RNA binding to human METTL3-METTL14 restricts N6-deoxyadenosine methylation of DNA in vitro
Figures
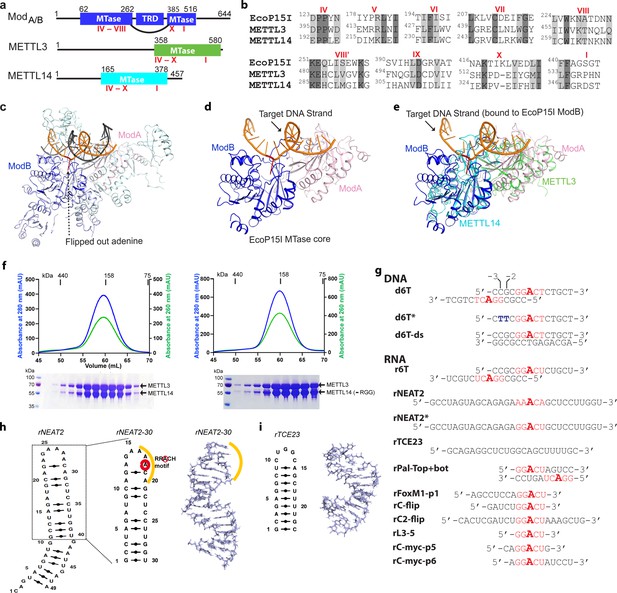
Structural similarity, purification of methyltransferases, and substrate designing.
(a–b) Domain architecture of Mod subunit of EcoP15I, human methyltransferase like-3 (METTL3), and human METTL14 methyltransferases (MTases). All three members belong to the β-class of SAM-dependent MTases and exhibit a sequential arrangement of motifs (IV-X followed by motif I) (Bujnicki et al., 2002; Woodcock et al., 2020; Malone et al., 1995). Motif IV (D/EPPY/W/L) and I are associated with the recognition of target adenine base (Gupta et al., 2015) and co-factor (SAM) binding (Wang et al., 2016b; Sledz and Jinek, 2016; Wang et al., 2016a), respectively. (c) Crystal structure of EcoP15I-DNA complex (PDB ID: 4ZCF) (Gupta et al., 2015): ModA MTase (cyan), ModB MTase (blue), non-methylating DNA strand (gray), target (methylating) DNA strand (orange), flipped adenine base (red stick). The methyltransferase cores of ModB and ModA are shown in dark blue and light pink, respectively. The Res subunit of EcoP15I was omitted for clarity. (d) MTase domains of two Mod subunits (ModA/B) of EcoP15I with target DNA strand. The regions flanking the methyltransferase core (e.g., CTD and TRD) are omitted for clarity. Only the region encompassing the MTase core (aa 90–132, 169–261, and 385–511) of EcoP15I Mod was selected for the alignment with the methyltransferase core of METTL3 (aa 358–580) and METTL14 (aa 165–378). (e) An overlay of MTase domains of EcoP15I and METTL14 (PDB ID: 5IL0) shows structural similarity within MTase folds (rmsd = 2.55 Å over 278 Cα atoms). RNA strand here was modeled based on the respective methylating strand in the EcoP15I structure.(f) Chromatogram of final size exclusion chromatography (SEC) step of purification showing METTL3-METTL14 complexes (left, full-length; right, METL3-METTL14[-RGG]) co-eluted as single homogenous species. Blue, absorbance at 280 nm; green, absorbance at 260 nm (A260). Coomassie stained gels (lower panels) confirm high purity of METTL3-METTL14 proteins in the SEC peak fractions. (g) Sequence of DNA and RNA oligonucleotides used in this study. All oligos have a covalently attached 5’-fluorescein (not shown). (h) Secondary structure of rNEAT2 and its 3D model as predicted by MC-SYM (Parisien and Major, 2008). rNEAT2 harbors a potential DRACH motif (yellow line). (i) Secondary structure of rTCE23 RNA and its solution NMR structure (PDB ID: 2ES5) (Oberstrass et al., 2006).
