Ribonucleotide reductase, a novel drug target for gonorrhea
Figures
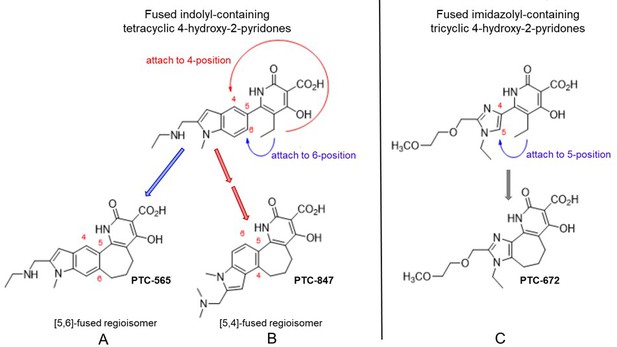
Structures of PTC-847 and PTC-672 with 4-hydroxypyridone nucleus.
Three methylene units link the 4-hydroxy-2-pyridone core through the indole C-5 and C-4 position yielding two fused seven-membered ring regioisomers with restricted conformations, shown as compounds A and B. The [5,6]-fused regioisomer (PTC-565, A) is a broad spectrum DNA gyrase and topoisomerase IV inhibitor targeting Gram-negative pathogens (Gerasyuto et al., 2018). The [5,4]-fused regioisomer with a basic dimethylamine group appended to the indole C-2 (PTC-847, B) is a potent class Ia RNR inhibitor selective for Ng. PTC-672 (C), a second selective Ng class Ia RNR inhibitor, has an imidazolyl moiety fused to the 4-hydroxy-2-pyridone through a seven-membered ring.
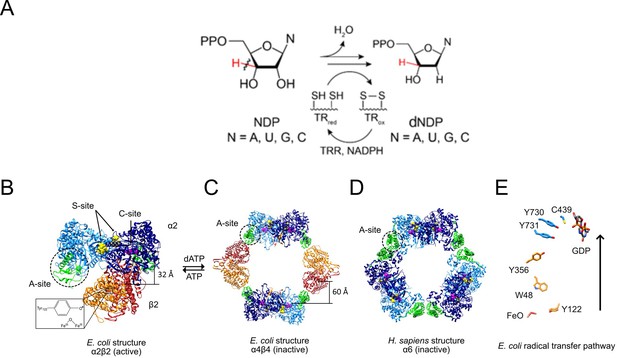
RNR reaction (A), structures of active and inactive states of Ia RNRs (B–D), and the radical transfer pathway (E).
(A) Each of the four NDPs are converted to their corresponding dNDPs, which involves cleavage of the 3′-C-H bond (red) and oxidation of two cysteines to a disulfide. Further turnover requires disulfide reduction by protein reductants thioredoxin (TR) and thioredoxin reductase (TRR). (B) A cryo-EM structure of an active Ec RNR generated from a double mutant of β2 (F3Y122•/E52Q) incubated with α2, substrate GDP (C-site) and allosteric effector TTP (S-site) (Kang et al., 2020). (C) The inactive dATP-inhibited α4β4 state of Ec RNR (Ando et al., 2011). (D) The inactive dATP inhibited α6 state of the human RNR (Brignole et al., 2018). (E) The essential diferric-tyrosyl radical cofactor and the components of the radical transfer pathway. A 32 Å distance is shown for the radical transfer pathway in B and 60 Å in C. In B-D, α2 is shown in blue with its N-terminal cone domain in green (A-site) surrounded by a dashed black circle. β2 is shown in red and orange.
