Selection for increased tibia length in mice alters skull shape through parallel changes in developmental mechanisms
Figures
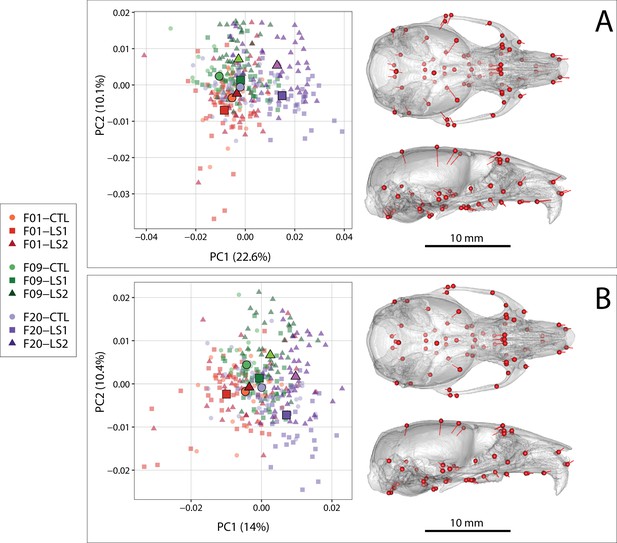
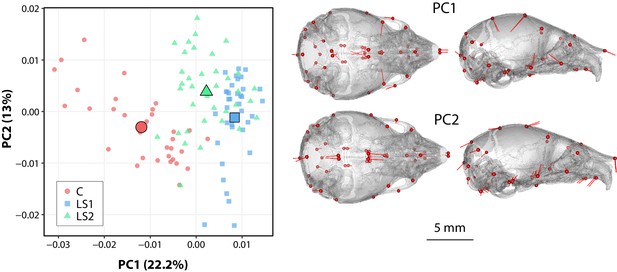
Scatter plots of the first two principal components (PC) of Procrustes shape variables in adult Longshanks and Controls throughout the selection process.
(A) Plot of sex-adjusted Procrustes shape variables (left), and vectors of shape change at each cranium landmark (magnified two times for visualization) showing shape transformations along PC1 from negative to positive scores (right). Large symbols indicate mean PC1 and PC2 scores for each respective cohort. (B) Plot of Procrustes shape variables additionally corrected for size.
-
Figure 1—source data 1
Adult morphometric and landmark data.
- https://cdn.elifesciences.org/articles/67612/elife-67612-fig1-data1-v2.xlsx
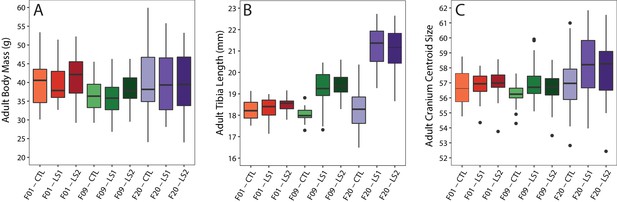
Boxplots of adult Longshanks and Control metrics.
(A–C) Boxplots of adult body mass, tibia length, and cranial centroid size between groups. See also Supplementary file 1.
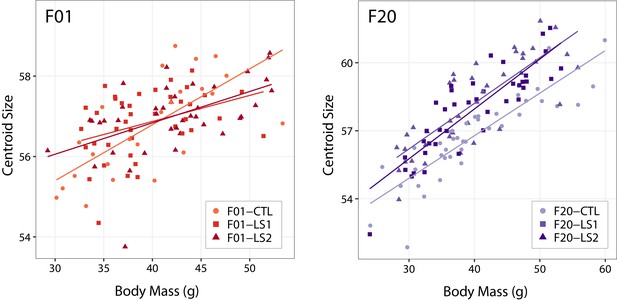
Scatter plots with regression lines by group showing the relationships between body mass and cranium size (centroid size) in adult founder mice (F01) and after 20 generations of selection (F20).
