The GNU subunit of PNG kinase, the developmental regulator of mRNA translation, binds BIC-C to localize to RNP granules
Figures
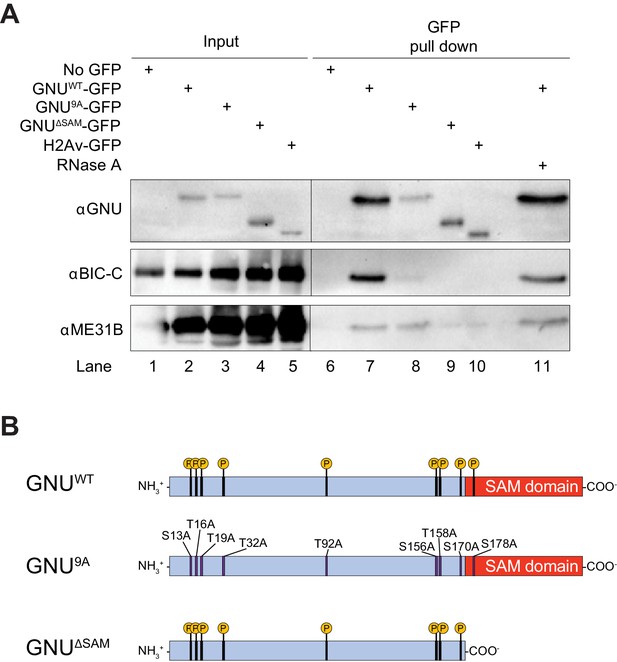
GNU physically associates with BIC-C and ME31B in mature oocytes.
(A) Immunoblot analysis of GNU-GFP immunoprecipitation of extracts from mature oocytes. Anti-GFP magnetic beads were used to perform pull-downs of GNU-GFP from extracts prepared from isolated mature oocytes expressing gnuwt-gfp, gnu9A-gfp, or gnuΔSAM-gfp transgenes. GFP immunoprecipitations from no transgene (no GFP) and h2av-gfp control extracts controlled for interactions with the beads or GFP tag. GNUWT-GFP pull-down results in immunoprecipitation of BIC-C and ME31B (lane 7). Neither BIC-C nor ME31B is immunoprecipitated in no transgene controls (lane 6), but some ME31B is immunoprecipitated with H2Av-GFP (lane 10). Both ME31B and BIC-C are immunoprecipitated by GNU9A-GFP (lane 8), whereas some ME31B, but not BIC-C, is immunoprecipitated by GNUΔSAM-GFP (lane 9). Treatment of extracts with 100 μM RNase A does not affect immunoprecipitation of BIC-C or ME31B by GNUWT-GFP (lane 11). Quantitation of the immunoblot can be found in Figure 1—figure supplement 3. (B) Schematic of GNU mutant proteins. The hypophosphorylated mutant GNU, GNU9A, has alanine substitutions at all CDK1 phosphorylation sites. The SAM domain of GNU (amino acids 172–240) has been deleted in the GNUΔSAM mutant protein, while the CDK1 phosphorylation sites remain unaffected. The fusion of GFP to the C-terminus used in the experiments is not shown.
-
Figure 1—source data 1
Raw immunoblots from Figure 1A and figure with labeled bands.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig1-data1-v2.zip
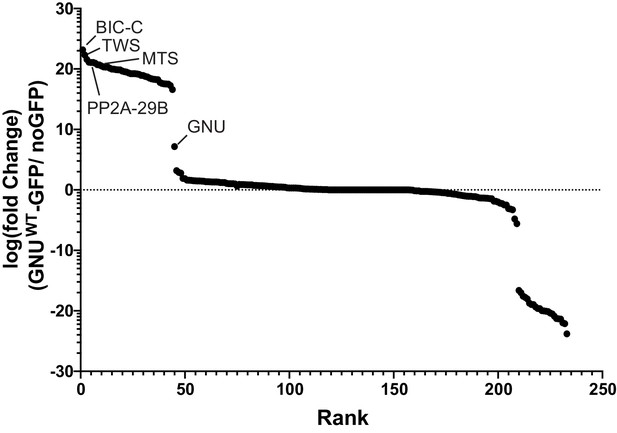
GNU interacts with BIC-C and PP2A subunits.
Enrichment plot showing fold enrichment of proteins immunoprecipitated from gnuwt-gfp or no GFP oocyte extracts. Immunoprecipitated proteins were identified by mass spectrometry. Label-free quantification of the mass spectrometry data was analyzed using Scaffold to determine fold enrichment. Proteins with at least a sevenfold enrichment in GNUWT-GFP pull-downs versus no GFP pull-downs were considered interactors of GNU. BIC-C was the most enriched protein in GNUWT-GFP pull-downs. Subunits of PP2A phosphatase (TWS, MTS, and PP2A-29B) were also significantly enriched. At least two independent replicate runs were performed for each IP-mass spectrometry analysis. Supplementary file 1 is a table that lists the interactors with GNU ranked by enrichment over the no GFP control.
-
Figure 1—figure supplement 1—source data 1
Total spectrum counts resulting from proteins identified through IP-MS of GNU-WT-GFP and no GFP control.
Results are shown in Figure 1—figure supplement 1 and supplementary file 1.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig1-figsupp1-data1-v2.xlsx
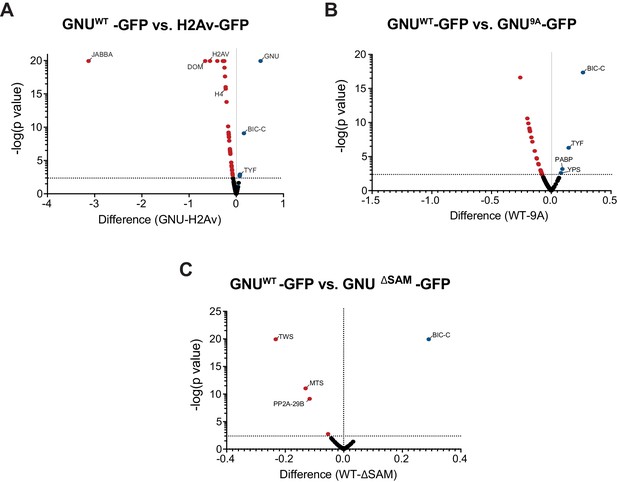
Specificity of GNU interactions and effect of the CDK1 phosphorylation mutants and deletion of the SAM domain of GNU.