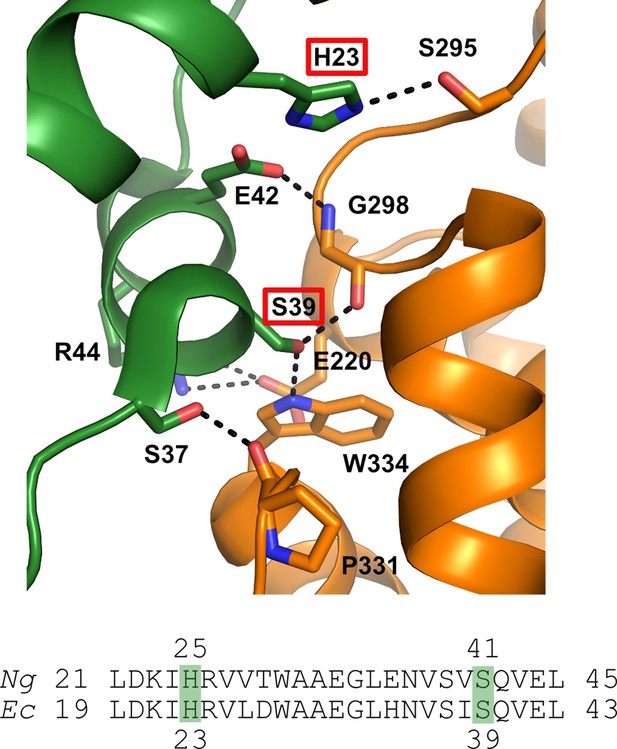
Structure of the α / β interface in the inactive a4b4 form of the E. coli class Ia RNR where β is in orange and α is in green.
Represented are the location of mutations identified in the Ng Ia RNR by isolating resistant strains of Ng and mapping the mutation sites (H25R and S41L in α). The structure for the Ec class Ia RNR is shown with the residues in Ec (H23, S39) that correspond to the mutation sites in Ng (residues H25 and S41) identified by red boxes (top) (Adapted from Figure 2, Chen et al., 2018). Also shown is the sequence alignment of α in this region for Ec and Ng.
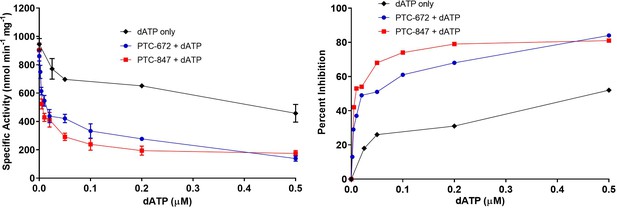
Inhibition of RNR by dATP and its potentiation in the presence of PTC-847 and PTC-672.
A 1:1 mixture of α and β subunits was incubated with various concentrations of dATP in the presence or absence of 25 nM PTC-672 or 100 nM PTC-847 and assayed for activity spectrophotometrically at 37 °C. The results show that low concentrations of PTC-672 (shown in blue) or PTC-847 (shown in red) potentiate the dATP (shown in black) effect. The activity assay used 0.1 µM α2, 0.1 µM β2, 1 mM GDP, 0.25 mM TTP, 100 µM Ec TR, 1 µM Ec TRR, 0.2 mM NADPH and various concentrations of dATP (2 nM to 1 µM) with or without 25 nM PTC-647 or 100 nM PTC-847 (n = 2 replicates at each concentration).
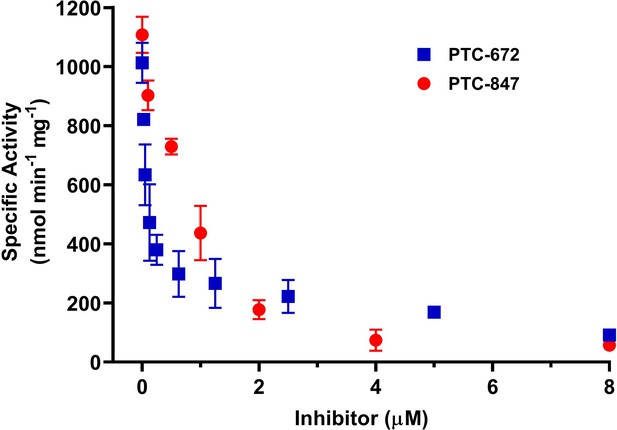
Inhibition of Ng class Ia RNR by PTC-847 and PTC-672.
A 1:1 mixture of α and β subunits was incubated with each compound and assayed for activity spectrophotometrically at 37 °C with PTC-847 (red) and PTC-672 (in blue). The assay used 0.1 µM α2, 0.1 µM β2, 1 mM GDP, 0.25 mM TTP, 100 µM Ec TR, 1 µM Ec TRR, 0.2 mM NADPH and various concentrations of PTC-847 (0.1–8 µM) or PTC-672 (0.025–8.0 µM). (n = 2 for each concentration).
