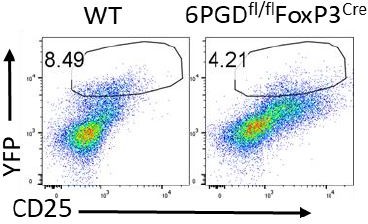
6-Phosphogluconate dehydrogenase (6PGD), a key checkpoint in reprogramming of regulatory T cells metabolism and function
Figures
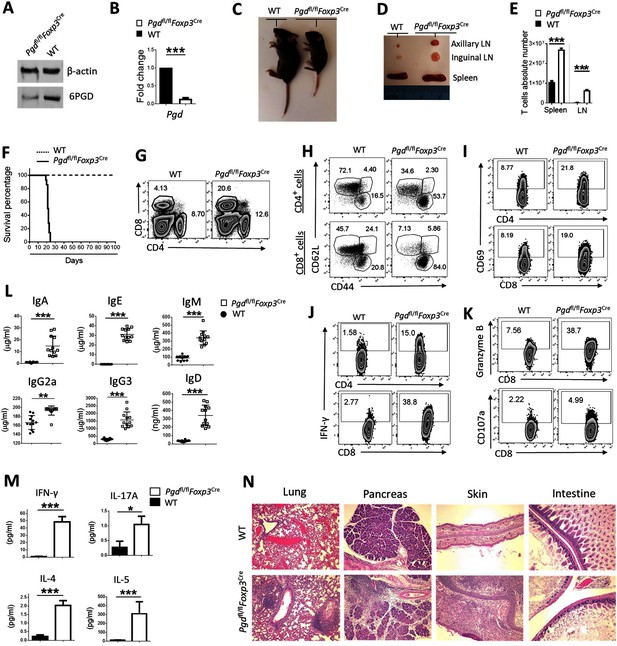
Deletion of 6-phosphogluconate dehydrogenase (6PGD) in regulatory T cells (Tregs) induces a fatal autoimmune phenotype.
(A–B) YFP+ cells were sorted from Pgd+/+Foxp3Cre (wild-type [WT]) and Pgdfl/flFoxp3Cre mice and deletion of Pgd was confirmed by western blot (A) and real-time PCR (B). (C) Representative image of 21-day-old WT and Pgdfl/flFoxp3Cre mice. (D) Representative image of lymphadenopathy in Pgdfl/flFoxp3Cre compared to WT mice. (E) T cells absolute number per spleen and peripheral lymph nodes (pLNs) of WT and Pgdfl/flFoxp3Cre mice are shown (N = 12 mice per group). (F) Survival curve of WT and Pgdfl/flFoxp3Cre mice. Representation of 15 mice per group. (G) Splenocytes from 20-day-old WT and Pgdfl/flFoxp3Cre mice were harvested and distribution of CD4+ versus CD8+ T cells were evaluated. (H) Deletion of 6PGD induces enhanced effector phenotype (CD44high CD62Llow) both in CD4+ T cells (top panel) and in CD8+ T cells (bottom panel) in 20-day-old Pgdfl/flFoxp3Cre compared to WT mice. (I) Both CD4+ and CD8+ T cells express higher levels of CD69 activation marker in Pgdfl/flFoxp3Cre compared to WT mice. (J) Splenocytes from 20-day-old WT and Pgdfl/flFoxp3Cre mice were stimulated with PMA (50 ng/ml)/ionomycin (1 μg/ml) plus GolgiPlug (1 μl/ml) for 4 hr and expression of IFN-γ was evaluated by flow cytometry. (K) Granzyme B (top panel) and CD107a degranulation activation marker (bottom panel) expression was assessed on splenocytes of 20-day-old WT and Pgdfl/flFoxp3Cre mice by flow cytometry. (L–M) Serum was collected from 20-day-old WT and Pgdfl/flFoxp3Cre mice and levels of serum antibodies (L) and IFN-γ, IL-17A, IL-4, and IL-5 (M) were detected as described in supplemental information. Results are representative of 12 mice per group. (N) Hematoxylin and eosin staining of lung: original magnification (X10), pancreas (X10), skin (X10), and intestine (X10) of WT and Pgdfl/flFoxp3Cre mice. *p < 0.05; **p < 0.01; ***p < 0.001.
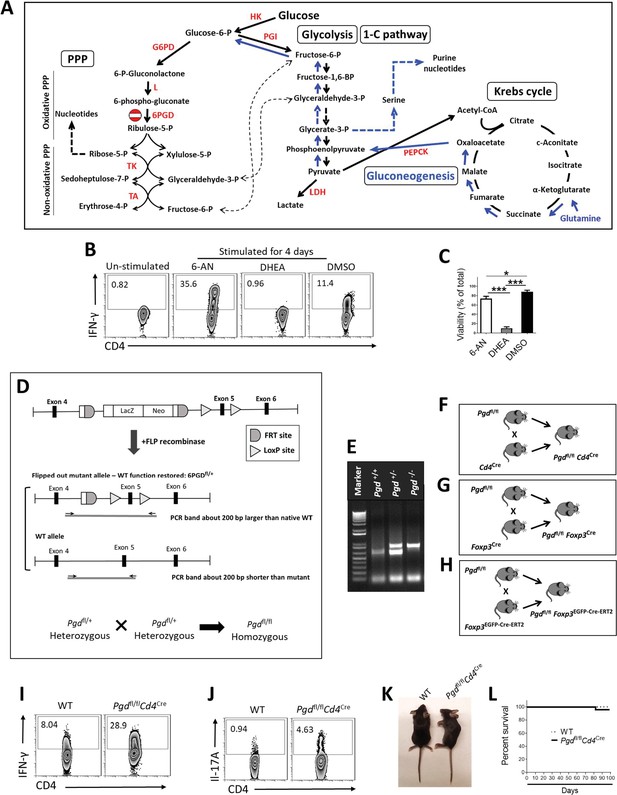
Effect of G6PD/6PGD inhibition and generation of Pgdfl/fl Foxp3cre mice.
