nasal processes. Threshold repression by both Gli2R and Gli3R prevents midfacial widening. Ciliary disruption, or lack of both Gli2/3 causes midfacial widening, which is prevented by forced Gli3R expression. Loss of Gpr161 ciliary pools in Gpr161mut1/mut1 phenocopies Gpr161 deletion in craniofacial mesenchyme using Wnt1-Cre; Gpr161f/f (Wnt1-Gpr161 cko) by showing midfacial widening and increased levels of Shh pathway targets. Gli2 loss is unable to rescue midfacial widening and increased Shh pathway targets in Gpr161 ko background, suggesting reduced GliR contributing to these phenotypes. Abbreviations: FEZ, frontonasal ectodermal zone; LNP, lateral nasal process; MNP, medial nasal process, Mx, maxillary process. (D) Model of Gpr161 ciliary and extraciliary pools in Hh pathway. Both ciliary and extraciliary pools of Gpr161 contribute to GliR formation by PKA-mediated phosphorylation. Complete loss of Gpr161 prevents Gli2/3 phosphorylation by PKA. Gli2 is less efficient in repressor formation than Gli3. Unphosphorylated Gli2 is dissociated from Sufu and accumulates in ciliary tips. GliA formation is dependent on lack of PKA phosphorylation but likely occurs downstream of Gli2 ciliary accumulation. Smo might further promote GliA formation. Gli2 accumulation in ciliary tips and GliA formation also occurs in Ankmy2 knockout where trafficking of ACs to cilia is affected. Loss of ciliary Gpr161 restricts ciliary cAMP-mediated PKA activation but does not hinder extraciliary Gpr161 from cAMP production. Such extraciliary production might happen in the endomembrane compartment, but PKA regulatory subunits involved are not known. Restricted phosphorylation limits Gli3R formation but is still sufficient to dissociate Gli2 from Sufu causing accumulation in ciliary tips. However, GliA formation is impacted. Tissue regions that depend on GliR are specifically affected from loss of ciliary pools of Gpr161. Abbreviations: RI/RII, type I/II PKA regulatory subunits.
Tables
Results of breeding animals having Gpr161 ko and/or mut1 alleles.
| Breeding between Gpr161 ko/+ parents | ||||
|---|---|---|---|---|
| Age | Litters | wt/wt | ko/wt | ko/ko |
| E8.5 | 16 | 36 (29%) | 54 (44%) | 33 (27%) |
| E9.5 | 26 | 51 (27%) | 85 (45%) | 54 (28%) |
| E10.25 | 8 | 16 (28%) | 27 (47%) | 15 (26%) |
| E10.5 | 6 | 14 (32%) | 30 (68%) | 0 (0%) ** |
| Breeding Gpr161 ko/+ with Gpr161 mut1/+ parents | ||||
| Age | Litters | wt/wt | ko/wt or mut1/wt | mut1/ko |
| E9.5 | 12 | 24 (26%) | 45 (49%) | 22 (24%) |
| E10.25 | 9 | 20 (31%) | 28 (44%) | 16 (25%) |
| E10.5 | 2 | 3 (21%) | 7 (50%) | 4 (29%) |
| E11.5 | 2 | 3 (21%) | 7 (50%) | 4 (29%) |
| E12.5 | 5 | 14 (35%) | 23 (58%) | 3 (7%)¥ |
| E13.5 | 4 | 8 (36%) | 14 (64%) | 0 (0%)†, * |
| Breeding between Gpr161 mut1/+ parents | ||||
| Age | Litters | wt/wt | mut1/wt | mut1/mut1 |
| E9.5 | 20 | 41 (30%) | 61 (45%) | 34 (25%) |
| E10.25 | 4 | 8 (25%) | 16 (50%) | 8 (25%) |
| E10.5 | 8 | 21 (30%) | 34 (49%) | 15 (21%) |
| E11.5 | 2 | 6 (43%) | 4 (29%) | 4 (29%) |
| E12.5 | 7 | 16 (29%) | 27 (49%) | 12 (22%) |
| E13.5 | 9 | 16 (27%) | 29 (48%) | 15 (25%) |
| E14.5 | 7 | 13 (30%) | 23 (52%) | 8 (18%)‡ |
| E14.75 | 6 | 13 (35%) | 24 (65%) | 0 (0%)§, ** |
| E15.5 | 5 | 10 (38%) | 16 (62%) | 0 (0%)¶, *** |
-
¥3 dead mut1/ko embryos were not counted.
†7 dead mut1/ko embryos were not counted.
-
‡7 dead mut1/mut1 embryos were not counted.
§ 13 dead mut1/mut1embryos were not counted.
-
¶ Dead or resorbed embryos were not counted.
* p<0.05 by Chi Square test.
