High-resolution, genome-wide mapping of positive supercoiling in chromosomes
Figures
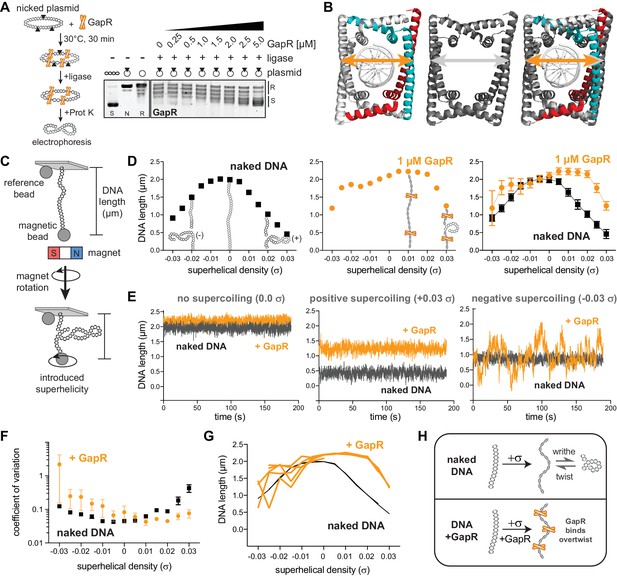
GapR interacts stably with overtwisted, positively supercoiled DNA.
(A) GapR DNA topology assay. GapR was incubated with nicked plasmid before treatment with T4 DNA ligase and subsequent quenching, deproteinization, and electrophoresis (schematic). Gel analysis of plasmid topology with positively supercoiled (S), nicked (N), and relaxed (R) standards. (B) Comparison of GapR-DNA crystal structures. Left, 6GC8 (Guo et al., 2018); middle, 6OZX (Tarry et al., 2019); right, overlay. Diameter of 6GC8 (orange arrow) and 6OZX (gray arrow) indicated. (C) Schematic of magnetic tweezer (MT) experiment. See also Figure 1—figure supplement 1C. (D) Behavior of naked DNA (left), DNA incubated with 1 µM GapR (middle), and overlay (right) in a rotation-extension experiment with the corresponding DNA conformation superimposed. Data indicate mean ± SD, n = 200 at each σ, in a single MT experiment. (E) DNA ± 1 µM GapR behavior over time from D under no supercoiling (σ = 0.0, left), positive supercoiling (σ = +0.03, middle), and negative supercoiling (σ = −0.03, right). (F) Coefficient of variation of force-extension experiments of DNA ± 1 µM GapR. Data indicate mean ± SEM, n ≥ 3. (G) Hysteresis of force-extension experiments. Traces indicate multiple rotation-extension measurements from one DNA molecule ± 1 µM GapR. (H) Model of GapR binding to overtwisted DNA.
-
Figure 1—source data 1
Raw gels associated with Figure 1A.
- https://cdn.elifesciences.org/articles/67236/elife-67236-fig1-data1-v2.zip
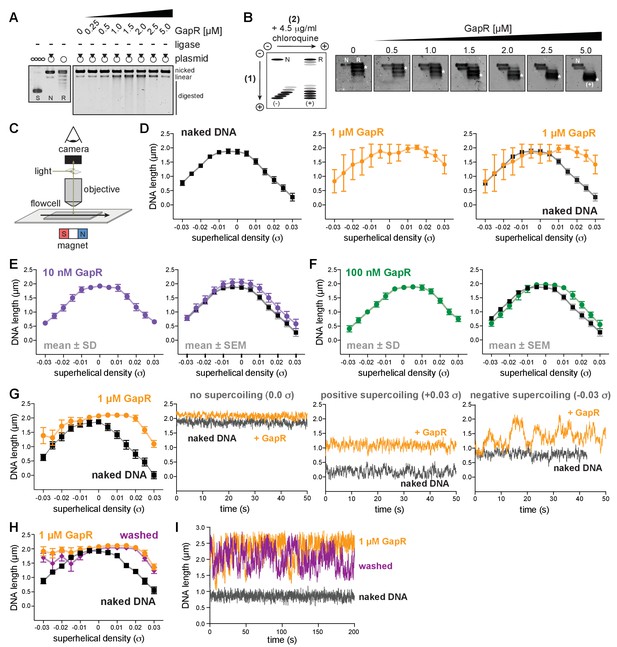
GapR binding to supercoiled DNA in a magnetic tweezer (MT) experiment.