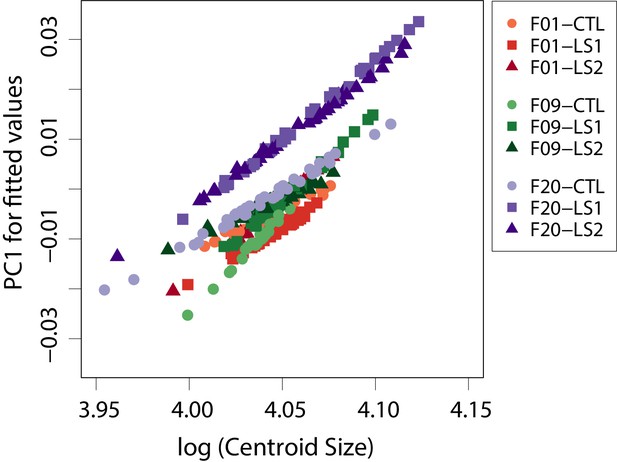
Scatter plot of fitted PC1 scores (shape scores predicted by regression of shape on size) vs log (centroid size) showing within group patterns of cranium allometry.
At any given cranium size, LS1 and LS2 are predicted to have positive shape scores (longer and narrower) shapes.
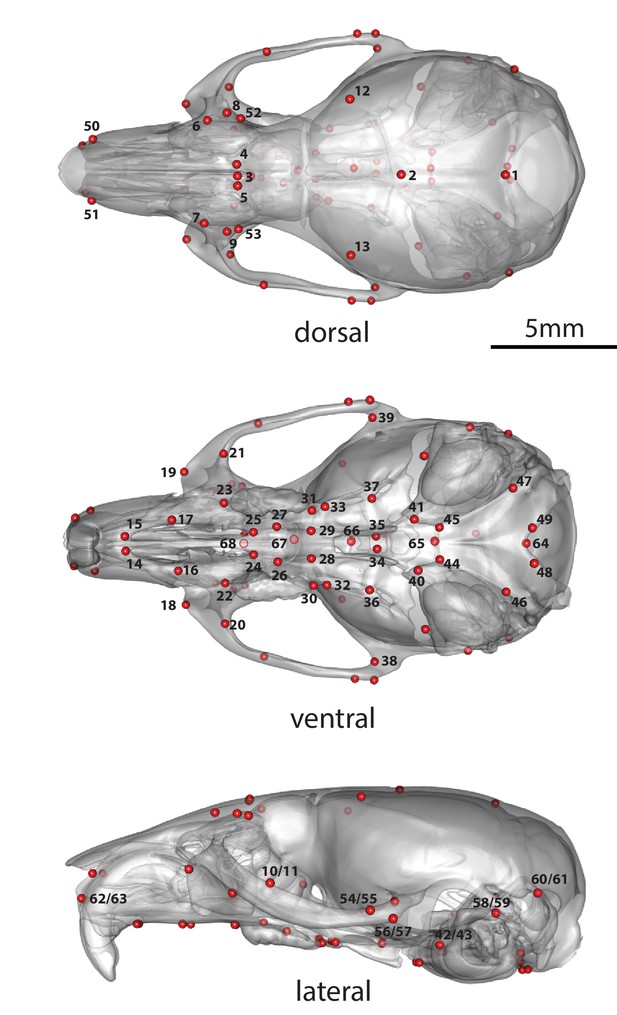
Adult cranium landmarks used in this study in lateral, dorsal, and ventral landmark views.
For landmark anatomical definitions, see Supplementary file 4.
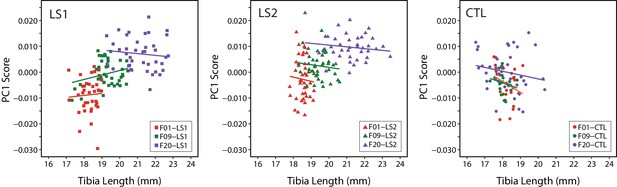
Scatter plots with regression lines by cohort showing the relationships between PC1 score (cranium shape) and tibia length in adult mice throughout selection.
Cranium shape data is adjusted for the effects of sex and size related allometry.
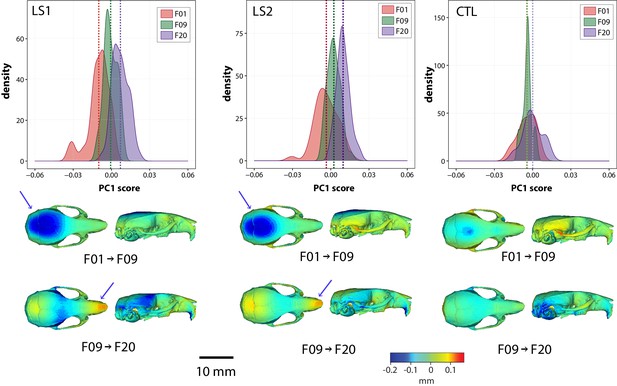
Intergenerational shape changes within both Longshanks lines and Controls throughout the selection process.