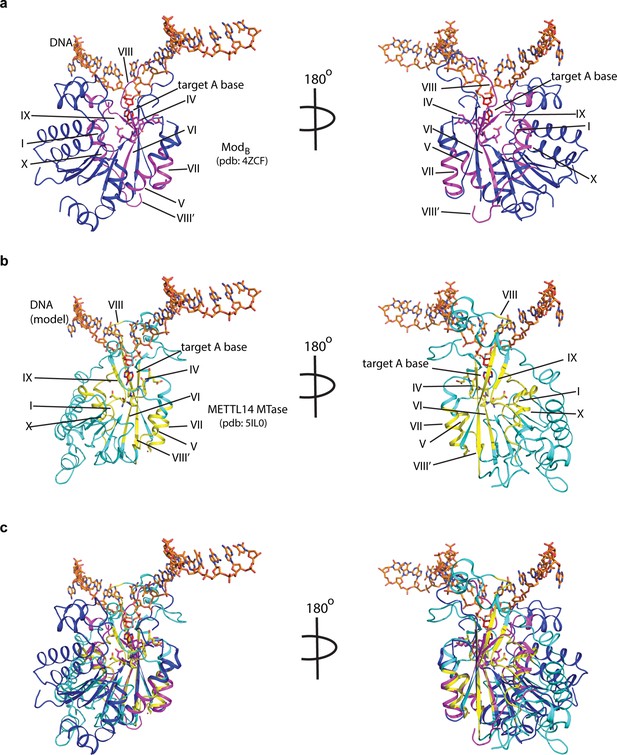
Potential mode of DNA recognition.
(a) Blue ribbons, the MTase core (ModB) of EcoP15I (aa 90–132, 169–261, 385–511 from PDB: 4ZCF) in complex with the target DNA strand (orange sticks). The conserved motifs in class β MTases are shown in magenta. The motif IV (D/EPPY/W/L) that surrounds the Watson-Crick edge of the flipped target adenine base (red) is shown in stick mode. (b) Cyan ribbons, MTase core of human METTL14 (PDB: 5IL0) with a modeled single-stranded DNA (ssDNA) as in ModB. The conserved motifs are colored in yellow. (c) An overlay of MTases of ModB and METTL14 was performed using the SSM method (secondary structure-based) that yielded an rmsd of 2.55 over 282aa of METTL14 aligned with 278aa of ModB. The canonical MTase motifs, including motif IV, in two MTases overlay well. Due to lack of structures of METTL3 or ModA (in complex with a flipped adenine base), we chose ModB and METTL14 for this superposition. We could not model RGG motifs due to the lack of predicted structure for this region.
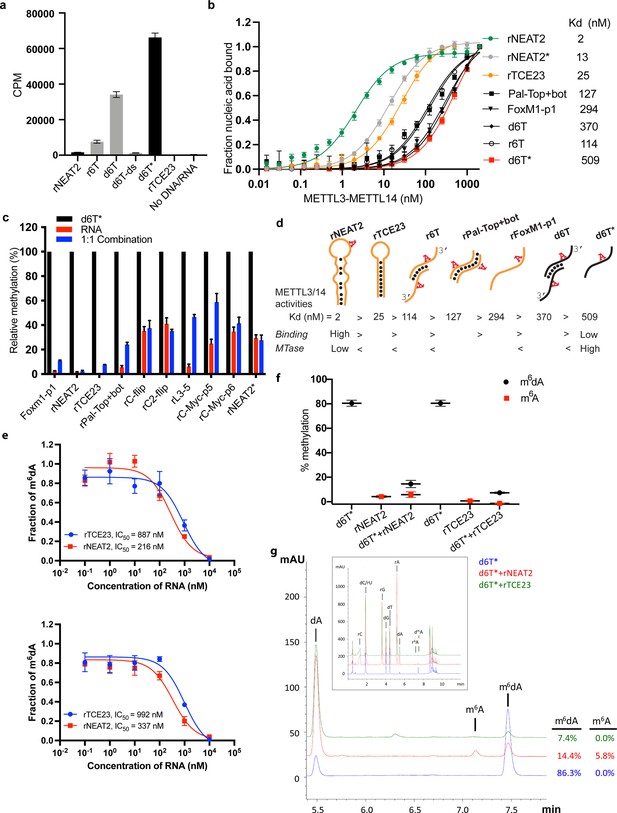
RNA-mediated restriction of methyltransferase like-3 (METTL3)-METTL14 activity.