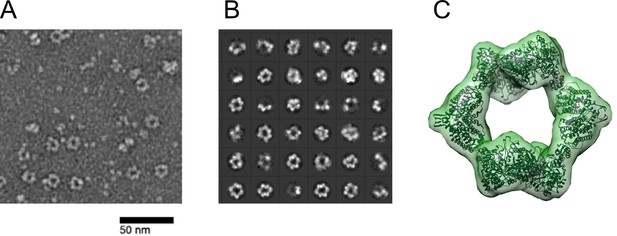
Ng RNR forms rings under inactivating conditions, as observed with Ec RNR.
(A) Representative negative stain image taken at ×84,000 nominal magnification (1.79 Å/pixel) of 1:1.5 α:β (15 ng/µL final concentration; 0.1 µM α2 and 0.15 µM β2) in the presence of 1 mM dATP and 1 mM CDP. Sample was prepared at 4 °C before staining. (B) Representative 2D class averages (36 shown of 200 total) from 27,109 picked particles from grids prepared under the same conditions as (A). (C) Negative stain reconstruction at 21 Å resolution of Ng class Ia RNR α4β4 with Ec class Ia RNR α4β4 crystal structure fit to the density (PDB 5CNS; Zimanyi et al., 2016).Recently (Levitz et al., 2021) Levitz et al reported refinement of the Ng a4b4 structure to 4.3 Ang.
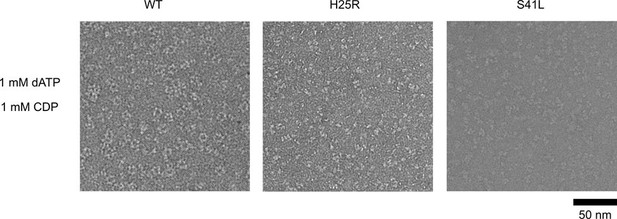
Ng RNR S41L and H25R variants do not form rings under dATP inhibiting conditions.
WT, PTC-672H25R, or PTC-847S41L α subunits were combined with WT β in a 1:1.5 ratio (15 ng/µL final concentration; 0.1 µM α and 0.15 µM β) in the presence of 1 mM dATP and 1 mM CDP and imaged on an F30 (FEI) microscope. Samples were prepared at 4 °C prior to staining.
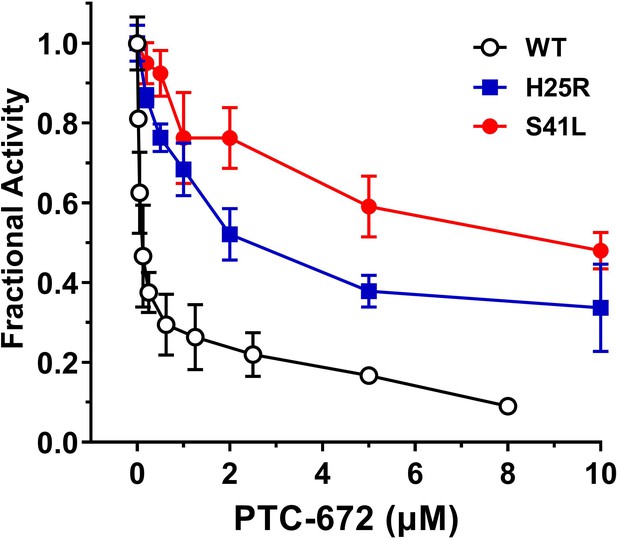
Inhibition of RNR of the mutants by PTC-672.
A 1:1 mixture of α and β subunits was incubated with each compound and assayed for activity spectrophotometrically at 37 °C with PTC-672. The assay used 0.1 µM α2, 0.1 µM β2, 1 mM GDP, 0.25 mM TTP, 100 µM Ec TR, 1 µM Ec TRR, 0.2 mM NADPH and various concentrations of PTC-672 (0.2–10 µM) in duplicates.