(A) No ligase control for GapR DNA topology assay. GapR was incubated with nicked (N) plasmid and treated identically as in Figure 1A. Gel analysis of plasmid topology with supercoiled (S) and relaxed (R) standards. No digestion is observed at 2.5–5 µM GapR due to GapR protection of DNA. (B) Analysis of ligation products from Figure 1A with 2D-chloroquine electrophoresis. Migration of different plasmid forms are diagrammed (left): N, nicked; R, relaxed; L, linear; (-), negatively supercoiled; (+), positively supercoiled. Migration of the most abundant species indicated (asterisk). (C) Schematic of the microscope and flow cell setup in an MT experiment. (D) Behavior of naked DNA (left), DNA incubated with 1 µM GapR (middle), and overlay (right) in a typical rotation-extension experiment. Data indicate mean ± SEM for three MT experiments. (E) Behavior of DNA incubated with 10 nM GapR in a typical rotation-extension experiment. Data indicate mean ± SD, n = 200 at each σ, in a single MT experiment (left) and mean ± SEM compared to naked DNA (black), for three MT experiments (right). (F) Behavior of DNA incubated with 100 nM GapR in a typical rotation-extension experiment as in (E). (G) Rotation-extension curve (top left; data indicate mean ± SD, n = 200 at each σ) and time-course measurements for a single GapR binding experiment. Time courses indicate behavior of naked DNA (black) and DNA bound by 1 µM GapR (orange) under positive supercoiling (σ = +0.03, top right), negative supercoiling (σ = −0.03, bottom left), and no supercoiling (σ = 0.0, bottom right). (H) Behavior of naked DNA (black), DNA incubated with 1 µM GapR (orange) and after GapR washout (purple) in a typical rotation-extension experiment as in (E). (I) Time-course measurements of naked DNA (black), DNA incubated with 1 µM GapR (orange), and after GapR washout (purple) under negative supercoiling (σ = −0.03).
-
Figure 1—figure supplement 1—source data 1
Raw gels associated with Figure 1—figure supplement 1A.
- https://cdn.elifesciences.org/articles/67236/elife-67236-fig1-figsupp1-data1-v2.zip
-
Figure 1—figure supplement 1—source data 2
Raw gels associated with Figure 1—figure supplement 1B.
- https://cdn.elifesciences.org/articles/67236/elife-67236-fig1-figsupp1-data2-v2.zip
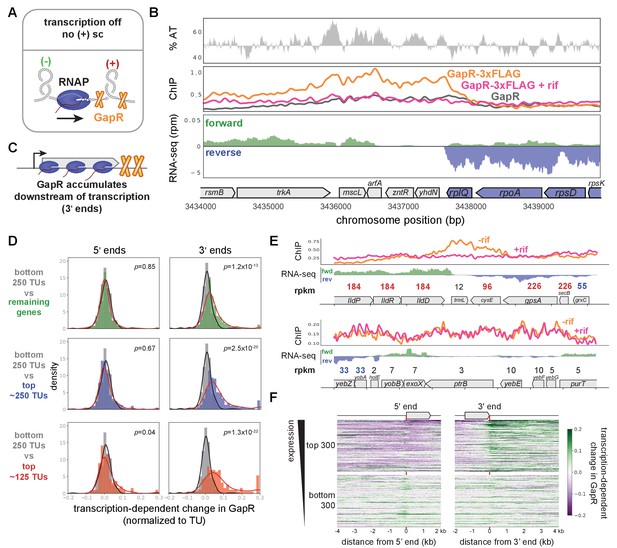
GapR is associated with positive supercoiling in E. coli.
(A) Positive supercoiling is generated downstream of RNA polymerase during transcription as predicted by the ‘twin-domain’ model. (B) GapR chromatin immunoprecipitation (ChIP) profiles at a highly expressed operon. AT content (top), with AT content below the genomic average (50%) plotted in reverse. ChIP-seq (middle) of untreated (orange) or rifampicin-treated (pink) GapR-3xFLAG cells and untreated GapR cells (gray). Transcription from the forward (green) and reverse (blue) strands with the position of annotated genes indicated (bottom). (C) GapR and positive supercoiling accumulates at the 3' end of genes, not within genes. (D) Transcription-dependent change in GapR ChIP at 5' (left) or 3' (right) ends normalized by binding within the transcription unit (TU) at different expression thresholds. Student’s t-test p-value shown. (E) Examples of GapR-3xFLAG ChIP without (orange) or with (pink) rifampicin treatment. Transcription of the forward (green) and reverse (blue) strands with annotated genes indicated. Expression values are colored using the same rpkm cutoffs as in D. (F) Heatmap showing transcription-dependent change in GapR around 5' and 3' ends for the top and bottom 300 long TUs sorted by expression.