Anti-GFP magnetic beads were used to perform pull-downs of GNU-GFP from extracts prepared from isolated mature oocytes expressing gnuwt-gfp, gnu9A-gfp, or gnuΔSAM-gfp transgenes. GFP immunoprecipitations from h2av-gfp control extracts controlled for interactions with the beads or GFP tag. Proteins were identified by mass spectrometry. At least two independent replicate runs were performed for each IP-mass spectrometry analysis. (A) Volcano plot showing significant differences between GNUWT-GFP and H2Av-GFP. Total spectrum counts of each identified protein were normalized to the total spectrum count for GFP in each replicate sample. Multiple t-test analysis was then performed on the normalized values to determine significant changes for each protein. As expected, the H2Av-GFP-immunoprecipitated proteins were significantly enriched for H2Av and other histones, as well as chromatin remodeling enzymes, histone chaperones, and histone-modifying enzymes. GNU, BIC-C, PABP, TYF, and TWS were significantly higher in GNUWT-GFP-immunoprecipitated proteins. Notably, most of the interactors with GNU were absent in H2Av-GFP-immunoprecipitated proteins. (B) Volcano plot showing significant differences between GNUWT-GFP and GNU9A-GFP, analyzed as in (A). The interaction of GNU with BIC-C and TYF was significantly reduced in GNU9A-GFP as compared to GNUWT-GFP (****p<0.000001 and *p=0.004, respectively). (C) Volcano plot showing significant differences between GNUWT-GFP and GNUΔSAM-GFP, analyzed as in (A). Only the BIC-C interaction was significantly reduced in GNUΔSAM-GFP as compared to GNUWT-GFP (****p<0.000001). In contrast, only the interactions of GNU with subunits of the PP2A phosphatase were significantly higher in GNUΔSAM-GFP as compared to GNUWT-GFP (TWS, ****p<0.000001; MTS, ***p=0.00048; PP2A-29B, **p=0.0018).
-
Figure 1—figure supplement 2—source data 1
Total spectrum counts for GNU-WT-GFP, GNU-deltaSAM-GFP, and H2Av-GFP pull-downs.
Results are shown in Figure 1—figure supplement 2 and Table 1.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig1-figsupp2-data1-v2.xlsx
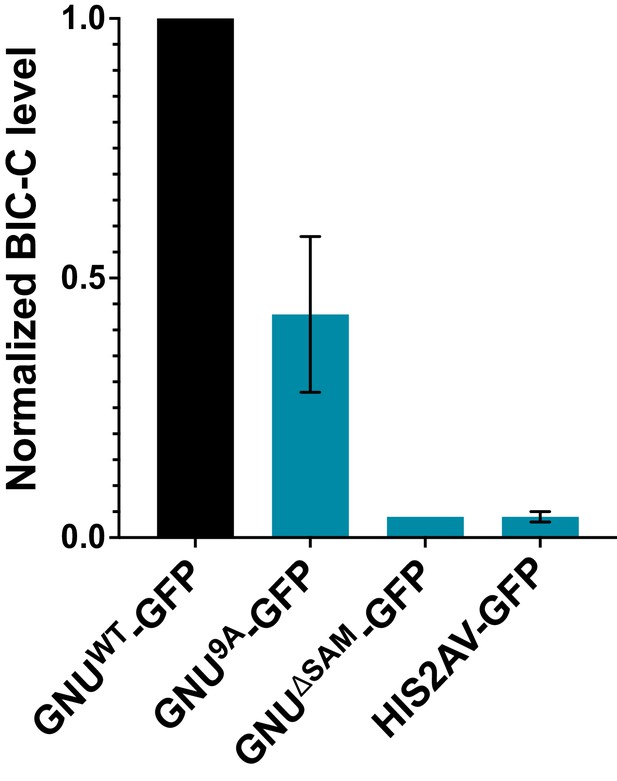
Quantitation of immunoprecipitated BIC-C bands in Figure 1A.
BIC-C levels were quantified and normalized to GNU levels in each precipitated sample. The graph shows the BIC-C levels for each GFP-IP relative to the levels of BIC-C precipitated with GNUWT-GFP. The error bars represent the SEM and correspond to three biological replicates. Significantly less BIC-C protein immunoprecipitated with the GNU9A-GFP than with GNUWT-GFP (unpaired t-test, *p=0.0292). BIC-C protein was immunoprecipitated with GNUΔSAM-GFP to a significantly lower degree than with GNUWT-GFP (unpaired t-test, ****p<0.0001). The level of BIC-C immunoprecipitated with GNU9A-GFP did not significantly differ from the level immunoprecipitated with GNUΔSAM-GFP (unpaired t-test, p=0.0884) or His2Av-GFP (unpaired t-test, p=0.846). BIC-C was immunoprecipitated at comparable levels with GNUΔSAM-GFP and His2Av-GFP (unpaired t-test, p=0.4371).
-
Figure 1—figure supplement 3—source data 1
Quantification of immunoblots shown in Figure 1A.
Bar graph of quantification is in Figure 1—figure supplement 3.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig1-figsupp3-data1-v2.xlsx
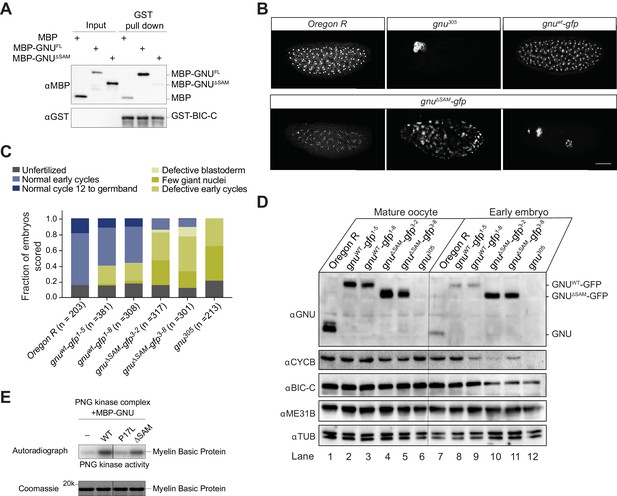
Deletion of the SAM domain of GNU reduces the interaction with BIC-C and confers partial GNU function.