(A) Schematic diagram interconnecting pentose phosphate pathway (PPP), glycolysis, gluconeogenesis, and the Krebs cycle. (B–C) IFN-γ production (B) and cell viability (C) by naïve CD4+ T cells in the presence of 10 µM 6PGD inhibitor (6-aminonicotinamide [6-NA]), 10 µM G6PD inhibitor (dehydroepiandrosterone [DHEA]), or the vehicle DMSO. (D) Strategy for Pgd knockout by inserting the SA–ßgeo–pA (LacZ-Neo) cassette into the intron between exons 4 and 5 of 6PGD, which disrupts gene function. The targeted Pgd exon 5 is flanked by loxP sites. (FRT, target site for FLP recombinase; loxP, target site for Cre-recombinase). Excision of the SA–ßgeo–pA cassette by FLP recombinase generated the heterozygous status of modified Pgd (Pgdfl/+). Heterozygous mice were crossed to generate homozygous Pgdfl/fl. (E) PCR validation of the Pgdfl/+ and Pgdfl/fl mice. (F–H) Pgdfl/fl mice were crossed with Cd4Cre, (F) Foxp3YFP-Cre, (G) and Foxp3EGFP-Cre-ERT2 and (H) mice. (I–J) Enhanced effector cells phenotype in spleen of Pgdfl/flCd4Cre mice. (K–L) Pgdfl/flCd4Cre mice show normal body phenotype (K) and survival rate (L). HK: hexokinase; G6PD: glucose-6-phosphate dehydrogenase; L: lactonase; 6PGD: 6-phosphogluconate dehydrogenase; TK: transketolase; TA: transaldolase; PGI: phosphoglucose isomerase; LDH: lactate dehydrogenase; PEPCK: phosphoenolpyruvate carboxykinase.
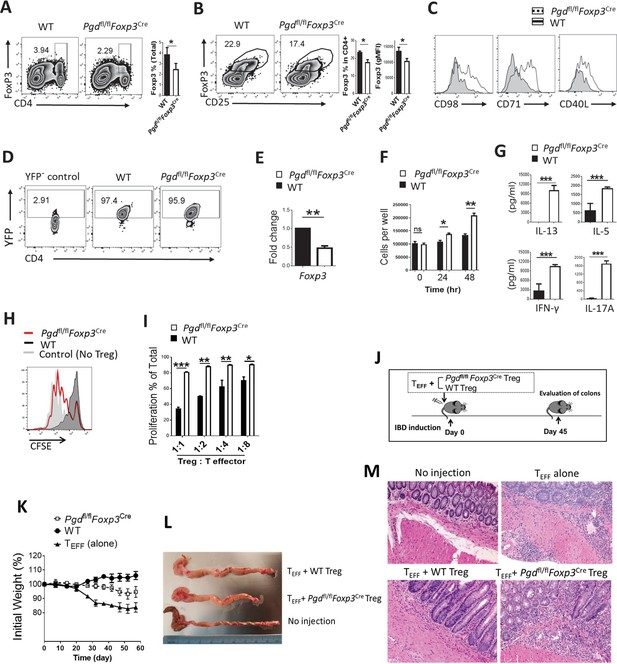
6-Phosphogluconate dehydrogenase (6PGD) blockade results in loss of suppressive function in regulatory T cells (Tregs).
(A–B) Frequency of CD4+Foxp3+ cells in spleen of 20-day-old Pgd+/+Foxp3Cre (wild type [WT]) was evaluated and demonstrated (A) in total cells or (B) in gated CD4+ T cells. Bar graphs show respective statistical differences for percentage of Foxp3+ cells and Foxp3 gMFI. (C) Expression of CD98, CD71, and CD40L on Tregs from WT and Pgdfl/flFoxp3Cre mice. (D–E) YFP+ cells were isolated from 20-day-old WT and Pgdfl/flFoxp3Cre mice. (D) Purified YFP+ cells are shown and (E) Foxp3 mRNA levels were evaluated by real-time PCR. (F) Isolated Tregs (YFP+) were cultured in vitro in presence of IL-2 (700 IU/ml) and anti-CD3/anti-CD28 coaled beads (Treg:beads ratio 1:3) and cells number was assessed at 24 and 48 hr time points. Results are representative of three independent experiments with N = 4. (G) Cytokine release in media as cultured in above culture condition. (H–I) Isolated Tregs (YFP+) from Pgdfl/flFoxp3Cre mice showed lower suppressive activity in suppression assay, as described in Materials and methods. Representative histogram of CFSE dilution pattern (Treg:Teff = 1:1 ratio) (H) and bar graph of T cell proliferation as a function of serial Treg:Teff ratios (I) are shown. Results are from three independent experiments with N = 4. (J) Tregs (YFP+) from WT and Pgdfl/flFoxp3Cre mice and T effector (CD4+CD45RBhigh) cells were isolated, mixed, and transferred to Rag1-/- mice as inflammatory bowel disease (IBD) model. Colons were evaluated 45 days after cells injection. (K) Weight change in IBD mouse model. (L) On day 45 post IBD induction Rag1-/- mice colon were measured for length and thickness. (M) Representative hematoxylin and eosin staining of Rag1-/- mice colon on day 45 after IBD induction. Results are from two independent experiments with N = 8 per group. *p < 0.05; **p < 0.01; ***p < 0.001.