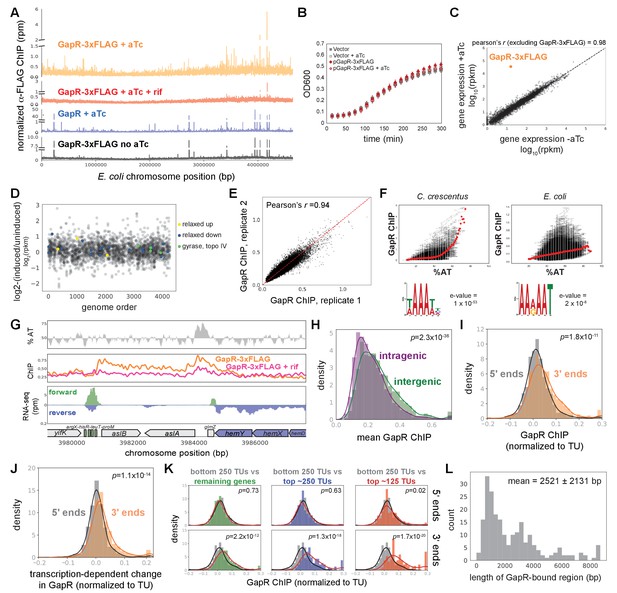
E. coli GapR chromatin immunoprecipitation-sequencing (ChIP-seq).
(A) ChIP-seq of GapR-3xFLAG with (orange) and without induction with anhydrous tetracycline (aTc, gray), after rifampicin treatment (rif, pink), and untagged GapR with induction (blue). (B) Growth of cells expressing GapR-3xFLAG plasmid or empty vector. Data indicate mean ± SEM, n = 6. (C) Transcriptional profiles of cells ± GapR-3xFLAG expression. (D) Supercoiling-sensitive genes are unaffected by GapR-3xFLAG expression. Log2-fold change in expression (rpkm) vs genome order (excluding GapR and poorly expressed genes). Supercoiling-sensitive genes (yellow/blue) and topoisomerase genes (light green). 15 genes that change greater than fourfold represent mostly proteins of unknown function or membrane proteins. (E) Correlation between two independent GapR-3xFLAG ChIP experiments. (F) GapR-3xFLAG ChIP versus AT content in C. crescentus (left) and E. coli (right). Mean ChIP at a given % AT (red dots). Motif from DREME (below). (G) GapR-3xFLAG ChIP profiles. AT content (top), with AT content below the genomic average (50%) plotted in reverse. Normalized ChIP-seq (middle) of GapR-3xFLAG cells that were either untreated (orange) or rifampicin-treated (pink), and untagged GapR expressing cells that were untreated (gray). Transcription from the forward (green) and reverse (blue) strands with annotated genes indicated (bottom). (H) Comparison of GapR ChIP at intragenic (purple) or intergenic (green) positions. Student’s t-test p-value is reported. (I) GapR accumulates at the 3' end of transcription units (TUs). GapR ChIP at 5' (gray) or 3' (orange) ends of TUs normalized by binding within the TU. Student’s t-test p-value is reported. (J) GapR binding at 3' ends depends on transcription. Transcription-dependent change in GapR ChIP at the 5' (gray) or 3' ends (orange) of TUs normalized by binding within the TU. Student’s t-test p-value is reported. (K) GapR binding at 5' (top) or 3' (bottom) ends of long TUs normalized by binding within the TU at different expression levels. Student’s t-test p-value is reported. (L) Histogram showing the length of transcription-dependent GapR binding events.
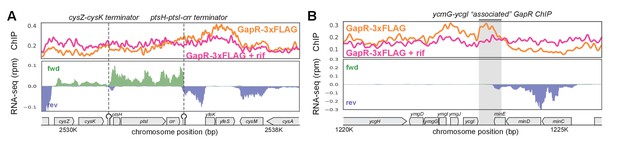
GapR full-length chromatin immunoprecipitation-sequencing (ChIP-seq).
(A) GapR ChIP-seq profiles in the absence (orange) and presence (pink) of the transcriptional inhibitor rifampicin (top). Transcription on the forward (green) and reverse (blue) strands with annotated genes indicated (bottom). Terminators on the top strand are indicated. (B) GapR profiles as in (A). Shaded region shows GapR ChIP generated by the minCDE transcript that is also at the 3' end of the ycmJ-ycgI locus.
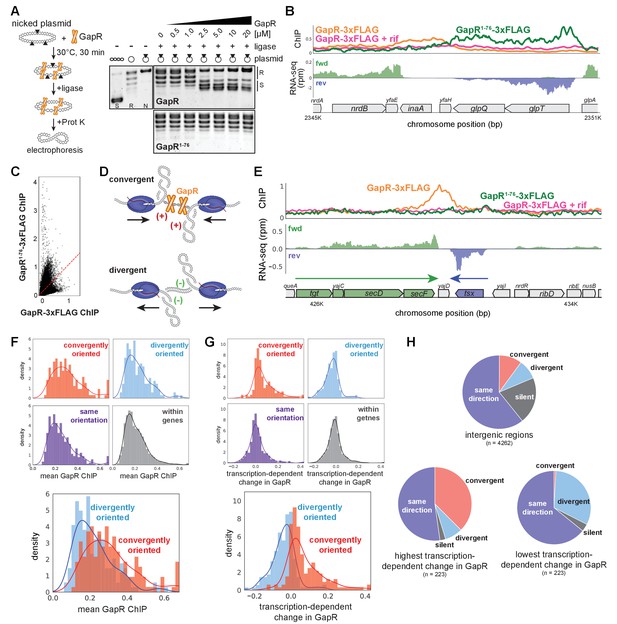
GapR recognizes DNA supercoiling and is associated with convergent transcription.