(A) Immunoblot analysis of in vitro pull-down of GNU by BIC-C. Recombinantly expressed and purified MBP-tagged full-length GNU or GNUΔSAM was incubated with GST-BIC-C followed by a GST pull-down. As a control, GST pull-down after incubation with MBP was performed. In contrast to the robust pull-down of MBP-GNU, only slight amounts of MBP-GNUΔSAM were pulled down by GST-BIC-C. The levels of GST-BIC-C pulled down are comparable between all samples. (B, C) Fertilized embryos were collected for 2 hr from wild-type (Oregon R), gnuwt-gfp (gnuwt-gfp; gnu305/ gnu305), gnuΔSAM-gfp (gnuΔSAM-gfp; gnu305/gnu305), or gnu305 (gnu305/gnu305) females. Embryos were fixed and stained with DAPI. (B) Representative images of embryonic phenotypes. The embryos from wild-type and gnuwt-gfp mothers show normal early nuclear division cycles, whereas the gnu305 embryo shows only a few giant nuclei. These nuclei are the consequence of DNA replication in the absence of nuclear division; the number of separate nuclei depends on whether polyploid polar bodies fuse and whether any mitotic divisions occur (Freeman and Glover, 1987; Lee et al., 2003). The gnuΔSAM-gfp embryos show (from left to right panels) normal early cycles, defective blastoderm, and a few giant nuclei. Scale bar represents 100 μm. (C) Quantification of the embryonic phenotypes. Two independent transgenic lines were analyzed for gnuwt-gfp (gnuwt-gfp1-5 and gnuwt-gfp1-8) and gnuΔSAM-gfp (gnuΔSAM-gfp3-2 and gnuΔSAM-gfp3-8). At least 300 embryos were scored for each transgenic line and at least 200 for the Oregon R and gnu305 controls. (D) Immunoblot analysis of protein levels in mature oocytes and embryos from gnu mutants. Extracts were made from mature oocytes and embryos collected for 1 hr from Oregon R, gnuwt-gfp, gnuΔSAM-gfp, and gnu305 females, and the levels of GNU, CYCB, BIC-C, and ME31B were examined by immunoblot. αTUB was used as a loading control. Two independent transgenic gnuwt-gfp and gnuΔSAM-gfp lines were examined. 30 oocytes or embryos were collected for each sample, and the equivalent of 10 oocytes was loaded into the gel per sample. Shown is one of two biological replicates. (E) In vitro assay of PNG kinase activity. Purified MBP-tagged GNUWT, GNUΔSAM, or GNUP17Lwas incubated with the recombinant PNG kinase complex and Myelin Basic Protein (an in vitro phosphorylation target of PNG). Levels of phosphorylation of Myelin Basic Protein by PNG with radiolabeled phosphate were measured by autoradiography. MBP-GNUP17L was used as a negative control, as this amino acid change affects the ability of GNU to activate PNG kinase. In contrast, both GNUWT and GNUΔSAM activate PNG kinase. The levels of Myelin Basic Protein are comparable across samples, as assessed by Coomassie staining.
-
Figure 2—source data 1
Raw immunoblots from Figure 2A and figure with labeled bands.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig2-data1-v2.zip
-
Figure 2—source data 2
Total embryo counts for gnu rescue experiment results in Figure 2C.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Raw immunoblots from Figure 2D and figure with labeled bands.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig2-data3-v2.zip
-
Figure 2—source data 4
Raw Coomassie-stained gel and autoradiograph from Figure 2E.
Figure with labeled bands.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig2-data4-v2.zip
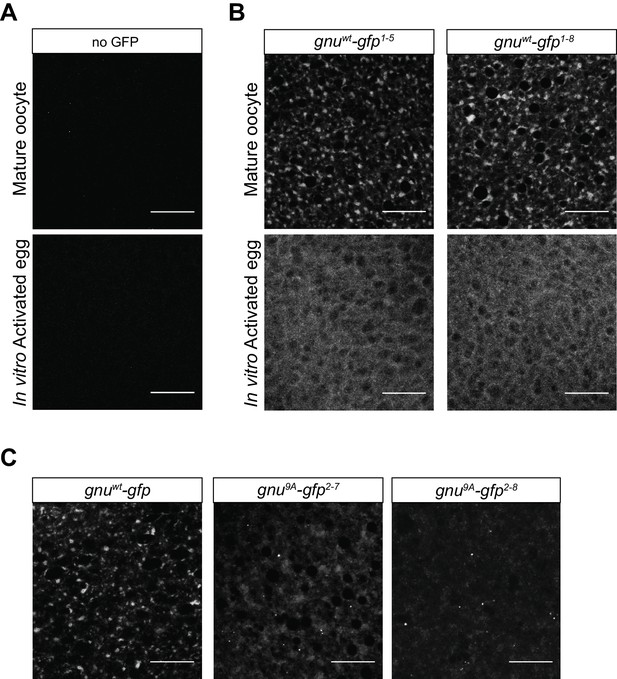
GNU localizes to granular cytoplasmic structures in mature oocytes.
Mature oocytes were isolated from gnuwt-gfp or gnu9A-gfp transgenic females. Oocytes were fixed and the vitelline membrane was removed manually before staining with the anti-GFP booster. A no GFP transgene control was analyzed for comparison. For imaging of activated eggs, mature oocytes were isolated and activated in vitro by incubation in the hypotonic buffer for 20 min. Successfully activated eggs were selected by bleach treatment, fixed with methanol, and stained with the anti-GFP booster and Hoechst 33342. Each image is a maximum intensity projection from five stacks of a z-series of the cytoplasm of one mature oocyte or activated egg. Scale bars represent 20 μm. (A) Representative images of no GFP transgene oocyte (top panel) and activated egg (bottom panel), stained with an anti-GFP booster. (B) Representative images of gnuwt-gfp transgenic oocyte (top panels) and activated egg (bottom panels), stained with an anti-GFP booster. Two different gnuwt-gfp transgenic lines (gnuwt-gfp1-5 and gnuwt-gfp1-8) were analyzed. A granular cytoplasmic localization pattern is observed for GNUWT-GFP. (C) Representative images of gnu9A-gfp transgenic oocytes, stained with an anti-GFP booster. Two different gnu9A-gfp transgenic lines (gnu9A-gfp2-7 and gnu9A-gfp2-8) were analyzed. A representative image of gnuwt-gfp oocytes is shown for comparison. A diffuse cytoplasmic signal and bright puncta are observed for GNU9A-GFP in oocytes from both gnu9A-gfp lines.
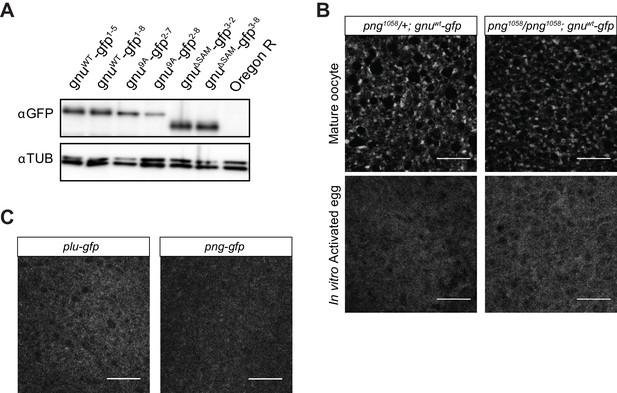
Comparison of GFP-tagged GNU levels in gnu-gfp transgenic lines, localization of PLU and PNG in mature oocytes, and GNU-GFP localization in png mutant oocytes.