-
** p<0.01 by Chi Square test.
*** p<0.001 by Chi Square test.
Phenotypes in Gpr161 mutants.
| Phenotype (age) | Gpr161ko/ko | Gpr161ko/mut1 | Gpr161mut1/mut1 |
|---|---|---|---|
| Lethality of embryos | E10.5 | E13.5 | E14.75 |
| Exencephaly (>E10.25) | 100% (102/102) | 100% (54/54) | 100% (12/12) |
| Spina bifida (>E10.25) | 100% (15/15) | 100% (16/16) | N/D |
| Midface widening (>E12.5) | N/A | N/A | 100% (23/23) |
| Microphthalmia (>E12.5) | N/A | 100% (3/3) | 100% (35/35) |
| Pericardial effusion (E12.5–13.5) | N/A | 92% (11/12) | N/D |
| Limb development | |||
| Forelimb | No forelimbs at E10.25 | Stunted at E12.5 | Polydactyly at E13.5 (6~7 digits) |
| Hindlimb | Present | Present | Polydactyly at E13.5 (6~7 digits) |
| Kinked tail (> E12.5) | N/A | 100% (3/3) | N/D |
-
N/A, not applicable; N/D, not determined.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| genetic reagent (M. musculus) | Gpr161mut1/+ | This study | See Methods | |
| genetic reagent (M. musculus) | Gpr161ko/+ | Hwang et al., 2018 | RRID:MGI:6357708 | |
| genetic reagent (M. musculus) | Gpr161f/f | Hwang et al., 2018 | RRID:MGI:6357710 | |
| genetic reagent (M. musculus) | Wnt1-Cre | Jackson Laboratory Lewis et al., 2013 | Jax strain No: 022501 | |
| genetic reagent (M. musculus) | Prx1-Cre | Jackson Laboratory Logan et al., 2002 | Jax strain No: 005584 | |
| genetic reagent (M. musculus) | Gli2tm1Alj | Mo et al., 1997 | RRID:MGI:1857509 | |
| cell line (Homo sapiens) | RPE hTERT | ATCC | ATCC CRL-4000; RRID:CVCL_4388 | |
| cell line (M. musculus) | NIH 3T3 Tet-on 3G | Clontech | 631197; RRID:CVCL_V360 | |
| cell line (M. musculus) | NIH-3T3 Flp-In | Thermo Fisher Scientific | R76107 | |
| cell line (M. musculus) | Gpr161 ko NIH-3T3 Flp-In | Pusapati et al., 2018 | Gift from Rajat Rohatgi | |
| cell line (M. musculus) | β-arrestin 1/2 double knockout MEFs | Kovacs et al., 2008 | Gift from R. Lefkowitz | |
| transfected construct (Homo sapiens) | INPP5E On-target plus siRNA | Dharmacon/ Horizon, Perkin Elmer Humbert et al., 2012 | J-020852–05 | 5′- GGAAUUAAAAGACGGAUUU-3’ |
| transfected construct (Homo sapiens) | BBS4 siGenome siRNA | Dharmacon/ Horizon, Perkin Elmer Loktev et al., 2008 | D-013649–04 | 5′- CGAUCUGACUUAUAUAAUG-3’ |
| antibody | Mouse monoclonal anti-Acetylated tubulin | Sigma | Cat# T6793 RRID:AB_477585 | IF (1:5000) |
| antibody | Mouse monoclonal anti-Arl13b | NeuroMab Facility | Cat# N295B/66 RRID:AB_2750771 | IF (1:500) |
| antibody | Mouse monoclonal anti-ɣ-tubulin | Sigma | Cat# T6557 RRID:AB_532292 | IF (1:500) |
| antibody | Mouse monoclonal anti-α-tubulin | Sigma | Cat# T6199 RRID:AB_477583 | WB (1:5000) |
| antibody | Rabbit polyclonal anti-FoxA2 | Abcam | Cat# ab40874 RRID:AB_732411 or Cat# ab108422 RRID:AB_11157157 | IF (1:2000) |
| antibody | Mouse monoclonal anti-Nkx6.1 | DSHB | Cat# F55A10-s RRID:AB_532378 | IF (1:500) |
| antibody | Rabbit polyclonal anti-Pax6 | Biolegend | Cat# 901302 RRID:AB_2749901 | IF (1:50) |
| antibody | Mouse monoclonal anti-Pax7 | DSHB | RRID:AB_528428 | IF (1:50) |
| antibody | Guinea pig anti-Gli2 | Gift from Jonathan Eggenschwiler Cho et al., 2008 | IF (1:500) | |
| antibody | Chicken anti-GFP | Abcam | Cat# ab13970 RRID:AB_300798 | IF (1:10000) |
| antibody | Rabbit polyclonal anti-Gpr161 | Custom-made Pal et al., 2016 | IF (1:250) | |
| antibody | Rabbit polyclonal anti-Tulp3 | Gift from Jonathan Eggenschwiler Norman et al., 2009 | IF (1:500) | |
| antibody | Goat polyclonal anti-Gli3 | R and D Systems | AF3690 | WB (1:1000) |
| antibody | Goat polyclonal anti-Gli2 | R and D Systems | AF3635 | WB (1:1000) |
| antibody | Mouse monoclonal anti-Gli1 | Cell Signaling RRID:AB_2294746 | Cat# 2643 | WB (1:1000) |