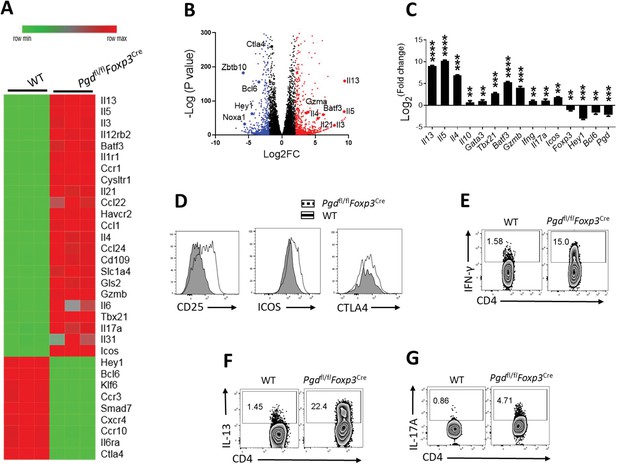
6-Phosphogluconate dehydrogenase (6PGD) deficiency in regulatory T cells (Tregs) induces shift toward CD4+ T helper subsets.
(A–B) Heat map and volcano plot representing gene expression in Tregs (YFP+) isolated from 20-day-old wild-type (WT) and Pgdfl/flFoxp3Cre mice. (C) Real-time PCR assay was done on selected genes from Tregs isolated from WT and Pgdfl/flFoxp3Cre mice. (D) Expression of CD25, ICOS, and CTLA-4 in Tregs (YFP+) from WT and Pgdfl/flFoxp3Cre mice. (E–G) CD4+ T cells in Pgdfl/flFoxp3Cre mice spleen produced higher amount of IFN-γ, IL-13, and IL-17A. Results are representative of two independent experiments with N = 3 per group. **p < 0.01; ***p < 0.001; ****p < 0.0001.
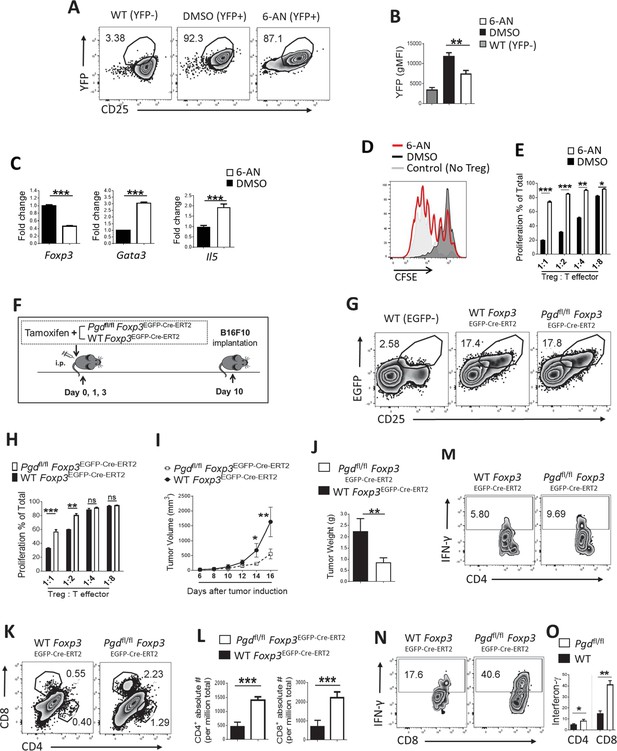
6-Phosphogluconate dehydrogenase (6PGD) blockade in regulatory T cells (Tregs) prevents their suppressive function and induces potent anti-tumor responses.