(A) Immunoblot analysis of mature oocytes from gnu-gfp transgenic lines. Extracts prepared from mature oocytes isolated from Oregon R, gnuwt-gfp, gnu9A-gfp, and gnuΔSAM-gfp females were immunoblotted, examining the levels of GFP and αTUB (as a loading control). Two independent lines were analyzed for each transgene. The levels of GFP-tagged GNU are comparable across most transgenes, with slightly lower levels in gnu9A-gfp2-8 oocytes. (B) Comparison of GNU-GFP localization in png1058/+ or png1058/png1058 mature oocytes or activated eggs. Mature oocytes were isolated from either png1058/+; gnuwt-gfp or png1058/png1058; gnuwt-gfp females, fixed, and the vitelline membrane removed manually before staining with an anti-GFP booster. For imaging of activated eggs, mature oocytes were isolated and activated in vitro by incubation in the hypotonic buffer for 20 min, before fixation with methanol and staining with anti-GFP booster and DAPI. No difference in GNU-GFP localization in oocytes and activated eggs is observed between png1058/+ and png1058/png1058. Scale bars represent 20 μm. The image shown is a maximum intensity projection of five stacks in a z-series from one oocyte. (C) Representative images of plu-gfp and png-gfp transgenic oocytes stained with an anti-GFP booster. Mature oocytes were isolated, fixed, and the vitelline membrane removed manually before staining with an anti-GFP booster. A diffuse signal with no specific localization pattern is observed for both PLU-GFP and PNG-GFP. Scale bars represent 20 μm. The image shown is a maximum intensity projection of five stacks a z-series from one oocyte.
-
Figure 3—figure supplement 1—source data 1
Raw immunoblots from Figure 3—figure supplement 1 and figure with labeled bands.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig3-figsupp1-data1-v2.zip
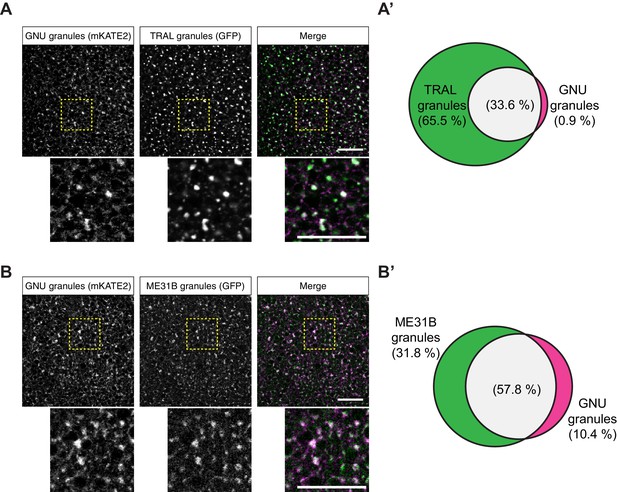
GNU-mKATE2 co-localizes with TRAL-GFP and ME31B-GFP granules in mature oocytes.
Mature oocytes were isolated from gnuwt-mkate2;tral-gfp or me31b-gfp;gnuwt-mkate2 females, fixed, and the vitelline membrane removed manually. Oocytes were stained with the anti-GFP booster and imaged by confocal microscopy for fluorescence at 488 nm to detect GFP and 568 nm to detect mKATE2. mKATE2 signal was detected without the use of a booster. Co-localization was measured by quantification of overlap between GFP+ granules and mKATE2+ granules using the surface-surface co-localization algorithm in Imaris (Bitplane). (A) Representative image of gnuwt-mkate2;tral-gfp oocytes. Co-localizing GNU-mKATE2 (magenta) and TRAL-GFP (green) granules are colored in white. The images shown are single slices of confocal z-stacks from one oocyte. Bottom images show the insets of each panel (dashed yellow box). Scale bar represents 20 μm. (A’) Venn diagram of quantified co-localization between GNU and TRAL granules. GNU and TRAL co-localize in 33.6±5.2% of all granules quantified. GNU-containing TRAL granules represent approximately a third of TRAL granules scored. Values are averaged across eight oocytes. (B) Representative image of me31b-gfp;gnuwt-mkate2 oocytes. Co-localizing GNU-mKATE2 (magenta) and ME31B-GFP (green) granules are colored in white. The images shown are single slices of confocal z-stack from one oocyte. Bottom images show the insets of each panel (dashed yellow box). Scale bar represents 20 μm. (B’) Venn diagram of quantified co-localization between GNU and ME31B granules. GNU and ME31B co-localize in 57.8±4.6% of all granules quantified. GNU-containing ME31B granules represent approximately half of ME31B granules scored. Values are averaged across eight oocytes.
-
Figure 4—source data 1
Quantification data for co-localization experiments in Figure 4 and Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig4-data1-v2.xlsx
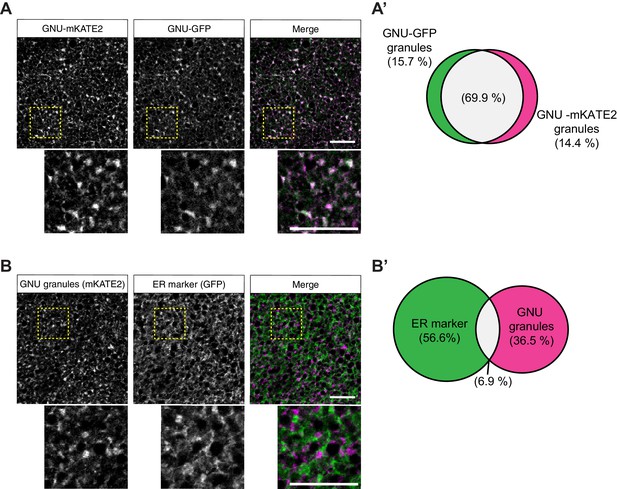
GNU-mKATE recapitulates mature oocyte localization of GNU-GFP.
Mature oocytes were isolated from gnuwt-mkate2;gnuwt-gfp or gnuwt-mkate2;pdi-gfp females, fixed, and the vitelline membrane removed manually. Oocytes were stained and imaged as in Figure 4. (A) Representative image of gnuwt-mkate2;gnuwt-gfp oocytes. Co-localizing GNU-mKATE2 (magenta) and GNU-GFP (green) granules are colored in white. (A’) Venn diagram of quantified co-localization between GNU-mKATE2 and GNU-GFP granules. GNU-mKATE2 and GNU-GFP co-localize in 69.9±4.1% of all granules quantified. (B) Representative image of gnuwt-mkate2;pdi-gfp oocyte. Co-localizing GNU-mKATE2 (magenta) and ER (based on the ER-resident protein PDI-GFP in green) are colored in white. (B’) Venn diagram of quantified co-localization between GNU and ER. GNU and the ER marker (PDI-GFP) co-localize in 6.9±3.2% of all granules quantified. In (A) and (B), the images shown are maximum intensity projections of a z-series from one oocyte. Scale bar represents 20 μm. In (A’) and (B’), values are averaged across eight oocytes.