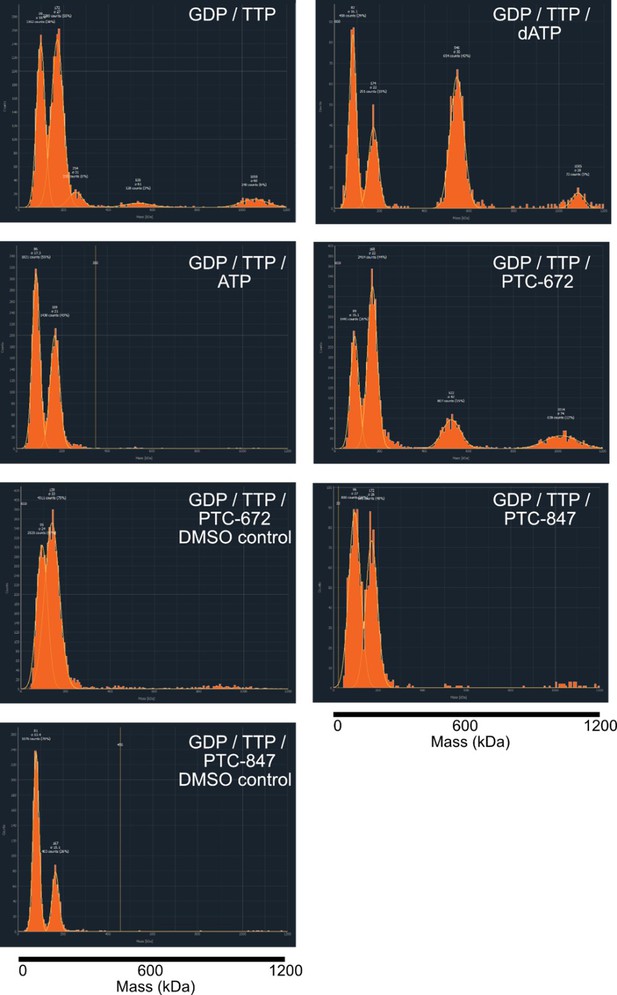
Representative histograms of mass photometry experiments under differing nucleotide and drug conditions.
Concentrations of additives were as follows: 1 mM GTP, 0.25 mM TTP, 0.2 mM dATP, 3 mM ATP, 25 µM PTC-672 (0.45% DMSO), 8 µM PTC-847 (0.04% DMSO). All experiments contained 2.5 ng/µL protein at a 1:1 α:β ratio. The mass of the α4β4 complex is 528 kDa.
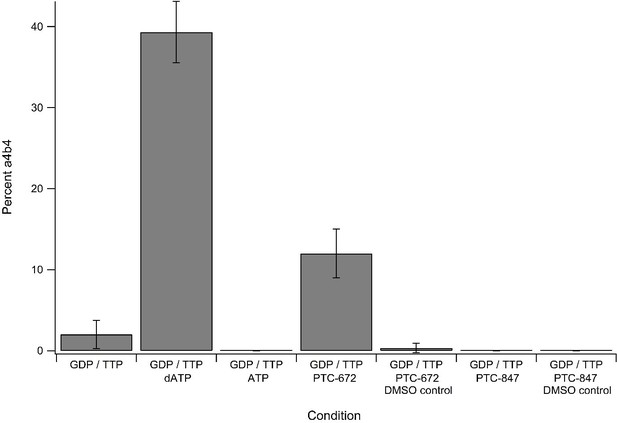
Percent±standard deviation of α4β4 seen over three replicates of mass photometry experiments under differing nucleotide and drug conditions.
Concentrations of additives were as follows: 1 mM GTP, 0.25 mM TTP, 0.2 mM dATP, 3 mM ATP, 25 µM PTC-672 (0.45% DMSO), 8 µM PTC-847 (0.04% DMSO). All experiments contained 2.5 ng/µL protein at a 1:1 α:β ratio.