(A–B) Wild-type (WT) Foxp3Cre naïve CD4+ T cells (CD62Lhigh CD44low) were driven toward Tregs in vitro (as inducible Treg [iTreg]) along with treatment with 6-aminonicotinamide (6-AN) or vehicle DMSO and Treg drive efficacy was evaluated as CD25+YFP+ cells. YFP+ percentage in CD4+ T cells (A) and YFP gMFI (B) are demonstrated. (C) Real-time PCR analysis on derived Tregs demonstrate lower expression of Foxp3 transcription factor and higher expression of Gata3 transcription factor and IL-5 under treatment with 6-AN versus vehicle DMSO. (D–E) 6-AN treatment of driven Tregs demonstrate lower suppressive capacity evaluated in in vitro suppression assay. YFP+ cells were sorted for the suppression assay. (F) Tamoxifen treatment schedule and tumor induction by implanting B16F10 cells in Pgdfl/fl Foxp3EGFP-Cre-ERT2 and WT mice. (G) Flow cytometry analysis show the same frequency of Tregs (EGFP+) in CD4+ cells in spleen of both mouse groups 10 days post tamoxifen treatment. (H) Suppression assay on sorted EGFP+ Tregs as described in supplemental information. (I–J) 6PGD blockade in Tregs resulted in lower tumor volume (I) and tumor weight (J). (K–L) Tumor of the mouse models from (F) were evaluated for infiltration of both CD4+ and CD8+ T cells on day 16 post implantation, which were higher in 6PGD blocked mice, as shown by representative flowgram (K) and bar graphs (L). (M–O) Tumor infiltrating CD4+ (M) and CD8+ (N) T cells showed higher capacity of IFN-γ production on day 16. Results are from two independent experiments with N = 8 per group. *p < 0.05; **p < 0.01; ***p < 0.001.
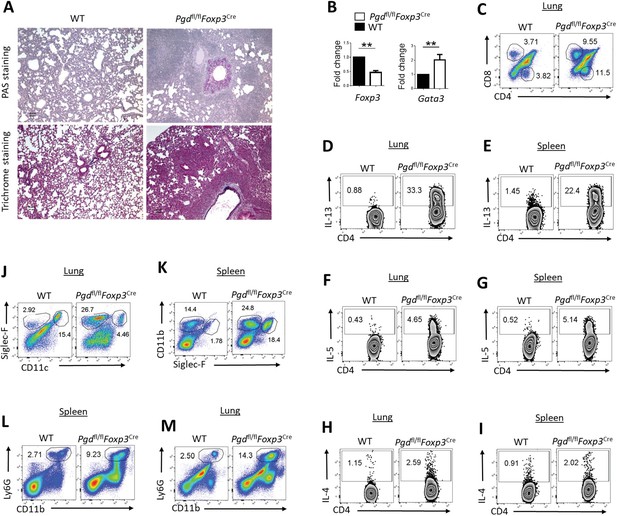
6-Phosphogluconate dehydrogenase (6PGD) function in regulatory T cells (Tregs) is required for suppressing allergic (Th2) responses.
(A) Histochemical PAS (top) and Trichrome staining (bottom) of lungs from 20-day-old wild-type (WT) and Pgdfl/flFoxp3Cre mice. In the lung of Pgdfl/flFoxp3Cre mice, there was higher collagen deposit (top) and higher mucus accumulation (bottom) presented as blue colors. (B) Sorted YFP+ Tregs from lungs expressed lower levels of Foxp3 and higher amount of Gata3 transcription factors in Pgdfl/flFoxp3Cre mice determined by real-time PCR. (C) Lung of 20-day-old Pgdfl/flFoxp3Cre mice demonstrated higher infiltration of T cells (both CD4+ and CD8+ cells). (D–I) Intracellular cytokine staining on lung and spleen for IL-13 (D–E), IL-5 (F–G), and IL-4 (H–I) in CD4+ T cells. (J) Representative frequency of eosinophils (CD11clow Siglec-F+) in the mouse’s lung. (K) Flow cytometry analysis of splenic eosinophils (CD11b+ Siglec-F+) and macrophages (CD11b+ Siglec-F-). (L–M) Frequency of CD11b+ Ly6G+ cells in spleen and lung of 20-day-old Pgdfl/flFoxp3Cre compared to WT mice. Results are representative of three independent experiments with N = 2–3 mice per group. **p < 0.01.
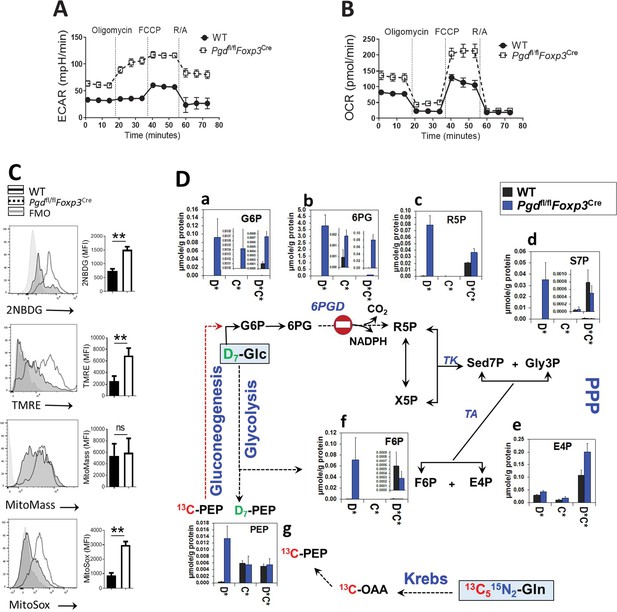
Blocking 6-phosphogluconate dehydrogenase (6PGD) in regulatory T cells (Tregs) induces reprogramming of glycolysis, mitochondrial respiration, and non-oxidative pentose phosphate pathway (PPP).