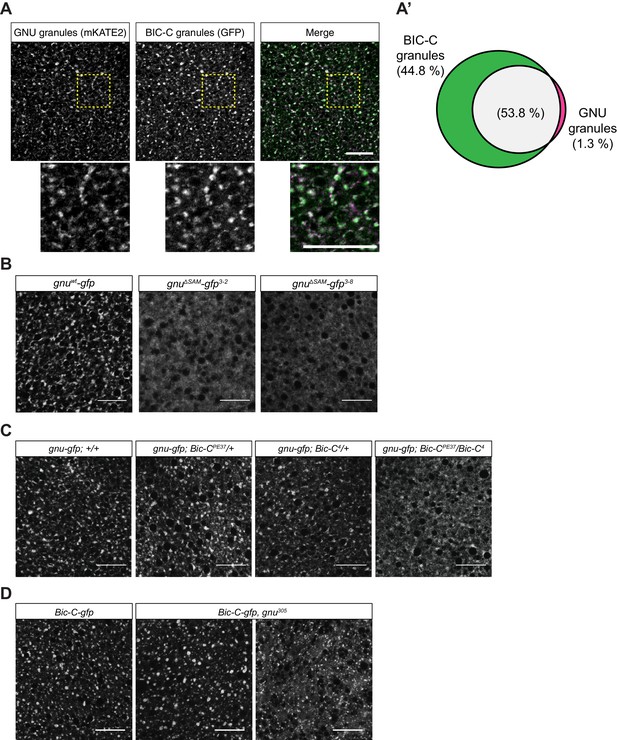
GNU localization to granules is dependent on its SAM domain, and BIC-C and GNU are co-dependent for localization.
(A, A’) Co-localization between GNU-mKATE2 and BIC-C-GFP. Mature oocytes were isolated from gnuwt-mkate2;Bic-C-gfp females and imaged and analyzed as in Figure 4. (A) Representative image of gnuwt-mkate2;Bic-C-gfp oocyte. Co-localizing GNU (magenta) and BIC-C (green) granules are colored in white. Bottom images show the insets of each panel (dashed yellow box). (A’) Venn diagram of quantified co-localization between GNU and BIC-C granules. GNU and BIC-C co-localize in 53.8±4.8% of all granules quantified. GNU-containing BIC-C granules represent approximately half of all BIC-C granules scored. Values are averaged across eight oocytes. (B) Representative images of gnuΔSAM-gfp transgenic oocytes, stained with an anti-GFP booster. Two different gnuΔSAM-gfp transgenic lines (gnuΔSAM-gfp3-2 and gnuΔSAM-gfp3-8) were analyzed. A representative image of gnuwt-gfp oocytes is shown for comparison. A diffuse cytoplasmic localization is observed for GNUΔSAM-GFP in oocytes from both gnuΔSAM-gfp transgenic lines. (C) Representative images of GNUWT-GFP localization in Bic-C mutant mature oocytes. Localization of GNUWT-GFP is comparable between gnuwt-gfp; +/+, and heterozygous oocytes for Bic-C loss-of-function alleles (gnuwt-gfp; Bic-CPE37/+ and gnuwt-gfp; Bic-C4/+). GNUWT-GFP localization is more diffuse in gnuwt-gfp; Bic-CPE37/Bic-C4 mutants. (D) Representative images of BIC-C-GFP localization in gnu mutant mature oocytes. BIC-C-GFP localizes to granules in control Bic-C-gfp oocytes. Localization of BIC-C-GFP in Bic-C-gfp;gnu305 looks comparable to Bic-C-gfp control in 70% of oocytes scored (n=20), with 30% of scored oocytes exhibiting a dispersed localization with punctate granules for BIC-C-GFP. The gnu305 allele is a protein null allele of gnu (Renault et al., 2003). In (A–D), scale bars represent 20 μm. In (B–D), the images shown are a maximum intensity projection of a z-series of five stacks from one oocyte, whereas in (A), the images shown are single slices of confocal z-stack from one oocyte.
-
Figure 5—source data 1
Quantification data for co-localization experiments shown in Figure 5A.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig5-data1-v2.xlsx
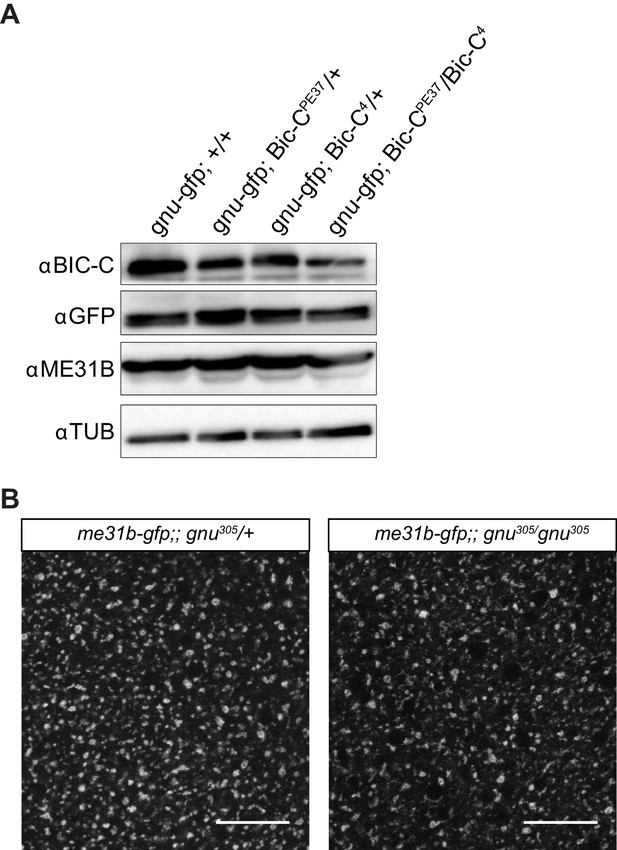
BIC-C levels are decreased in Bic-C mutant mature oocytes and ME31B localization is not affected in gnu mutants.