Intergenerational shape changes within both Longshanks lines and Controls throughout the selection process. Top: Density plots following intergenerational shifts in mean PC1 scores within LS1 (left), LS2 (center), and CTL (right) lines for size and sex adjusted shape data. Bottom: Heatmaps showing shape transformations between mean shapes in the first 9 generations of selection (F01 to F09) and the next 11 generations (F09 to F20) after correcting for size and sex effects. Blue indicates areas of relative reduction, red indicates areas of relative expansion, and green indicates neutral areas. Longshanks independently undergo vault height reduction between generations F01 and F09, followed by snout elongation between generations F09 and F20 (blue arrows).
Scatter plots of the first principal components (PC) in neonate Longshanks and Control cranium Procrustes shape variables at generation 32 (F32).
Left: Plot of litter size adjusted Procrustes shape variables (left), large symbols indicate mean PC1 and PC2 scores for each respective cohort. Shapes of individual points indicate Longshanks lines (circle = CTL, square = LS1, and triangle = LS2). Right: Neonate cranium with vectors of shape change at each cranium landmark (magnified four times for visualization) showing shape transformations along PC1 (top) and along PC2 (bottom) from negative to positive scores.
-
Figure 4—source data 1
Neonate morphometric and landmark data.
- https://cdn.elifesciences.org/articles/67612/elife-67612-fig4-data1-v2.xlsx
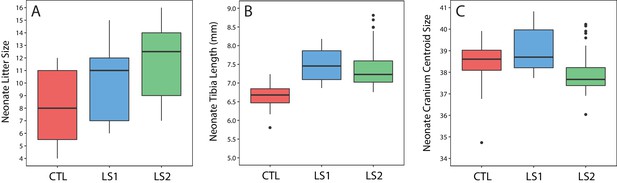
Boxplots showing differences in neonate Longshanks and Control metrics.
(A) Boxplot of neonate litter sizes. (B and C) Boxplots of litter size adjusted tibia length and cranium centroid size.
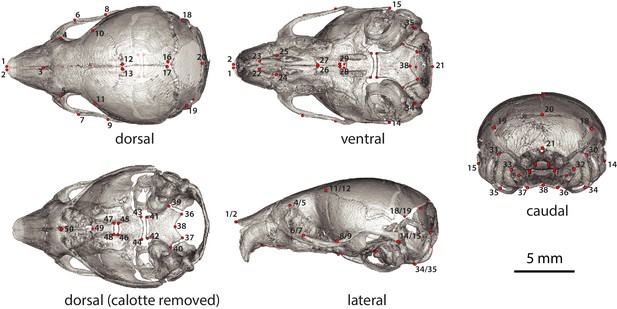
Neonate (P07) cranium landmarks used in this study in lateral, dorsal, dorsal cranial base, caudal, and ventral landmark views.
For landmark anatomical definitions, see Supplementary file 5.
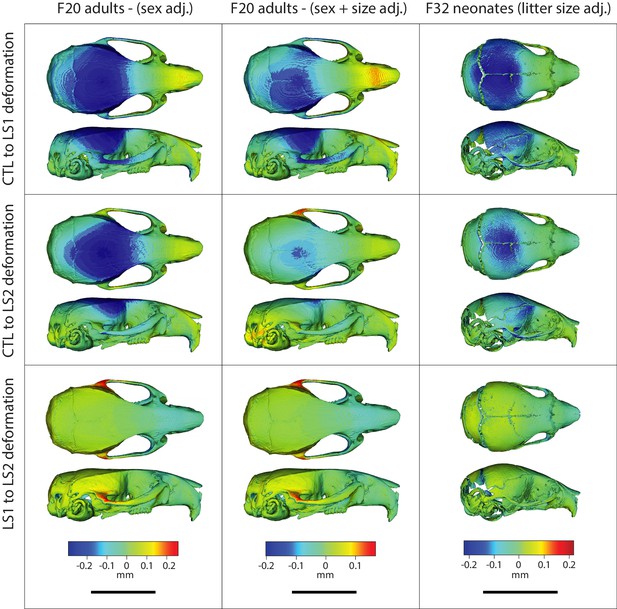
Comparison of adult and neonate cranial phenotypes through shape change heatmaps.