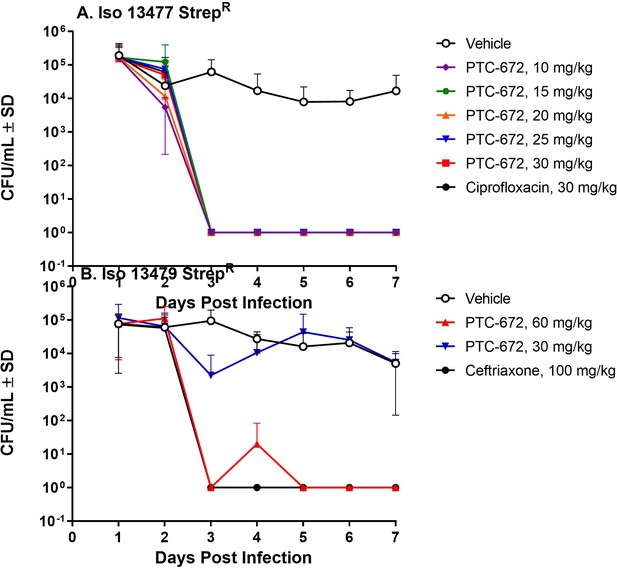
PTC-672 in vivo efficacy in a mouse model of gonorrhea.
Mice were vaginally inoculated with streptomycin resistant Ng strains and swabbed daily over a 1-week period. At each sampling time point, the number of bacteria (CFU/mL) were counted by plating onto selective plates and the mean CFU/mL ± SD values were determined. The mean ± SD values were plotted. (A) Symbols: vehicle open black, ciprofloxacin solid black, PTC-672 10 mg/kg purple, 15 mg/kg green, 20 mg/kg orange, 25 mg/kg blue, and 30 mg/kg red. (B) Symbols: vehicle open black, ceftriaxone solid black, PTC-672 30 mg/kg blue and 60 mg/kg red.
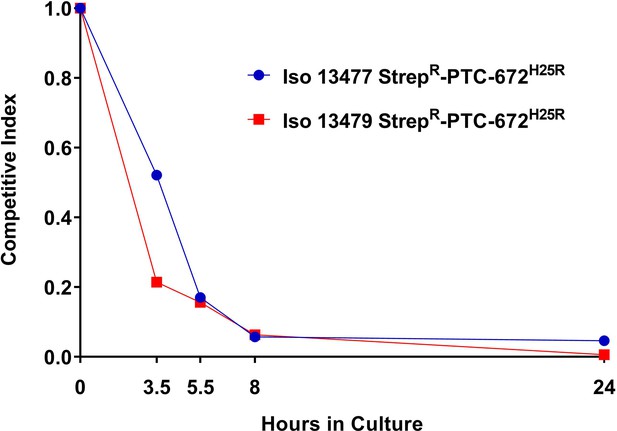
H25R mutation in Ng RNR reduces fitness in vitro.
Iso 13479 StrepR-PTC-672H25R (red) and Iso 13477 StrepR-PTC-672H25R (blue) were mixed in a 1:1 ratio, inoculated into fresh growth medium, and cultured. At each sampling time point, the numbers of total and resistant bacteria were determined by plating onto non-selective and PTC-672-supplemented plates, respectively. Relative fitness was expressed as the CI (see text).
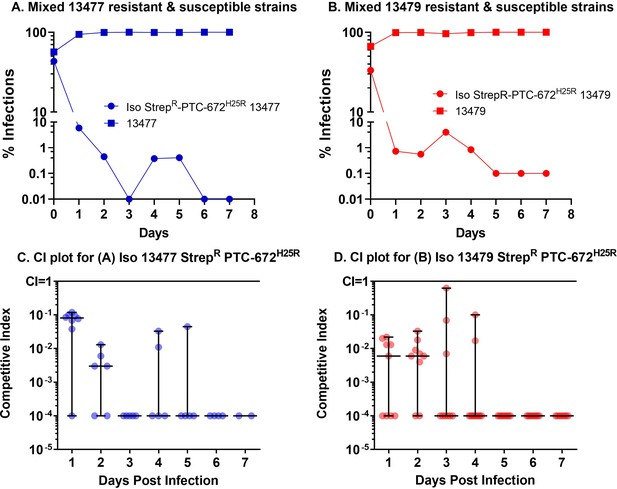
H25R mutation in Ng α of its Ia RNR reduces fitness in vivo.
Mice were vaginally inoculated with the mixed isogenic resistant and susceptible strains and swabbed daily over a one-week period. The CI (plots C and D) was determined at each sampling time point (taken from plots A and B) as described above. The medians ± ranges are shown as solid horizontal lines. (A) The H25R mutant strain of 13477 was found to be outcompeted when co-infected with the parent 13477 strain. (B) The H25R mutant strain of 13479 (MDR) was found to be outcompeted when co-infected with the parent 13479 strain. (C) Competitive index calculations determined from CFUs taken from graph A. (D) Competitive index calculations determined from CFUs plotted in graph B.