(A) Immunoblot analysis of extracts from mature oocytes from Bic-C mutant females. Mature oocytes were collected from gnuwt-gfp, gnuwt-gfp; Bic-CPE37/+, gnuwt-gfp; Bic-C4/+, and gnuwt-gfp; Bic-CPE37/Bic-C4 females and the levels of BIC-C, GNU (by GFP tag), ME31B, and αTUB examined by immunoblot. The levels of BIC-C protein are lower in gnuwt-gfp; Bic-CPE37/Bic-C4 than in heterozygotes for either Bic-C allele or control. Levels of ME31B and GNU-GFP are comparable across all genotypes. (B) Representative images of ME31B-GFP localization in gnu mutant mature oocytes. ME31B localization is unaffected in me31b-gfp;gnu305/gnu305 when compared to a me31b-gfp;gnu305/+ control. The gnu305 allele is a protein null allele of gnu. Scale bars represent 20 μm. The image shown is a maximum intensity projection of five stacks of a z-series from one oocyte.
-
Figure 5—figure supplement 1—source data 1
Raw immunoblots from Figure 5—figure supplement 1A and figure with labeled bands.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig5-figsupp1-data1-v2.zip
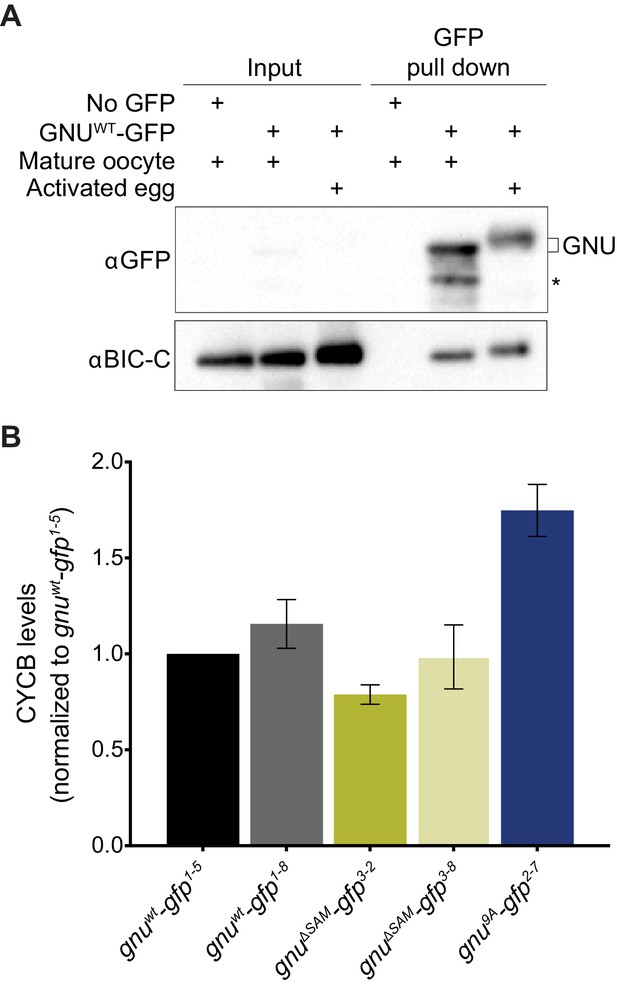
Experimental test for model that RNP granule localization of GNU prevents activation of PNG.
(A) BIC-C and GNU remain physically associated after egg activation. Anti-GFP magnetic beads were used to perform pull-downs of GNU-GFP from extracts prepared from isolated mature oocytes or in vitro activated eggs expressing gnuwt-gfp transgenes. For the analysis of activated eggs, mature oocytes were isolated and activated in vitro by incubation in hypotonic buffer for 20 min. GFP immunoprecipitations from no transgene (no GFP) mature oocyte extracts controlled for interactions with the beads or GFP tag. GNU-GFP pull-down from both mature oocyte or activated egg extracts results in immunoprecipitation of BIC-C. The asterisk marks a GNU-GFP degradation product we often observe in immunoprecipitations from mature oocytes. (B) Deletion of the SAM domain in GNU does not increase levels of CYCB in mature oocytes. Mature oocytes were isolated from gnu305 homozygous females expressing gnuwt-gfp, gnuΔSAM-gfp, or gnu9Α-gfp transgenes. The levels of CYCB and αTUB were examined by immunoblot. Two independent lines were analyzed for each transgene, except for gnu9Α-gfp for which only one line was analyzed. Levels of CYCB were quantified and normalized to TUB levels. The graph shows normalized levels of CYCB relative to gnuwt-gfp1-5 oocytes. Error bars correspond to SEM, and each bar represents five biological replicates. CYCB levels were not significantly different between oocytes from the two gnuwt-gfp lines (paired t-test, p=0.2855). No significant difference was observed between gnuwt-gfp and gnuΔSAM-gfp3-8 (paired t-test, p=0.9281), but the CYCB levels in gnuΔSAM-gfp3-2 oocyte were significantly lower than in gnuwt-gfp1-5 oocytes (paired t-test, *p=0.0137). Levels of CYCB in gnu9Α-gfp2-7 oocytes are significantly higher compared to gnuwt-gfp (paired t-test, **p=0.0053).
-
Figure 6—source data 1
Quantification of immunoblots shown in Figure 6A.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Raw immunoblots from Figure 6A and figure with labeled bands.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig6-data2-v2.zip
-
Figure 6—source data 3
Relative levels of CYCB in mature oocytes expressing GNU transgenes.
- https://cdn.elifesciences.org/articles/67294/elife-67294-fig6-data3-v2.xlsx
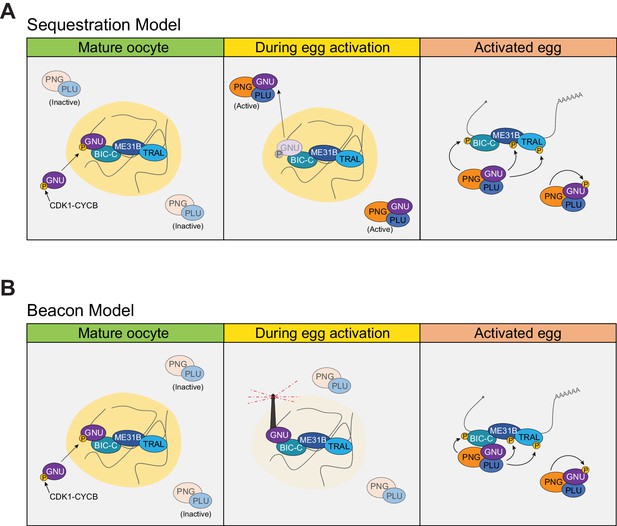
Models of regulation of PNG by GNU in RNP granules.