(A–B) Extracellular acidification and oxygen consumption analysis on YFP+ sorted cells from Pgdfl/fl Foxp3Cre and wild-type (WT) mice was performed using Seahorse XF24 metabolic analyzer as described in supplemental information. The extracellular acidification rate (ECAR) (A) and oxygen consumption rate (OCR) (B) are shown (N = 4). (C) YFP+ cells glucose (Glc) uptake capacity was determined by 2-NBDG uptake, mitochondrial potential (ΔΨm) by TMRE, mitochondrial mass by MitoTracker Deep Red FM and mitochondrial ROS by MitoSox Red. Results are representative of three independent experiments with N = 4 per group. (D) YFP+ cells were sorted from WT and Pgdfl/flFoxp3Cre mice and cultured in the presence of IL-2 (700 IU/ml) and anti-CD3/anti-CD28 coated beads (Treg:beads ratio 1:3) plus D7-Glc and 13C5,15N2-glutamine (Gln) for 48 hr. Isotope labeling patterns of metabolites of cell extracts were analyzed by IC-UHRMS. Data shown demonstrates D7-Glc and 13C5,15N2-Gln incorporation into the PPP metabolites via non-oxidative PPP and gluconeogenesis. Results were generated with N = 2. Legend in X-axis D* = sum of D1 to Dx or Glc-derived species; C* = sum of 13C1 to 13Cx or Gln-derived species; C*D* = sum of dual 13C1 to 13Cx and D1 to Dx or Glc and Gln-derived species; G6P, glucose-6-phosphate; 6PG, 6-phosphogluconate; R5P, ribose-5-phosphate; S7P, sedoheptulose-7-phosphate; Gly3P, glyceraldehyde-3-phosphate; X5P, xylulose-5-phosphate; E4P, erythrose-4-phosphate; F6P, fructose-6-phosphate; OAA, oxaloacetate; PEP, phosphoenolpyruvate.
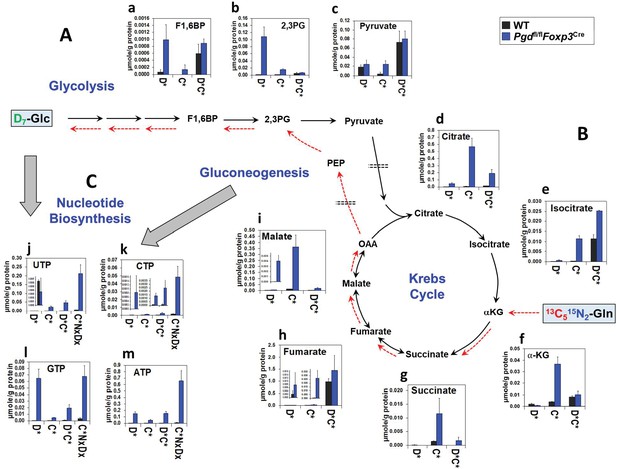
Glycolysis, Krebs cycle, and nucleotide biosynthesis are enhanced in 6-phosphogluconate dehydrogenase (6PGD)-deficient Tregs.
(A–C) The same cell extracts as in Figure 4 were analyzed for metabolites of glycolysis (A), the Krebs cycle (B), and nucleotide biosynthesis (C) by IC-UHRFTMS. Legend in X-axis: D* = sum of D1 to Dx or Glc-derived species; C* = sum of 13C1 to 13Cx or Gln-derived species; C*D* = sum of dual 13C1 to 13Cx and D1 to Dx or Glc and Gln-derived species; C*NxDx = sum of triply labeled (13C1 to 13Cx, D1 to Dx, and 15N1 to 15Nx) species. X is ≥1 for each metabolite isotopologue. Abbreviations used are: F1,6BP: fructose-1,6-bisphosphate; 2,3PG: 2,3-diphosphoglycerate; αKG: α-ketoglutarate; OAA: oxaloacetate; UTP: uridine triphosphate; CTP: cytidine triphosphate; GTP: guanosine triphosphate; ATP: adenosine triphosphate.
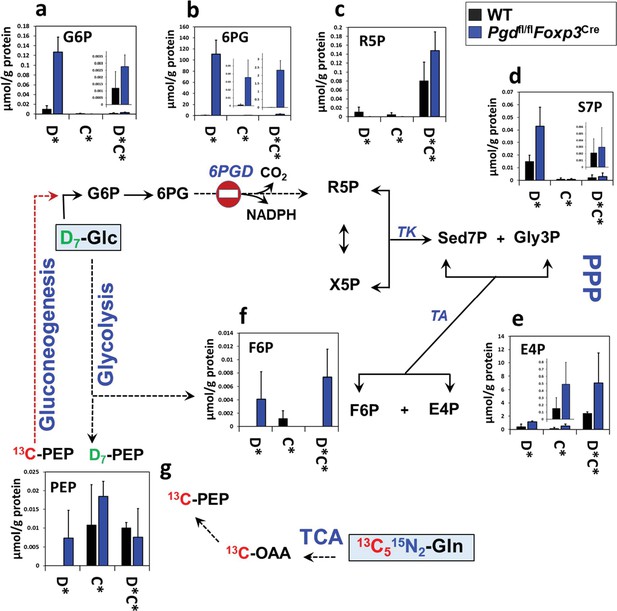
Pentose phosphate pathway (PPP) pathway reprogramming in 6-phosphogluconate dehydrogenase (6PGD)-deficient regulatory T cells (Tregs) generated in vitro.