| antibody | Alexa Fluor 488-, 555-, 594-, 647- conjugated secondary antibodies | Life Technologies, Carlsbad, CA | IF (1:2000) | |
| antibody | IRDye tagged secondary antibodies | LI-COR | IF (1:5000) | |
| antibody | Hoechst 33342 | Life Technologies | IF (1:5000) | |
| recombinant DNA reagent | pgLAP5 vector (plasmid) | Addgene | Plasmid #19706; RRID:Addgene_19706 | |
| recombinant DNA reagent | pDONR221_Gpr161 (M. musculus) | Mukhopadhyay et al., 2013 | Synthesized by DNA2.0 | NM_001081126.1 |
| sequence-based reagent | Gpr161 mut1 F | This paper | PCR primers | CAG AAA GCA ACA GCA AAG CA |
| sequence-based reagent | Gpr161 mut1 R | This paper | PCR primers | ACC CTG ACA CTG CCC TTA GC |
| sequence-based reagent | Gpr161 Ex4 F | Hwang et al., 2018 | PCR primers | CAA GAT GGA TTC GCA GTA GCT TGG |
| sequence-based reagent | Gpr161 floxed R | Hwang et al., 2018 | PCR primers | ATG GGG TAC ACC ATT GGA TAC AGG |
| sequence-based reagent | Gpr161 Neo cassette R | Hwang et al., 2018 | PCR primers | CAA CGG GTT CTT CTG TTA GTC C |
| sequence-based reagent | Cre-F | Hwang et al., 2018 | PCR primers | AAT GCT GTC ACT TGG TCG TGG C |
| sequence-based reagent | Cre-R | Hwang et al., 2018 | PCR primers | GAA AAT GCT TCT GTC CGT TTG C |
| sequence-based reagent | Gli2 sense | Mo et al., 1997 | PCR primers | AAA CAA AGC TCC TGT ACA CG |
| sequence-based reagent | Gli2 antisense | Mo et al., 1997 | PCR primers | CAC CCC AAA GCA TGT GTT TT |
| sequence-based reagent | pPNT | Mo et al., 1997 | PCR primers | ATG CCT GCT CTT TAC TGA AG |
| sequence-based reagent | Gli1-F | Sigma | Gli1 TaqMan probes for qRT-PCR | GCA GTG GGT AAC ATG AGT GTC T |
| sequence-based reagent | Gli1-R | Sigma | Gli1 TaqMan probes for qRT-PCR | AGG CAC TAG AGT TGA GGA ATT GT |
| sequence-based reagent | Gli1-Probe (FAM/TAMRA) | Sigma | Gli1 TaqMan probes for qRT-PCR | CTC TCC AGG CAG AGA CCC CAG C |
| sequence-based reagent | Ptch2-F | Sigma | Ptch2 TaqMan probes for qRT-PCR | CAG AGT GAC TAC CTC CAT GAC TGT |
| sequence-based reagent | Ptch2-R | Sigma | Ptch2 TaqMan probes for qRT-PCR | GCT GGG TGG ACG TAT GCT |
| sequence-based reagent | Ptch2-probe (FAM/TAMRA) | Sigma | Ptch2 TaqMan probes for qRT-PCR | CTC CAC CCA CCA CCT CTG CCT |
| sequence-based reagent | Taqman probes for Gpr161 | Applied Biosystems | Mm01291057_m1 | |
| sequence-based reagent | Taqman probes for Gapdh | Applied Biosystems | Mm99999915 | |
| commercial assay or kit | TR-FRET cAMP assay | Cisbio | 62AM4PEB | |
| commercial assay or kit | GenElute mammalian total RNA purification kit | Sigma | RTN350 | |
| commercial assay or kit | SYBR Green Quantitative RT-qPCR Kit | Sigma | QR0100 | |
| commercial assay or kit | Mycoplasma PCR Detection Kit | Genlantis | MY01050 | |
| commercial assay or kit | Kicqstart One-Step Probe RT-qPCR ReadyMix | Sigma | KCQS07 | |
| chemical compound, drug | Penicillin/Streptomycin | Sigma | P4333 | Cell culture |
| chemical compound, drug | Glutamine | Sigma | G7513 | Cell culture |
| chemical compound, drug | Polyfect | Qiagen | 301105 | Cell culture |
| chemical compound, drug | DMEM-High Glucose | Sigma | D5796 | Cell culture |
| chemical compound, drug | DMEM F12 | Sigma | D6421 | Cell culture |
| chemical compound, drug | BCS | Sigma | 12133C | Cell culture |
| chemical compound, drug | FBS | Sigma | F0926 | Cell culture |
| chemical compound, drug | Paraformaldehyde | Electron microscopy solutions | 15710 | |
| chemical compound, drug | Normal donkey serum | Jackson ImmunoResearch, West Grove, PA | Cat# 017-000-121; RRID:AB_2337258 | |
| chemical compound, drug | Fluoromount-G | Southern Biotech | 0100–01 | |
| chemical compound, drug | Permount | ThermoFisher Scientific | SP15-100 | |
| software, algorithm | ImageJ software | RRID:SCR_003070 | ||
| software, algorithm | GraphPad Prism | RRID:SCR_002798 |