(A) Sequestration Model. In this model, GNU is prevented from interacting with the PNG/PLU complex by spatial separation. In mature oocytes, GNU is sequestered to granules in a CDK1 phosphorylation and SAM-domain-dependent manner (left panel), where it interacts with BIC-C, ME31B, and TRAL. PNG and PLU are localized throughout the oocyte cytoplasm. Upon egg activation, RNP granules disassemble, thus releasing GNU as it is being dephosphorylated (middle panel). In activated eggs, GNU is no longer prevented from binding PNG by sequestration of RNP granules, and PNG mediates the phosphorylation of its targets as well as autoregulation through phosphorylation of its subunits (right panel). (B) Beacon model. In this model, localization of GNU to RNP granules functions to localize PNG activity to these granules. In mature oocytes, GNU is recruited onto granules via SAM domain interactions, with CDK1 phosphorylation stabilizing its interactions within the granules (left panel). Inactive PNG is diffuse throughout the cytoplasm. As egg activation occurs, GNU is dephosphorylated and brings PNG to the disassembling granules, where it can phosphorylate TRAL, ME31B, BIC-C, and potentially other translation regulators (middle panel). In fully activated eggs, PNG activity would not be restricted to granules previously marked by GNU, and the complex is able to phosphorylate targets throughout the activated egg prior to inactivation of the complex (right panel). The beacon model is more consistent with our data than the sequestration model.
Tables
Interactors with GNU in mature oocytes identified through IP-MS.
| Identified protein | Total spectrum count | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No GFP | GNUWT-GFP | GNU9A-GFP | GNUΔSAM-GFP | H2Av-GFP | GNUWT-GFP (+RNase A) | ||||||||
| Rep 1 | Rep 2 | Rep 1 | Rep 2 | Rep 3 | Rep 1 | Rep 2 | Rep 1 | Rep 2 | Rep 1 | Rep 2 | Rep 3 | ||
| GFP | – | – | 319 | 376 | 307 | 62 | 52 | 353 | 520 | 358 | 49 | 31 | 467 |
| GNU | – | – | 179 | 178 | 220 | 81 | 90 | 152 | 109 | 3 | – | – | 205 |
| BIC-C | – | – | 92 | 13 | 73 | 5 | – | – | – | – | – | – | 90 |
| TYF | – | – | 64 | 6 | 11 | 9 | – | 45 | 5 | 11 | 3 | – | 38 |
| YPS | – | – | 40 | 11 | 23 | 10 | – | 38 | 2 | 28 | – | – | – |
| TWS | – | – | 39 | 9 | 22 | 10 | 3 | 92 | 14 | – | – | – | 32 |
| TRAL | 3 | 5 | 37 | 7 | 17 | 5 | 5 | 22 | 2 | 20 | 2 | – | 11 |
| PP2A-29B | – | – | 27 | 4 | 30 | 8 | 4 | 52 | 7 | 5 | – | – | 24 |
| CUP | – | – | 20 | 5 | 7 | 7 | 4 | 9 | – | 4 | – | – | 9 |
| ATX-2 | – | – | 20 | 6 | 9 | – | – | 23 | 6 | 6 | – | – | 16 |
| ME31B | 2 | 2 | 11 | 3 | 6 | 6 | 8 | 6 | 2 | 9 | – | – | 6 |
| MTS | – | – | 10 | – | 10 | 3 | – | 42 | 5 | – | – | – | 8 |
| RPL36A | – | – | 7 | 5 | 5 | – | – | 3 | 2 | – | – | – | 7 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | gnu | FLYB: FBgn0001120 | ||
| Gene (D. melanogaster) | png | FLYB: FBgn0000826 | ||
| Gene (D. melanogaster) | plu | FLYB: FBgn0003114 | ||
| Gene (D. melanogaster) | Bic-C | FLYB: FBgn0000182 | ||
| Gene (D. melanogaster) | tral | FLYB: FBgn0041775 | ||
| Gene (D. melanogaster) | me31b | FLYB: FBgn0004419 | ||
| Strain, strain background (D. melanogaster) | WT: Oregon R | N/A | ||
| Genetic reagent (D. melanogaster) | gnu-wt-gfp[1-5] | Hara et al., 2017 | Genotype: w;P{w[+mC], gnu-wt-gfp}; gnu[305]/ TM3, Sb | |
| Genetic reagent (D. melanogaster) | gnu-wt-gfp[1-4] | Hara et al., 2017 | Genotype: w, P{w[+mC], gnu-wt-gfp};;gnu[305]/TM3, Sb | |
| Genetic reagent (D. melanogaster) | gnu-wt-gfp[1-8] | Hara et al., 2017 | Genotype: w;P{w[+mC], gnu-wt-gfp}; gnu[305]/ TM3, Sb | |
| Genetic reagent (D. melanogaster) | gnu-9A-gfp[2-7] | Hara et al., 2017 | Genotype: w;P{w[+mC], gnu-9A-gfp}; gnu[305]/ TM3, Sb | |
| Genetic reagent (D. melanogaster) | gnu-9A-gfp[2-8] | Hara et al., 2017 | Genotype: w;P{w[+mC], gnu-9A-gfp}; gnu[305]/ TM3, Sb | |
| Genetic reagent (D. melanogaster) | tral-gfp[89] | The Flytrap Project; Morin et al., 2001; PMID:11742088 | Flytrap:G00089; DGRC:110584; RRID:DGGR_110658 | Genotype: w[*];P{w[+mC]=PTT-un1} G00089 |
| Genetic reagent (D. melanogaster) | me31b-gfp | Nakamura et al., 2001 | Genotype: w, P{[W[+mC]], me31b-gfp} | |
| Genetic reagent (D. melanogaster) | Bic-C-gfp[v318872] | Sarov et al., 2016 | RRID:VDRC_v318872 | Transgene carries dsRed marker. Genotype: w;;PBac{fTRG01264.sfGFP-TVPTBF}VK00033 |
| Genetic reagent (D. melanogaster) | Bic-C-gfp[v318334] | Sarov et al., 2016 | RRID:VDRC_v318334 | Transgene carries dsRed marker. Genotype: w;;PBac{fTRG01264.sfGFP-TVPTBF}VK00033 |
| Genetic reagent (D. melanogaster) | gnu-ΔSAM-gfp[3-2] | This paper | Genotype: w;P{w[+mC], gnu-ΔSAM-gfp}; gnu[305]/ TM3, Sb | |
| Genetic reagent (D. melanogaster) | gnu-ΔSAM-gfp[3-8] | This paper | Genotype: w;P{w[+mC], gnu-ΔSAM-gfp}; gnu[305]/ TM3, Sb | |
| Genetic reagent (D. melanogaster) | gnu-mKATE2 | This paper | Genotype: w;P{w[+mC], gnu-mKATE2}; gnu[305]/ TM3, Sb | |
| Genetic reagent (D. melanogaster) | BiC-C[4] | Schüpbach and Wieschaus, 1991 | RRID:BDSC_3248 | Genotype: Bic-C[4], cn[1], exu[1], bw[1]/CyO |