Tables
MIC of Ng strains (A) and Gram-negative pathogens and gut microorganisms (B).
(A) WHO F does not carry any antimicrobial resistance elements and is considered an antibiotic susceptible strain. WHO G-M strains are resistant to quinolones. WHO K-O strains carry penA, ponA, porB1, and mtrR mutations associated with decreased susceptibility to cephalosporins. (Unemo et al., 2016). N. meningitidis (NMSB) ATCC 13090 is the suggested reference strain for serogroup B (Clinical and Laboratory Standards Institute, 2004). (B) This panel was constructed to test for inhibition of Gram-negative pathogens and normally occurring intestinal organisms, where WT = wild type, MDR = multiple drug resistant, and QuinR = quinolone-resistant strains. The Nm, Gram-negative, and normal gut organism strains were obtained from the American Type Culture Collection (ATCC), or were kindly provided by MicroMyx, LLC, (MMX), Kalamazoo, MI, or the laboratory of Lynn Zechiedrich (LZ), Baylor College of Medicine, Houston, Texas. PTC-compound susceptibility testing was performed in accordance with the Clinical and Laboratory Standards Institute (CLSI) M07-A9 guideline (Clinical and Laboratory Standards Institute, 2012). Genus names: Acinetobacter (A), Klebsiella (K), Pseudomonas (P), Staphylococcus (S), Yersinia (Y).
| Panel A - N. gonorrhoeae and N. meningitidis MIC (µg/mL) | ||||
|---|---|---|---|---|
| Strain | PTC-565 | PTC-847 | PTC-672 | |
| N. gonorrhoeae (WHO F) | 13477 | 0.1 | 0.05 | 0.05 |
| N. gonorrhoeae (WHO G) | 13478 | 0.39 | 0.1 | 0.05 |
| N. gonorrhoeae (WHO K) | 13479 | 0.78 | 0.1 | 0.2 |
| N. gonorrhoeae (WHO L) | 13480 | 0.19 | 0.05 | 0.2 |
| N. gonorrhoeae (WHO M) | 13481 | 0.39 | 0.05 | 0.2 |
| N. gonorrhoeae (WHO O) | 13483 | ND | 0.1 | 0.4 |
| N. meningitidis (NMSB) | ATCC 13090 | 0.2, 0.39(two values) | 0.2 | 0.1 |
| Panel B - Gram-negative pathogens and normal gut organisms MIC (µg/mL) | ||||
| Strain | PTC-565 | PTC-847 | PTC-672 | |
| A. baumannii WT | ATCC BAA747 | 0.78 | > 62.5 | ND |
| A. baumannii MDR | MMX2240 | 3.1 | 62.5 | > 50 |
| E. coli WT | ATCC 25922 | 0.78 | > 62.5 | > 62.5 |
| E. coli QuinR | LZ3111 | 0.39 | 31.25 | > 12.5 |
| K. pneumoniae WT | ATCC 35657 | 0.78 | 62.5 | ND |
| K. pneumoniae MDR | MMX1232 | 15.6 | > 62.5 | ND |
| P. aeruginosa WT | ATCC 27853 | 12.5 | > 62.5 | > 62.5 |
| S. aureus WT | ATCC 29213 | 12.5 | 62.5 | > 62.5 |
| S. aureus MDR | ATCC 700789 | 0.78 | > 12.5 | > 12.5 |
| Y. pseudotuberculosis | ATCC 13979 | 1.95 | > 25 | > 25 |
PTC-847 does not inhibit Ng gyrase or topoisomerase IV in vitro.