Wild-type (WT) and Pgdfl/flFoxp3Cre naïve CD4+ T cells were differentiated into Tregs (as inducible Tregs [iTreg]) in the presence of IL-2+ TGF-β for 4 days, in vitro. In last 48 hr of polarization D7-glucose (Glc) and 13C5,15N2-glutamine (Gln) were replaced in the media. Isotope labeling patterns of metabolites of cell extracts were analyzed by IC-UHRMS. Data shown demonstrates D7-Glc and 13C5,15N2-Gln incorporation into the PPP metabolites via non-oxidative PPP and gluconeogenesis. Results were generated with N = 2. Abbreviations are the same as Figure 4.
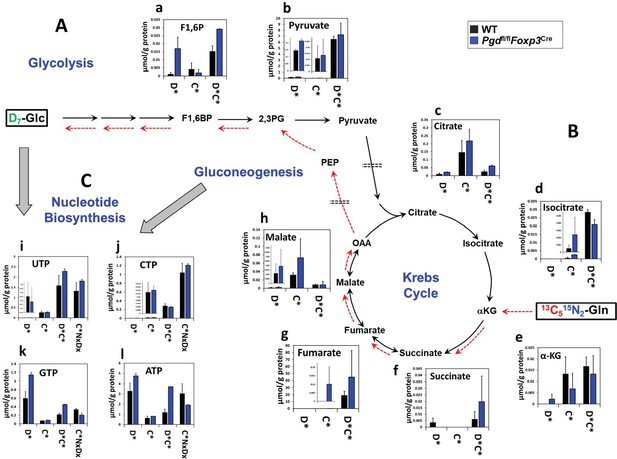
Glycolysis, Krebs cycle, and nucleotide biosynthesis reprogramming in 6-phosphogluconate dehydrogenase (6PGD)-deficient Tregs generated in vitro.
(A–C) The same cell extracts as in Figure 4—figure supplement 2 were analyzed for metabolites of glycolysis (A), the Krebs cycle (B), and nucleotide biosynthesis (C) by IC-UHRFTMS. These results were from in vitro generated Tregs as inducible Tregs (iTreg). Legend and abbreviations are the same as Figure 4—figure supplement 1.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-mouse CD3ε-PerCP (clone: 145–2 C11); (Armenian hamster monoclonal) | BioLegend | Cat No#100326; RRID: AB_893317 | FACS (1:100) |
| Antibody | Anti-mouse CD4-APC (clone: GK1.5); (rat monoclonal) | BioLegend | Cat No#100412; RRID: AB_312697 | FACS (1:100) |
| Antibody | Anti-mouse CD4-PE (clone: GK1.5); (rat monoclonal) | BioLegend | Cat No#100408; RRID: AB_312693 | FACS (1:100) |
| Antibody | Anti-mouse CD4-Pacific Blue (clone: GK1.5); (rat monoclonal) | BioLegend | Cat No#100428; RRID: AB_ 493,647 | FACS (1:100) |
| Antibody | Anti-mouse CD8a-FITC (clone: 53–6.7); (rat monoclonal) | BioLegend | Cat No#100706; RRID: AB_312745 | FACS (1:100) |
| Antibody | Anti-mouse CD8a-Pacific Blue(clone: 53–6.7); (rat monoclonal) | BioLegend | Cat No#100725; RRID: AB_493425 | FACS (1:100) |
| Antibody | Anti-mouse CD25-BV711 (clone: PC61); (rat monoclonal) | BioLegend | Cat No#102049; RRID: AB_2564130 | FACS (1:100) |
| Antibody | Anti-mouse CD11b-Pacific Blue (clone: M1/70); (rat monoclonal) | BioLegend | Cat No#101224; RRID: AB_755986 | FACS (1:100) |
| Antibody | Anti-mouse CD11c-PE-Cy7 (clone: N418); (Armenian hamster monoclonal) | BioLegend | Cat No#117318; RRID: AB_493568 | FACS (1:100) |
| Antibody | Anti-mouse CD25-PE-Cy7 (clone: PC61); (rat monoclonal) | BioLegend | Cat No#102016; RRID: AB_312865 | FACS (1:100) |
| Antibody | Anti-mouse CD40L-APC (clone: MR1); (Armenian hamster monoclonal) | BioLegend | Cat No#106510; RRID: AB_2561561 | FACS (1:100) |
| Antibody | Anti-mouse CD44-PE-Cy7 (clone: IM7); (rat monoclonal) | BioLegend | Cat No#103030; RRID: AB_830787 | FACS (1:100) |
| Antibody | Anti-mouse CD45.2-PE-Cy7 (clone: 104); (mouse monoclonal) | BioLegend | Cat No#109830; RRID: AB_1186098 | FACS (1:100) |
| Antibody | Anti-mouse CD71-APC (clone: RI7217); (rat monoclonal) | BioLegend | Cat No#113820; RRID: AB_2728135 | FACS (1:100) |
| Antibody | Anti-mouse CD98-PE-Cy7 (clone: RL388); (rat monoclonal) | BioLegend | Cat No#128214; RRID: AB_2750547 | FACS (1:100) |
| Antibody | Anti-mouse CD45.2-PerCP (clone: 104); (mouse monoclonal) | BioLegend | Cat No#109826; RRID: AB_893349 | FACS (1:100) |
| Antibody | Anti-mouse CD62L-Pacific Blue (clone: MEL-14); (rat monoclonal) | BioLegend | Cat No#104424; RRID: AB_493380 | FACS (1:100) |
| Antibody | Anti-mouse CD69-PE (clone: H1.2F3); (Armenian hamster monoclonal) | BioLegend | Cat No#104508; RRID: AB_313111 | FACS (1:100) |
| Antibody | Anti-mouse CD107a-APC-Cy7 (clone: 1D4B); (rat monoclonal) | BioLegend | Cat No#121616; RRID: AB_10643268 | FACS (1:100) |
| Antibody | Anti-mouse Foxp3-PE (clone: MF-14); (rat monoclonal) | BioLegend | Cat No#126404; RRID: AB_1089117 | FACS (1:100) |