(A) GapR1-76 does not recognize DNA topology. Full-length GapR and GapR1-76 were incubated with nicked plasmid before treatment with T4 DNA ligase and subsequent quenching, deproteinization, and electrophoresis (schematic). Gel analysis of plasmid topology with supercoiled and relaxed standards as in Figure 1A. (B) GapR1-76-3xFLAG chromatin immunoprecipitation (ChIP) (green) and GapR-3xFLAG ChIP without (orange) and with (pink) rifampicin treatment (top). Transcription of the forward (green) and reverse (blue) strands with annotated genes indicated. (C) Correlation between GapR-3xFLAG and GapR1-76-3xFLAG ChIP experiments. (D) Positive supercoils are trapped by convergent transcription. (E) ChIP of GapR1-76-3xFLAG (green) and GapR-3xFLAG without (orange) and with (pink) rifampicin treatment at convergently oriented transcription units (TUs). Transcription of the forward (green) and reverse (blue) strands with annotated genes indicated. (F) GapR ChIP in gene bodies (dark gray), in divergent regions (blue), convergent regions (red), and where transcription is in the same orientation (purple). Overlay of divergent and convergent regions (bottom). (G) Transcription-dependent changes in GapR plotted as in (F). (H) Regions with high transcription-dependent change in GapR are more frequently between convergent genes. Pie charts summarize the orientation of flanking genes for all intergenic regions (top) and intergenic regions with highest (bottom left) or lowest (bottom right) transcription-dependent change in GapR.
-
Figure 3—source data 1
Raw gels associated with Figure 3A.
- https://cdn.elifesciences.org/articles/67236/elife-67236-fig3-data1-v2.zip
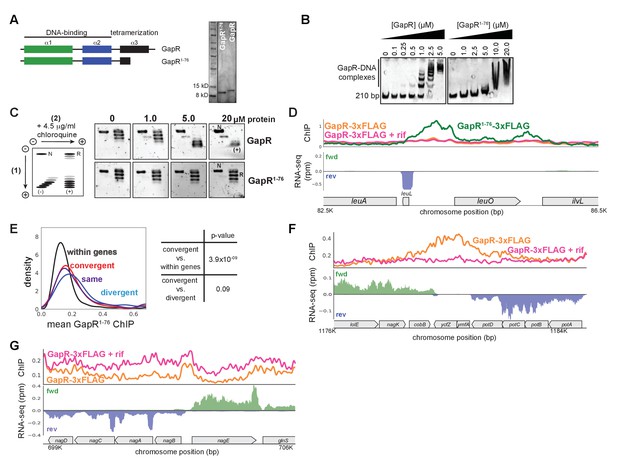
GapR full-length and truncation variant chromatin immunoprecipitation-sequencing (ChIP-seq).
(A) Schematic and purification gel of full-length and tetramerization-deficient GapR1-76. (B) Electrophoretic mobility shift assays of full-length and GapR1-76 binding to 210 bp DNA. (C) Analysis of ligation products from Figure 3A with 2D-chloroquine electrophoresis. Migration of different plasmid forms is diagrammed (left): N, nicked; R, relaxed; L, linear; (-), negatively supercoiled; (+), positively supercoiled. (D) GapR1-76 ChIP-seq (green) and GapR ChIP-seq profiles in the absence (orange) and presence (pink) of the transcriptional inhibitor rifampicin (top). Transcription on the forward (green) and reverse (blue) strands with annotated genes indicated (bottom). (E) Mean GapR1-76 ChIP in the middle of gene bodies (black), divergently transcribed regions (blue), convergently transcribed regions (red), and where transcription is in the same orientation (purple). (F) GapR profiles as in (D) showing GapR accumulation at a representative region of convergent transcription. (G) GapR profiles as in (D) showing GapR de-enrichment at a representative region of divergent transcription.
-
Figure 3—figure supplement 1—source data 1
Raw gels associated with Figure 3—figure supplement 1A.
- https://cdn.elifesciences.org/articles/67236/elife-67236-fig3-figsupp1-data1-v2.zip
-
Figure 3—figure supplement 1—source data 2
Raw gels associated with Figure 3—figure supplement 1B.
- https://cdn.elifesciences.org/articles/67236/elife-67236-fig3-figsupp1-data2-v2.zip
-
Figure 3—figure supplement 1—source data 3
Raw gels associated with Figure 3—figure supplement 1C.
- https://cdn.elifesciences.org/articles/67236/elife-67236-fig3-figsupp1-data3-v2.zip
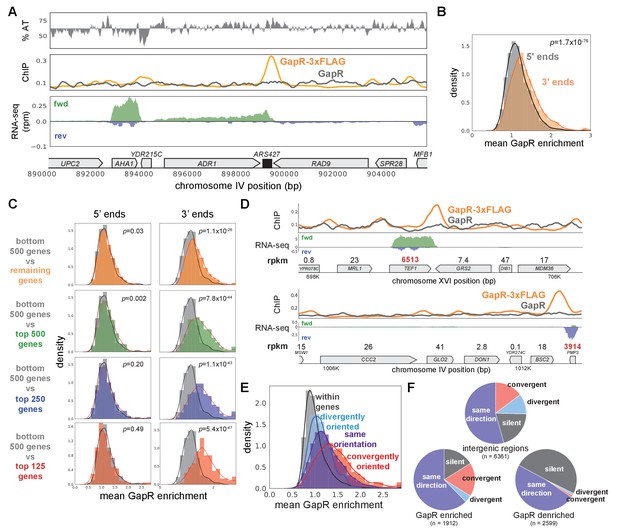
GapR is associated with positive supercoiling in yeast.