Microplate-based supercoiling assays for DNA gyrase and topoisomerase IV were performed as previously described (Maxwell et al., 2006). The sequence homology between the Ng and Ec enzymes are 70% and 75% in gyrase subunit A and B, respectively, and 64% and 65% in topoisomerase IV subunits A and B, respectively. The assays thus followed the Ec protocols and are described in detail for the Ng enzymes in the Methods. Decatenation assays using purified gyrase AB and topoisomerase IV proteins were performed following the protocols for Profoldin’s topoisomerase II and IV DNA decatenation assays (ProFoldin, Hudson, MA) also in Materials and methods.
| Compound | IC50 (µM) | ||||
|---|---|---|---|---|---|
| E. coli | N. gonorrhoeae | Human | |||
| Topo IV | Gyrase | Topo IV | Gyrase | Topo II | |
| Ciprofloxacin | 7 | 0.6 | 18 | 0.4 | ─ |
| PTC-847 | > 32 | > 32 | > 32 | 19 | > 100 |
Inhibition of class Ia RNRs in vitro.
Various concentrations of PTC-672 or PTC-847, or 1 mM N3CDP (positive control) were added to 37 °C reaction mixtures containing α, β, ATP, TR, TRR, and NADPH. The mixtures were incubated for either 30 s, 5 min or 15 min prior to initiation of the reaction with 5-[3H]-CDP. Aliquots were quenched at 1, 2, 3 and 4 min in 2% HClO4. All samples were neutralized by the addition of 0.5 M KOH and processed following a standard protocol (Ravichandran et al., 2020). Percent inhibition was calculated relative to a 1% DMSO negative control.
| Ia RNR | PTC-672 % Inhibition | PTC-847 % Inhibition |
|---|---|---|
| Ng | 78% at 2.5 µM | 93% at 4 µM |
| Ec | 92% at 16 µM | 81% at 14 µM |
| Human | 4% at 100 µM | 21% at 100 µM |
Additional files
-
Supplementary file 1
PTC-847 and PTC-672 MIC values across 206 N. gonorrhoeae strains collected at the Public Health England.
Two isolates gave elevated PTC-847 or PTC-672 MIC values. However, these isolates were sensitive to all other antibiotics. Due to the selectivity of the PTC compounds, these two strains will be genotyped to confirm they are Neisseria species. PTC-compound susceptibility testing was performed in accordance with the Clinical and Laboratory Standards Institute (CLSI) M07-A9 guideline (Clinical and Laboratory Standards Institute, 2012).
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp1-v2.docx
-
Supplementary file 2
Time -dependent kill of Ng upon PTC-847 (A) or PTC-672 treatment (B).
At PTC-847 or PTC-672 concentrations of ≥1 x MIC, a > 3 log reduction in measured Ng 13477 colony forming units per milliliter of culture (CFU/mL) was observed. The time-dependent kill assays were performed in accordance with the CLSI M26-A guideline (Clinical and Laboratory Standards Institute, 1999).
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp2-v2.docx
-
Supplementary file 3
Time-dependent post antibiotic effect observed with PTC-847.
Ng 13477 was pre-treated with PTC-847 at 4 X MIC for 2 h then allowed to grow in culture media supplemented with PTC-847 at 0, 0.25, 0.5, or 1 X MIC (Data for 0, 0.25 x MIC, 0.5 x MIC, and 1 x MIC are indicated in black, grey, purple, and blue respectively). A > 6 log reduction in measured Ng 13477 CFU/mL was observed at 20 h post pre-treatment in the absence of PTC-847. The post antibiotic effect assay was performed in accordance with the CLSI M26-A guideline (Clinical and Laboratory Standards Institute, 1999).
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp3-v2.docx
-
Supplementary file 4
PTC-847 inhibition of DNA synthesis in N. gonorrhoeae strain 13477.
We followed the incorporation of the radiolabeled precursors into total nucleic acids (black circles), DNA (red squares), and protein (blue triangles). In Ng, only radiolabeled uracil is incorporated into DNA or RNA, which was also shown to be the case for Nm (Jyssum and Jyssum, 1979).
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp4-v2.docx
-
Supplementary file 5
Frequency of resistance to PTC-847.
By plating at high cell density on agar plates containing PTC-847 at 4, 8, 16 and 32-fold MIC, Ng 13477 exhibited a spontaneous or acquired frequency of resistance to PTC-847 on the order of 10–8. The colonies obtained were passaged multiple times on PTC-847-containing plates to obtain a stable PTC-847-resistant strain (PTC-847R).
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp5-v2.docx
-
Supplementary file 6
PTC-847R strain exhibits no cross resistance to existing antibiotics.