| Antibody | Anti-mouse siglec-F (CD170)-APC (clone: S17007L); (rat monoclonal) | BioLegend | Cat No#155508; RRID: AB_2750237 | FACS (1:100) |
| Antibody | Anti-mouse Ly6C-APC (clone: HK1.4); (rat monoclonal) | BioLegend | Cat No#128016; RRID: AB_1732076 | FACS (1:100) |
| Antibody | Anti-mouse Ly6G-PE (clone: 1A8); (rat monoclonal) | BioLegend | Cat No#127608; RRID: AB_1186099 | FACS (1:100) |
| Antibody | Anti-mouse IL-4-PE (clone: 11B11); (rat monoclonal) | BioLegend | Cat No#504104; RRID: AB_315318 | FACS (1:100) |
| Antibody | Anti-mouse IL-5-APC (clone: TRFK5); (rat monoclonal) | BioLegend | Cat No#504306; RRID: AB_315330 | FACS (1:100) |
| Antibody | Anti-mouse IL-13-PE (clone: eBio13A); (rat monoclonal) | ThermoFisher | Cat No# 12-7133-82; RRID: AB_763559 | FACS (1:100) |
| Antibody | Anti-mouse IL-17A-PE (clone: eBio17B7); (rat monoclonal) | ThermoFisher | Cat No#12-7177-81; RRID: AB_763582 | FACS (1:100) |
| Antibody | Anti-mouse IFN-γ-APC (clone: XMG1.2); (rat monoclonal) | BioLegend | Cat No#505810; RRID: AB_315404 | FACS (1:100) |
| Antibody | Anti-mouse Granzyme B-PE (clone: NGZB); (rat monoclonal) | ThermoFisher | Cat No#12-8898-82; RRID: AB_10870787 | FACS (1:100) |
| Antibody | Anti-mouse CD3ε-Purified (clone: 145–2 C11); (Armenian hamster monoclonal) | BioLegend | Cat No#100340; RRID: AB_11149115 | Culture (1:1000) |
| Antibody | Anti-mouse CD28-Purified (clone: 37.51); (Syrian hamster monoclonal) | BioLegend | Cat No#102116; RRID: AB_11147170 | Culture (1:1000) |
| Antibody | Anti-mouse 6PGD; (rabbit polyclonal) | Sigma Aldrich | Cat No#HPA031314; RRID: AB_10610278 | Western blot (1:1000) |
| Antibody | Anti-β-actin (D6A8); (rabbit monoclonal) | Cell Signaling Technology | Cat No#8457; RRID: AB_10950489 | Western blot (1:1000) |
| Genetic reagent (species) | Pgd TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4351372; Assay ID:Mm01263703_m1 | |
| Genetic reagent (species) | Foxp3 TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID: Mm00475162_m1 | |
| Genetic reagent (species) | Gata3 TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID:Mm00484683_m1 | |
| Genetic reagent (species) | Il5 TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID:Mm00439646_m1 | |
| Genetic reagent (species) | Il13 TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID: Mm00434204_m1 | |
| Genetic reagent (species) | Ifng TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID: Mm01168134_m1 | |
| Genetic reagent (species) | Tbet (Tbx21) TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No# 4331182; Assay ID: Mm00450960_m1 | |
| Genetic reagent (species) | Il17a TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID: Mm00439618_m1 | |
| Genetic reagent (species) | Icos TaqMan Assay probe (FAM-MGB) | ThermoFisher | Cat No#4331182; Assay ID: Mm00497600_m1 | |
| Genetic reagent (species) | Il4 TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID:Mm00445259_m1 | |
| Genetic reagent | Il10 TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID: Mm00439614_m1 | |
| Genetic reagent (species) | Batf3 TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID: Mm01318274_m1 | |
| Genetic reagent (species) | Hey1 TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID: Mm00468865_m1 | |
| Genetic reagent (species) | Bcl6 TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID: Mm00477633_m1 | |
| Genetic reagent (species) | Gzb TaqMan Assay probe (FAM-MGB) (mouse) | ThermoFisher | Cat No#4331182; Assay ID: Mm00442834_m1 | |
| Genetic reagent (species) | 18 S rRNA TaqMan Assay probe (VIC-MGB) (mouse) | ThermoFisher | Cat No#4319413E | |
| Cell line (species) | B16-F10 (mouse) | ATCC | ATCC CRL-6475; RRID:CVCL_0159 | |