| Genetic reagent (D. melanogaster) | Bic-C[PE37] | Schüpbach and Wieschaus, 1991 | Genotype: Bic-C[PE37], bw[1]/CyO | |
| Genetic reagent (D. melanogaster) | gnu[305] | Freeman and Glover, 1987 | FLYB: FBal0005121 | Genotype: ru, gnu[305], th, st, roe, p(p), e(s), ca/TM3, Sb |
| Genetic reagent (D. melanogaster) | png[1058] | Shamanski and Orr-Weaver, 1991; PMID:1913810 | RRID:BDSC_38437 | Genotype: y[1], png[1058], w[*]/FM6 |
| Genetic reagent (D. melanogaster) | h2av-gfp | RRID:BDSC_24163 | Genotype: w[*]; P{w[+mC]=His2Av-EGFP.C}2/ SM6a | |
| Genetic reagent (D. melanogaster) | pdi-gfp | RRID:BDSC_6839 | Genotype: w[1118]; P{w[+mC]=PTT-GA}Pdi[G00198]/TM3, Sb[1] Ser[1] | |
| Genetic reagent (D. melanogaster) | me31b-gfp; Bic-C[305]/Xa | This paper | Generated from cross between me31b-gfp females and Bic-C[4] males. Genotype: w, P{[W[+mC]], me31b-gfp}; Bic-C[4]/Xa | |
| Genetic reagent (D. melanogaster) | gnu-gfp/+; Bic-C[305]/Sco | This paper | Generated from cross between gnu-wt-gfp[1-4] females and Bic-C[4] males. Genotype: w;P{w[+mC], gnu-wt-gfp}/+; Bic-C[4]/Sco | |
| Genetic reagent (D. melanogaster) | me31b-gfp;; gnu[305]/TM3 | This paper | Genotype: w, P{[W[+mC]], me31b-gfp};; gnu[305]/TM3 | |
| Genetic reagent (D. melanogaster) | png[1058]/ FM0; gnu-wt-gfp; gnu[305]/TM3 | This paper | Genotype: png[1058]/FM0; P{w[+mC], gnu-wt-gfp}/+; gnu[305]/TM3 | |
| Genetic reagent (D. melanogaster) | gnu-gfp/+; BicC[4]/Bic-C[PE37] | This paper | Generated from genetic cross between gnu-gfp/+; BicC[4]/Sco males and Bic-C[PE37] females. | |
| Genetic reagent (D. melanogaster) | plu-gfp[5-04] | This paper | PLU-GFP transgene; good expression; rescues homozygous plu. Genotype: y,w; P{plu C/ GFP E.1} | |
| Genetic reagent (D. melanogaster) | png-gfp[F-041] | This paper | PNG-GFP transgene; good expression; rescues homozygous png[1058]. Genotype: y,w; P{png-gfp} | |
| Genetic reagent (D. melanogaster) | twine/CyO | Other | RRID:BDSC_4274; RRID:Kyoto_107663 | Sterile males due to spermlessness. Genotype: twe[1] cn[1] bw[1]/CyO |
| Antibody | Guinea pig polyclonal anti-GNU | Lee et al., 2003 | PMID:14665672 | (1/5000) in TBS-T |
| Antibody | Rabbit polyclonal anti-BIC-C | P. Lasko (McGill University) | (1/2000) in TBS-T | |
| Antibody | Guinea pig polyclonal anti-GFP | M. Pardue (MIT) | (1/5000) in TBS-T | |
| Antibody | Rat monoclonal anti-αTUB YOL1/34 | AbD Serotec (Bio-Rad) | (1/1000) in TBS-T | |
| Antibody | Mouse monoclonal anti-CycB | Developmental Studies Hybridoma Bank | DSHB Cat#F2F4 RRID:AB_528189 | (1/100) in Hikari Solution B |
| Antibody | Rat monoclonal anti-MBP | Sigma-Aldrich | Sigma-Aldrich: SAB4200082 | (1/2000) in Hikari Solution |
| Antibody | Rabbit polyclonal anti-GST labeled with HRP | MBL | MBL:PM013-7; RRID:AB_10598029 | (1/5000) in Hikari solution |
| Antibody | Rabbit polyclonal anti-ME31B | Nakamura et al., 2001 | PMID:27791980; RRID:AB_2568986 | (1/10,000) in TBS-T |
| Antibody | Donkey polyclonal HRP-conjugated anti-guinea pig IgG | Jackson ImmunoResearch | Jackson Immuno Research:706-035-148: RRID:AB_2340447 | |
| Antibody | Goat polyclonal HRP-conjugated anti-mouse IgG | Jackson ImmunoResearch | Jackson Immuno Research:115-035-164: RRID:AB_2338510 | |
| Antibody | Goat polyclonal HRP-conjugated anti-rat IgG | Jackson ImmunoResearch | Jackson Immuno Research:112-035-062: RRID:AB_2338133 | |
| Antibody | Donkey polyclonal HRP-conjugated anti-rabbit IgG | Jackson ImmunoResearch | Jackson Immuno Research: 711-035-152; RRID:AB_10015282 | |
| Antibody | Recombinant Nanobody gba488-100 (GFP-Booster Atto488) | Chromotek | Booster for GFP fluorescence (1/400) in diluent buffer | |
| Recombinant DNA reagent | pCasPeR4_gnu-wt-gfp | Hara et al., 2017 | RRID:Addgene_113005 | |
| Recombinant DNA reagent | pCasPeR4_gnu-ΔSAM-gfp | This paper | Generated from pCasPeR4_gnu-wt-gfp | |
| Recombinant DNA reagent | pCasPeR4_gnu-mkate2 | This paper | Generated from pCas PeR4_gnu-wt-gfp | |
| Commercial assay or kit | gtm_20 anti-GFP magnetic beads | Chromotek | GFP immunoprecipitation | |
| Chemical compound, drug | HIKARI signal enhancer | Nacalai | Nacalai: 02270–81 | Signal Enhancer HIKARI for Western Blotting and ELISA |
| Software, algorithm | Imaris (Bitplane) | Bitplane | Imaging data visualization and analysis | |
| Software, algorithm | FIJI (ImageJ) | ImageJ | Imaging data visualization and analysis | |
| Software, algorithm | Scaffold (version 4) | Proteome Software | Proteomic data visualization and analysis |
Additional files
-
Supplementary file 1
Supplementary table showing interactors with GNU in mature oocytes identified by IP-MS in ranked order.
The fold enrichment in GNU-GFP over a no GFP control is shown. The source data for this table are in Figure 1—figure supplement 1—source data 1.
- https://cdn.elifesciences.org/articles/67294/elife-67294-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67294/elife-67294-transrepform-v2.docx