(A) Chromatin immunoprecipitation (ChIP) of S. cerevisiae grown in raffinose before GapR induction. AT content (top), ChIP-seq (middle) of GapR-3xFLAG (orange) or untagged GapR (gray) expressing cells. Transcription of the forward (green) and reverse (blue) strands with annotated genes indicated (bottom). (B) Mean GapR enrichment (GapR-3xFLAG ChIP normalized by untagged ChIP) in a 500 bp window at the 5' and 3' end of long genes. Student’s t-test p-value is shown. (C) Mean GapR enrichment at 5' and 3' ends of long genes at various transcriptional cutoffs. Student’s t-test p-value is shown. (D) Examples of GapR-3xFLAG (orange) and untagged GapR (gray) ChIP. Transcription of the forward (green) and reverse (blue) strands with annotated genes indicated. Expression values are colored using the same rpkm cutoffs as in (C). (E) GapR is enriched between convergently oriented genes. Student’s t-test, convergent versus all other regions, p<10−56. (F) GapR-bound regions are more frequently between convergent genes. Fisher’s exact test, GapR-enriched versus -denriched, p<10−13. Pie charts shown as in Figure 3H.
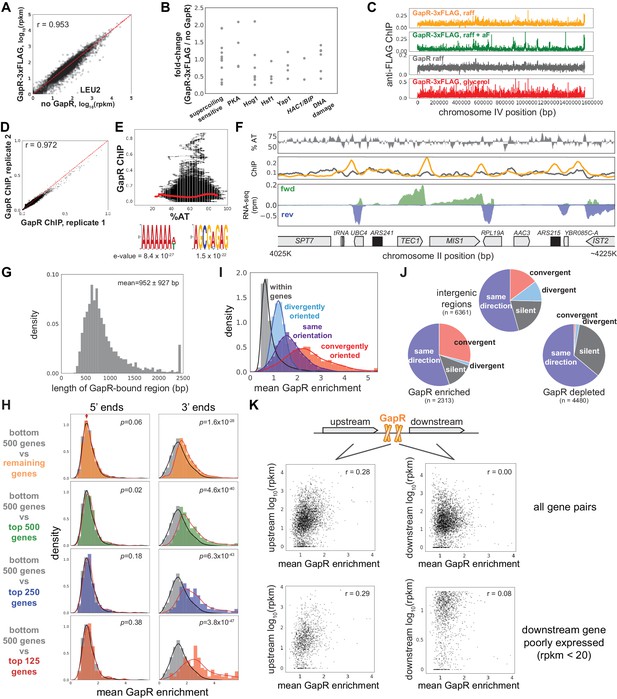
S. cerevisiae GapR chromatin immunoprecipitation-sequencing (ChIP-seq).
(A) Transcriptional profiles of cells with and without GapR-3xFLAG integrated at LEU2. The largest change in expression pattern is at the LEU2 locus. (B) Changes in gene expression upon GapR expression in supercoiling-sensitive genes and stress response pathways (see Materials and methods). (C) ChIP-seq profiles for GapR-3xFLAG (orange) and untagged GapR (gray) cells grown in glycerol. ChIP-seq profiles for GapR-3xFLAG cells grown in raffinose and in raffinose + α-factor. Cells were grown to OD 0.3–0.5 before addition of 2% galactose for 6 hr followed by fixation and ChIP. (D) Correlation between two independent GapR-3xFLAG ChIP-seq experiments in raffinose. (E) GapR-3xFLAG ChIP-seq in raffinose versus AT content. Mean enrichment at a given % AT (red dots). Motifs from DREME (below). (F) GapR ChIP profiles in raffinose. AT content (top), AT content below the genomic average (64%) is plotted in reverse. Normalized ChIP-seq (middle) of GapR-3xFLAG (orange) or untagged GapR (gray) expressing cells. Transcription (bottom) from the forward (green) and reverse (blue) strands with annotated genes indicated. (G) Histogram showing the length of GapR binding events. (H) Mean GapR enrichment in glycerol at 5' and 3' ends of long genes at various transcriptional cutoffs. Student’s t-test p-value is shown. (I) GapR enrichment in glycerol is highest between convergently oriented genes. (J) GapR-bound regions in glycerol are more frequently between convergent genes. Pie charts shown as in Figure 3H. (K) GapR enrichment between co-oriented genes is correlated with expression from the upstream gene and is uncorrelated with expression of the downstream gene.