| Chemical compound, drug | 6-Aminonicotinamide (6-AN) | Sigma Aldrich | Cat No#A68203 | |
| Chemical compound, drug | Tamoxifen | Sigma Aldrich | Cat No#T5648-1G | |
| Chemical compound, drug | Dehydroepiandrosterone (DHEA) | Cayman Chemical | Cat No#15728 | |
| Chemical compound, drug | Dimethyl sulfoxide (DMSO) | Sigma Aldrich | Cat No#D2438 | |
| Chemical compound, drug | Tetra-methylrhodamine ester (TMRE) | ThermoFisher | Cat No#T669 | |
| Chemical compound, drug | MitoSOX Red | ThermoFisher | Cat No#M36008 | |
| Chemical compound, drug | MitoTracker Deep Red FM | ThermoFisher | Cat No#M22426 | |
| Chemical compound, drug | 2-NBD-glucose (2-NBDG) | Cayman Chemical | Cat No#11046 | |
| Chemical compound, drug | D-GLUCOSE (1,2,3,4,5,6,6-D7, 97–98%) | Cambridge Isotope laboratories | Cat No#DLM-2062-PK | |
| Chemical compound, drug | 13C5,15N2-Glutamine | Cambridge Isotope laboratories | Cat No#CNLM-1275-H-PK | |
| Chemical compound, drug | Fetal bovine serum, heat inactivated | ThermoFisher | Cat No#16140071 | |
| Chemical compound, drug | NuPAGE 4–12%, Bis-Tris, 1.5 mm, Mini Protein Gel | ThermoFisher | Cat No#NP0335BOX | |
| Chemical compound, drug | NuPAGE MES SDS Running Buffer | ThermoFisher | Cat No#NP0002 | |
| Chemical compound, drug | Recombinant Mouse IL-2 | BioLegend | Cat No#575404 | |
| Chemical compound, drug | Recombinant Mouse TGF-β1 | BioLegend | Cat No#763104 | |
| Commercial assay or kit | Milliplex MAP Mouse Cytokine/Chemokine Magnetic Bead Panel – Premixed 32 Plex | Millipore | Cat No# MCYTMAG-70K-PX32 | |
| Commercial assay or kit | Mouse Ig Isotyping Array Q1 | RayBiotech | Cat No#QAM-ISO-1–2 | |
| Commercial assay or kit | CellTrace CFSE Cell Proliferation Kit | ThermoFisher | Cat No#C34554 | |
| Commercial assay or kit | Dynabeads Mouse T-Activator CD3/CD28 | ThermoFisher | Cat No#11456D | |
| Commercial assay or kit | Fixation/Permeabilization Solution Kit with BD GolgiPlug | BD Bioscience | Cat No#555028 | |
| Commercial assay or kit | eBioscience Foxp3/ Transcription Factor Staining Buffer Set | ThermoFisher | Cat No#00-5523-00 | |
| Commercial assay or kit | LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Aqua) | ThermoFisher | Cat No#L34957 | |
| Commercial assay or kit | EasySep Mouse CD4+ T Cell Isolation Kit | STEMCELL Technologies | Cat No#19852RF | |
| Commercial assay or kit | EasySep Mouse Naïve CD4+ T Cell Isolation Kit | STEMCELL Technologies | Cat No#19765 | |
| Commercial assay or kit | EasySep Mouse CD4+ CD25+ Regulatory T Cell Isolation Kit II | STEMCELL Technologies | Cat No#18783 | |
| Commercial assay or kit | RNeasy Mini Kit | Qiagen | Cat No#74104 | |
| Commercial assay or kit | Seahorse XF Cell Mito Stress Test Kit | Agilent | Cat No#103015–100 | |
| Commercial assay or kit | Pierce BCA Protein Assay Kit | ThermoFisher | Cat No#23225 | |
| Genetic reagent(Mus musculus) | Pgdfl/fl Foxp3YFP-Cre | This paper | N/A | 6PGD exon 5 floxed. Send reagent request to pseth@bidmc.harvard.edu |
| Genetic reagent(Mus musculus) | C57BL/6 J (B6 CD45.2+) | The Jackson Laboratory | Stock No: 000664 | |
| Genetic reagent(Mus musculus) | B6.SJL-Ptprca Pepcb/BoyJ (B6 CD45.1+) | The Jackson Laboratory | Stock No: 002014 | B6 Cd45.1 | |
| Genetic reagent(Mus musculus) | B6.129S7-Rag1tm1Mom/J | The Jackson Laboratory | Stock No: 002216 | |
| Genetic reagent(Mus musculus) | B6.129(Cg)-Foxp3tm4(YFP/icre)Ayr/J | The Jackson Laboratory | Stock No: 016959 | |
| Genetic reagent(Mus musculus) | Foxp3tm9(EGFP/cre/ERT2)Ayr/J | The Jackson Laboratory | Stock No: 016961 | |
| Software, algorithm | FlowJo_V10 | FlowJo | https://www.flowjo.com/; RRID:SCR_008520 | |
| Software, algorithm | GraphPad Prism_V6 | GraphPad | https://www.graphpad.com/; RRID:SCR_002798 |