(a) Methyltransferase activity of METTL3-METTL14 in the presence of various DNA or RNA substrates measured by radiometric assay. CPM, counts per minute. The highest activity was measured with the d6T* oligo. (b) FP-based binding assay for DNA and RNA oligos showing highest affinity for rNEAT2 RNA (green) and lowest affinity for d6T* oligo (red). Equilibrium dissociation constants (Kd) for each oligo are shown on the right side of the isotherms. The data were fit into one site-specific binding model (Y = Bmax*X/(Kd+ X)). See Materials and methods section and source data for details. (c) Methyltransferase activity of METTL3-METTL14 on the respective RNA oligos (red), d6T* DNA alone (black) and equimolar mixture of the two (blue), measured by radiometric assay. (d) Predicted secondary structures of each oligonucleotide. Yellow, RNA strand; black, DNA strand. The values of the equilibrium dissociation constants (Kd) shown for each oligonucleotide indicate an inverse relationship between binding affinity and methyltransferase activity. (e) Dose-dependent inhibition of METTL3-METTL14 activity by RNA oligos rNEAT2 or rTCE23, as measured by radiometric assay in a reaction buffer containing 5.0 mM NaCl (upper panel) and 50.0 mM NaCl (lower panel). IC50, concentration of RNA required to achieve 50% inhibition of the METTL3-METTL14 activity. (f) Attenuation of the methyltransferase activity in presence of rNEAT2 or rTCE23, as measured by oligonucleotide intact mass analysis. Quantitation of modified dA (black circle) or rA (red square) is shown in absence or presence of equivalent amounts of rNEAT2 or rTCE23. (g) Nucleoside composition analysis of the METTL3-METTL14 reactions. UHPLC chromatograms showing the reaction in absence (blue trace) or in the presence of either rNEAT2 (red trace) or rTCE23 (green trace). The quantitation of the fraction of modified bases in the nucleoside pool was consistent with the results from the oligonucleotide intact mass analysis shown in (f). The insert shows the full chromatographic trace with all detected nucleosides. Results presented in panels (a–c and e–f) are the average of three independent experiments (n = 3) with one standard deviation (s.d.) for each oligonucleotide (shown as error bars). Source data are provided as a Source Data file.
-
Figure 2—source data 1
MTase activity on individual RNA/DNA substrates.
- https://cdn.elifesciences.org/articles/67150/elife-67150-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Binding isotherms of METTL3-METTL14.
- https://cdn.elifesciences.org/articles/67150/elife-67150-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Comparison of MTase activity on different RNA/DNA substrates.
- https://cdn.elifesciences.org/articles/67150/elife-67150-fig2-data3-v2.xlsx
-
Figure 2—source data 4
Dose-dependent inhibition of MTase activity by RNA.
- https://cdn.elifesciences.org/articles/67150/elife-67150-fig2-data4-v2.xlsx
-
Figure 2—source data 5
RNA-mediated attenuation of MTase activity by intact mass analysis.