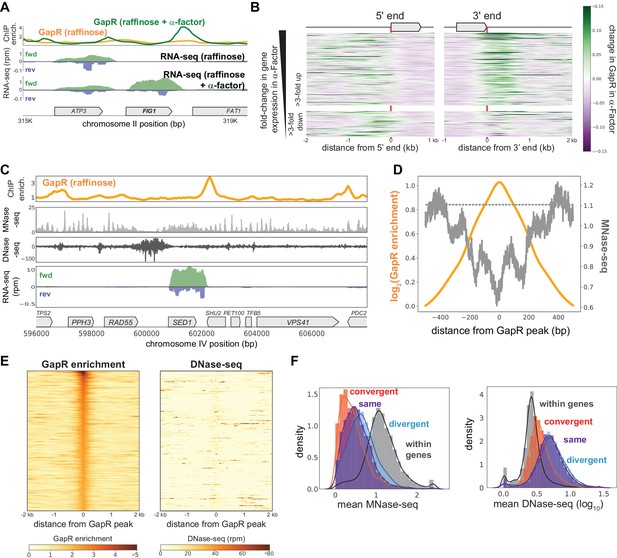
GapR binding in S. cerevisiae is responsive to transcription and is not restricted to open chromatin.
(A) GapR enrichment at Figure 1 in raffinose without (orange) or with (green) α-factor arrest before GapR induction (top). Transcription of the forward (green) and reverse (blue) strands in raffinose without (second panel) or with (third panel) α-factor arrest with annotated genes indicated. (B) Heatmap showing α-factor dependent change in GapR enrichment at the 5' and 3' ends of long transcription units (TUs) sorted by transcriptional change in α-factor. (C) GapR enrichment (orange) compared to nucleosome occupancy (MNase-seq, light gray) and chromatin accessibility (DNase-seq, dark gray). Transcription of the forward (green) and reverse (blue) strands with annotated genes indicated. (D) GapR chromatin immunoprecipitation (ChIP) peaks are de-enriched for nucleosomes. Log2(GapR enrichment) (orange, left y-axis), MNase-seq reads (dark gray, right y-axis), and mean genomic MNase-seq occupancy (dashed gray line). (E) Heatmap of GapR enrichment (left) and DNase accessibility (right) of the 500 most GapR-enriched loci. (F) Association between transcriptional orientation and MNase-seq and DNase-seq.
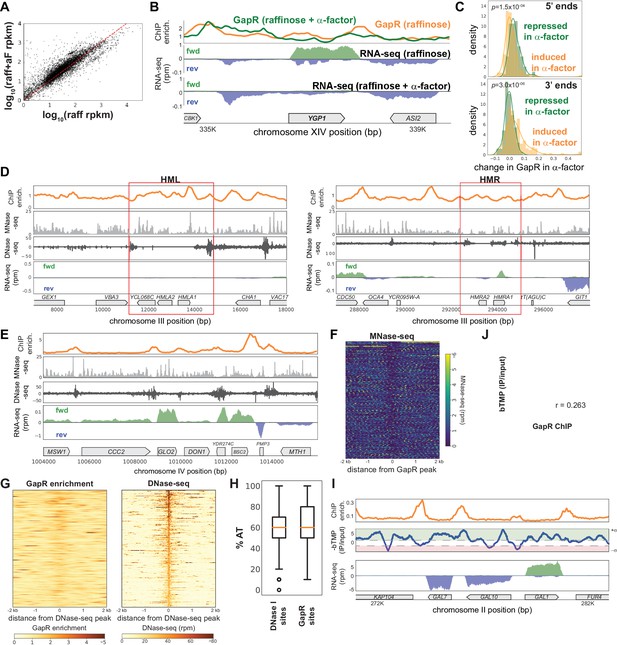
GapR chromatin immunoprecipitation (ChIP) compared to nucleosome occupancy and chromatin accessibility.
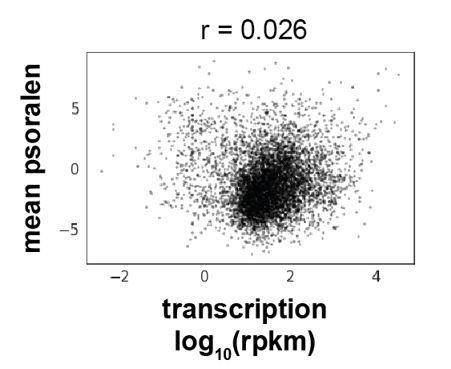
(A) Comparison of transcription in raffinose with and without α-factor treatment. (B) GapR enrichment at YGP1 in raffinose without (orange) or with (green) α-factor arrest before GapR induction (top). Transcription of the forward (green) and reverse (blue) strands in raffinose without (second panel) or with (third panel) α-factor arrest with annotated genes indicated. (C) α-factor-dependent GapR enrichment at 5' and 3' ends for induced versus repressed genes. Student’s t-test p-value is reported. (D) GapR enrichment in raffinose (orange) compared to nucleosome occupancy measured by MNase-seq (light gray) and chromatin accessibility measured by DNase-seq (dark gray) at the yeast mating loci, HML and HMR. Transcription from the forward (green) and reverse (blue) strands with annotated genes indicated (bottom). (E) GapR binding compared to nucleosome occupancy and DNase-seq at a locus on chromosome IV as in (D). (F) Heatmap showing MNase-seq at the 500 highest GapR-enriched regions. (G) Heatmap showing GapR enrichment in raffinose at the 500 most DNAase-accessible regions. (H) Boxplot showing the %AT of GapR-enriched and DNase I-sensitive sites showing mean, interquartile range, and outliers. (I) GapR enrichment in raffinose (orange) compared with psoralen tiling array data (blue, plotted in reverse). Psoralen data peaks above threshold (+1.5) were called as ‘negative supercoils’ (−σ, red) and peaks below threshold (−1.5) were called as ‘positive supercoils’ (+σ, green). Transcription from the forward (green) and reverse (blue) strands with annotated genes indicated (bottom). (J) Correlation between genomic psoralen enrichment (bTMP IP/input) and GapR-3xFLAG ChIP-seq calculated for every 100 bp.