Susceptibility of the WT Ng 13477 strain and the PTC-847R strain were measured for a wide variety of antibiotics having different modes of action. MICs were determined in accordance with the CLSI M07-A9 guideline (Clinical and Laboratory Standards Institute, 2012). The PTC-847R strain was equally sensitive to all classes of antibiotics as the susceptible WT Ng 13477 strain, except for resistance to PTC-847. The PTC-847 MIC for WT Ng 13477 was 0.05 µg/mL compared to 15.6 µg/mL for the PTC-847R strain.
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp6-v2.docx
-
Supplementary file 7
Nucleotide pools measured in WT Ng 13477.
The susceptible WT Ng 13477 strain was grown to log phase and treated with DMSO or PTC-847 at 1 X MIC for 1 h. Nucleotides were extracted in acidified acetonitrile/H2O (65:35) followed by centrifugation. The supernatant was lyophilized and subjected to LC/MS. 13C9,15N3─CTP was added for LC/MS analysis. Peak areas of NTP and dNTPs were normalized to the peak area of 13C9,15N3─CTP.
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp7-v2.docx
-
Supplementary file 8
Characterization of purified Ng RNR.
(A) Activity assay of Ng RNR. To optimize RNR activity, a 1:1 ratio of α2 and β2 subunits was examined over a physiological concentration range of 0.01–10 µM and its specific activity (SA) determined in a reaction mix of 100 µM Ec thioredoxin (TR), 1 µM Ec thioredoxin reductase (TRR), and 0.2 mM NADPH at 37 °C. The SA for 0.01–0.12 µM subunits was measured with 1 mM GDP, 0.25 mM TTP, and 0.2 mM NADPH using a spectrophotometric assay whereas the 0.2–10 µM SA was measured with 1 mM 5-[3H]-CDP, 3 mM ATP and 2 mM NADPH by radioactive assay. Fitting the data from 0.01 to 0.12 µM using gave Vmax = 1300 nmol min–1 mg–1 and Km = (0.030 ± 0.006) µM. (n = 2 replicates for each concentration). (B) EPR spectra of the tyrosyl radical. The X-band EPR spectrum of the tyrosyl radical of Ec β2 (red) and Ng β2 (black) at 77 K. Microwave power: 35 dB. Modulation: 1.5 Gauss. Modulation frequency: 100 kHz. Time constant: 20.48ms.
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp8-v2.docx
-
Supplementary file 9
dATP inhibition of Ng RNR.
The continuous spectrophotometric assay with GDP/TTP (Figure S4) was used for concentrations of dATP to 25 μM and NADPH concentration was 0.2 mM. The discontinuous radioactive assay with [5-3H] CDP (1 mM) and ATP (3 mM) was used for dATP at 50–200 μM and NADPH concentration at 2 mM (see ref 38). The assay with mutant αs (H25R and S41L) used the spectrophotometric assay with GDP/TTP.
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp9-v2.docx
-
Supplementary file 10
PTC-672 and PTC-847 do not inhibit a representative panel of normally occurring intestinal organisms.
The panel was constructed to test for inhibition of normally occurring intestinal organisms (Thursby and Juge, 2017). Susceptibility testing was performed in accordance with the Clinical and Laboratory Standards Institute (CLSI) M07-A9 guideline (Clinical and Laboratory Standards Institute, 2012). PTC-672 was tested against a more extensive panel of organisms.
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp10-v2.docx
-
Supplementary file 11
Commensal Neisseria are less sensitive to PTC-672 than Ng 13477.
Two commensal Neisseria ‘type strains’ swabbed from the oropharynx of healthy volunteers (†Berger, 1971 and ‡Riou and Guibourdenche, 1987) were tested for susceptibility to inhibition in accordance with the Clinical and Laboratory Standards Institute (CLSI) M07-A9 guideline (Clinical and Laboratory Standards Institute, 2012).
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp11-v2.docx
-
Supplementary file 12
The distribution of the identified RNR types in various bacteria (Lundin et al., 2009) that were used to assess antibacterial activity of the PTC compounds.
- https://cdn.elifesciences.org/articles/67447/elife-67447-supp12-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67447/elife-67447-transrepform1-v2.pdf