- https://cdn.elifesciences.org/articles/67150/elife-67150-fig2-data5-v2.xlsx
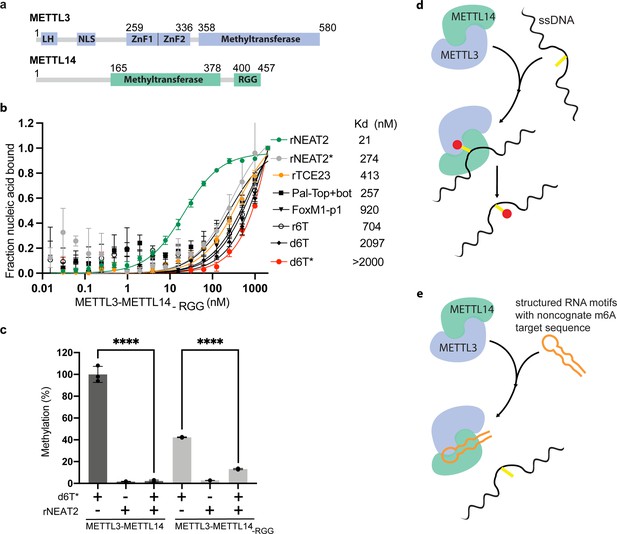
Role of RGG motifs and model of RNA-mediated regulation of methyltransferase activity.
(a) Domain architecture of methyltransferase like-3 (METTL3) and METTL14. LH, leader helix; NLS, nuclear localization signal; ZnF1/2, zinc-finger domain 1/2; RGG, arginine-glycine rich repeats motif. (b) FP-based-binding assay for DNA and RNA oligos showing the highest affinity of METTL3-METTL14(-RGG) for rNEAT2 RNA (green) and lowest affinity for d6T* (red). Equilibrium dissociation constants (Kd) for each oligo are shown. The data were fit into one site-specific binding model (Y = Bmax*X/(Kd+ X)). See Materials and methods section and source data for details. (c) Relative methyltransferase activity of full-length METTL3-METTL14 and the truncated enzyme devoid of the RGG motif in METTL14 ([METTL3-METTL14(-RGG)]) in the presence of d6T*, rNEAT2, or an equimolar mixture of these two oligos, as measured by radiometric assay. Results presented are the average of three independent experiments (n = 3) with one standard deviation (s.d.) for each oligonucleotide (shown as error bars). The results of two groups were analyzed and compared using two-tailed Student’s unpaired t-test (p-value < 0.0001). Details about Student’s t-test are provided in the Source Data file. (d, e) Proposed models showing that the METTL3-METTL14 complex can methylate a target adenine (yellow) in a single-stranded DNA region (black). Structured motifs present in ncRNA/mRNAs (orange) can block the methyltransferase activity by a shape-dependent binding of these RNAs to METTL3-METTL14.
-
Figure 3—source data 1
Binding isotherms of METTL3-METTL14 (-RGG).
- https://cdn.elifesciences.org/articles/67150/elife-67150-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Relative MTase activity of METTL3-METTL14 and METL3-METL14 (-RGG).
- https://cdn.elifesciences.org/articles/67150/elife-67150-fig3-data2-v2.xlsx
Tables
Equilibrium binding constants.
| Nucleic acid substrate | kd in nM (range of Kd in nM) | |
|---|---|---|
| Full-length METTL3-METTL14 | METTL3-METTL14-RGG | |
| RNA | ||
| rNEAT2 | 2.1 (1.8–2.3) | 21.2 (19.2–23.5) |
| rNEAT2* | 13.0 (12.3–13.8) | 274.1 (150.7–497.2) |
| rTCE23 | 25.6 (23.2–28.5) | 413 (341–502) |
| rPal-top+ bot | 127 (97.2–167) | 257 (132–491) |
| rFoxM1-p1 | 294 (245–354) | 920 (480–1940) |
| r6T | 114 (103–127) | 704 (323–1668) |
| DNA | ||
| d6T | 370 (343–401) | 2097 (1494 – >2000) |
| d6T* | 509 (470–553) | >2000 |