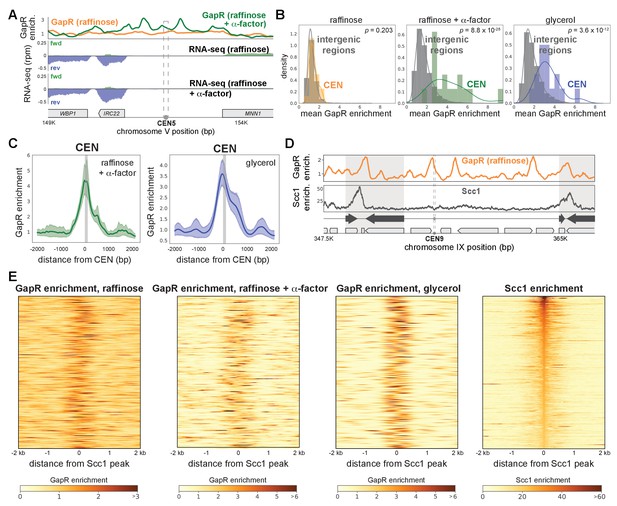
Positively supercoiled DNA is associated with centromeres and cohesin.
(A) GapR enrichment at CEN5 in cells without (orange) or with (green) α-factor arrest (top). Transcription of the forward (green) and reverse (blue) strands in cells grown in raffinose without (second panel) or with (third panel) α-factor arrest with annotated genes indicated. (B) GapR enrichment at centromeres in cells grown in raffinose, after α-factor arrest, and grown in glycerol. Student’s t-test p-value is shown. (C) GapR enrichment over all centromeres after α-factor arrest (green) or grown in glycerol (blue). Mean enrichment (solid line) with 95% confidence intervals (shaded area). Gray bar represents position of centromeres. (D) GapR (raffinose, top) and cohesin (Scc1 enrichment, bottom) are associated with convergent genes (arrows) at pericentromere boundaries (shaded areas). (E) Heatmaps of GapR (three left panels) and Scc1 (right) enrichment at the 500 most Scc1-bound loci.
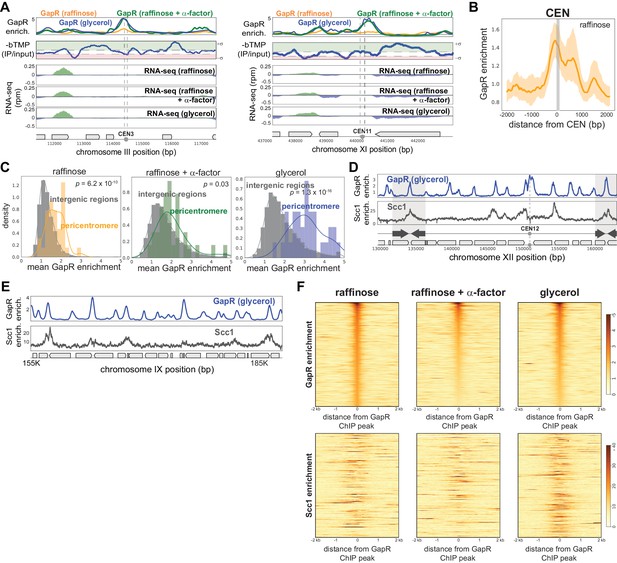
GapR chromatin immunoprecipitation (ChIP) at centromeres, pericentromeres, and cohesin-bound regions.
(A) GapR enrichment (top) in glycerol (blue), in raffinose (orange) and after α-factor arrest (green) at CEN3 (left) and CEN11 (right). Supercoiling based on psoralen tiling array data (blue, plotted in reverse). Psoralen data peaks above threshold (+1.5) were called as ‘negative supercoils’ (−σ, red) and peaks below threshold (−1.5) were called as ‘positive supercoils’ (+σ, green). Transcription from the forward (green) and reverse (blue) strands with annotated genes indicated (bottom). (B) GapR enrichment over all centromeres in raffinose. Mean enrichment (solid line) with 95% confidence intervals (shaded area). Gray bar represents position of centromeres. (C) GapR enrichment at pericentromeres in cells grown in raffinose, after α-factor arrest, and grown in glycerol. Student’s t-test p-value is shown. (D) GapR (glycerol, top) and cohesin (Scc1 enrichment, bottom) are associated with convergent genes (arrows) that mark the boundaries of pericentromeres (shaded areas). (E) GapR and cohesin binding outside of pericentromeres as in (D). (F) Heatmap showing GapR (top) and cohesin (bottom) enrichment for the 500 most GapR-enriched genomic regions in raffinose, after α-factor arrest, and in glycerol.
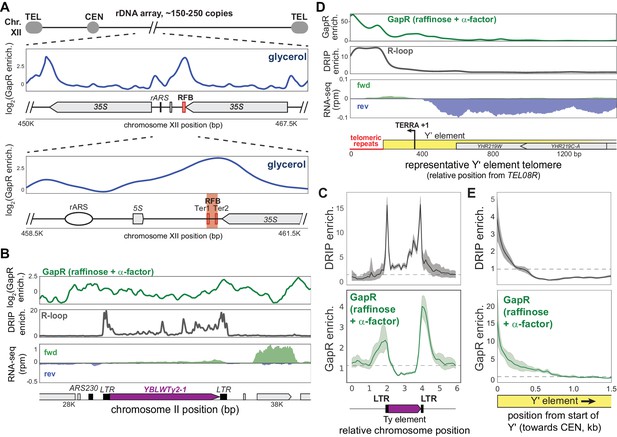
Positive supercoiling is associated with autonomously replicating sequences (ARS) and R-loops.
(A) GapR enrichment (glycerol) at the rDNA shown in two successive zoom-ins (top and bottom panels) with the replication fork barrier (RFB) and termination sequences (Ter1/Ter2) indicated. (B) Log2(GapR enrichment) (α-factor) at a Ty element (top). DNA-RNA hybrid formation by DRIP-seq (middle). Transcription of the forward (green) and reverse (blue) strands with annotated genes indicated (bottom). (C) Alignment of GapR and DRIP enrichment surrounding all yeast Ty elements. Data indicate mean (solid line) with 95% confidence intervals (shaded area), no enrichment (dotted line). (D) GapR enrichment at a telomere with a Y' element (top). DRIP enrichment (middle). Transcription of the forward (green) and reverse (blue) strands with the organization of the Y' element indicated (bottom). Position given is from end of TEL08R. (E) Alignment of GapR and DRIP enrichment surrounding all yeast telomeres with Y' elements as in (C). Telomeric repeats are removed from analysis.
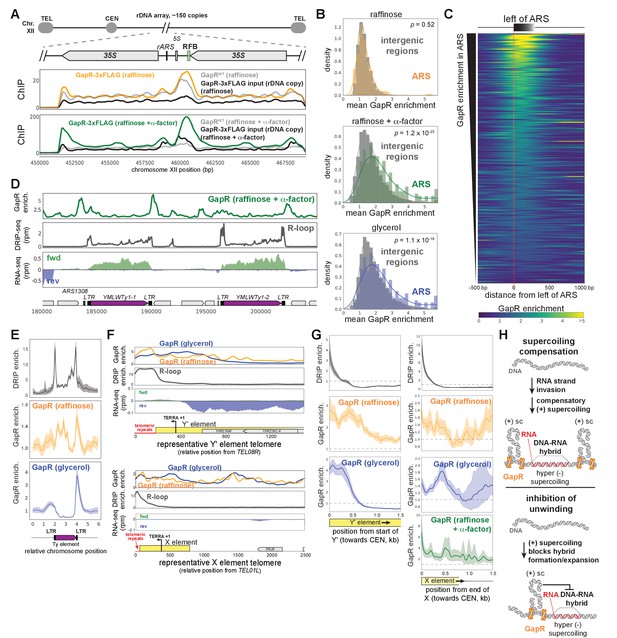
GapR chromatin immunoprecipitation (ChIP) at rDNA, autonomously replicating sequences, and R-loops.
(A) GapR ChIP with controls in the rDNA locus. GapR-3xFLAG ChIP, GapR-3xFLAG input, GapR ChIP, in raffinose (top), GapR-3xFLAG ChIP, GapR-3xFLAG input, GapR ChIP, in raffinose + α-factor (bottom). (B) GapR enrichment at autonomously replicating sequences (ARS) in cells grown in raffinose, after α-factor arrest, and grown in glycerol. Student’s t-test p-value is shown. (C) Heatmap showing GapR enrichment (glycerol) at all ARS sorted by GapR enrichment. (D) Examples of GapR enrichment near Ty elements (purple). GapR enrichment after α-factor arrest (top), DNA-RNA hybrids by DRIP-seq (dark gray). Transcription from the forward (green) and reverse (blue) strands with annotated genes indicated (bottom). (E) Alignment of GapR and DRIP enrichment surrounding all yeast Ty elements. Data indicate mean (solid line) with 95% confidence intervals (shaded area). (F) Examples of GapR enrichment near a telomere with (top) or without (bottom) a Y' element, plotted as in (D). (G) GapR and DRIP enrichment at all telomeres with (left) and without (right) Y' elements, aligned from the end of the telomeric repeat. Data indicate mean (solid line) with 95% confidence intervals (shaded area). X element represents both the ‘core’ X element and any X element combinatorial repeats. (H) Association of DNA-RNA hybrid (R-loop) forming regions and positive supercoiling.

Comparison of contaminating nuclease in GapR prep in the presence or absence of magnesium.
At high concentrations of GapR, GapR binding to plasmid DNA protects DNA from nuclease degradation.