Heatmaps show the deformations required to transform between the mean shape of a given cohort to the mean shape of another. Blue indicates areas of relative reduction, red indicates areas of relative expansion, and green indicates neutral areas. Scale bar = 10 mm.
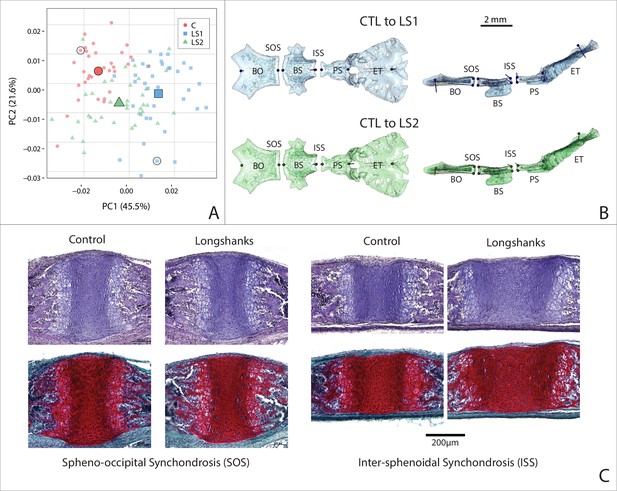
Cranial base contribution to the Longshanks phenotype.
(A) Scatter plot of the first two PCs of litter size-adjusted Longshanks and Control cranial base Procrustes shape variables in generation 32 (F32) neonates. Large symbols indicate mean PC1 and PC2 scores for each respective cohort. (B) Neonate cranial bases with vectors of shape change at midline cranial base landmarks (magnified six times for visualization) showing shape transformations to go from the mean Control cranial base to the mean LS1 (blue) and mean LS2 (green) cranial base shapes. Views in (B) are dorsal (left) and lateral (right). Basi-occipital bone (BO), spheno-occipital synchondrosis (SOS), basi-sphenoid bone (BS), intersphenoidal synchondrosis (ISS), presphenoid bone (PS) and ethmoid (ET). (C) Sagittal midline histological sections stained in H + E (top) and safranin-o (bottom) showing differences in synchondrosis morphology of two extreme specimens in CTL and LS1 of approximately equal centroid size (indicated by dashed circles in A).
-
Figure 6—source data 1
Adult morphometric and landmark data.
- https://cdn.elifesciences.org/articles/67612/elife-67612-fig6-data1-v2.xlsx
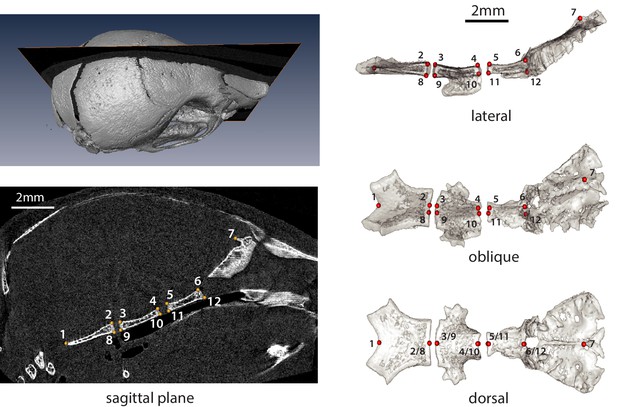
Neonate (P07) cranial base landmarks on the sagittal midline used in this study.
Numbered landmarks applied to CT scan reconstruction slices at the midline (left) and landmarks numbered on a 3D cranial base mesh (right). For landmark anatomical definitions, see Supplementary file 6.
Videos
Mean cranial deformation in Longshanks.
Top: deformation of an F20 Control mean cranial shape into a F20 Longshanks Line 1 (LS1) mean cranial shape, with both mean configurations scaled to the pooled mean centroid size of the total sample (n = 327). Bottom: as above, but the skulls in each group have been scaled to the mean centroid size of that group. In both cases, the most significant changes relate to the flattening of the frontal and parietal regions, decreasing height of the braincase, and relative elongation of the snout in Longshanks.
Extreme cranial deformation in Longshanks.
Top: deformation trajectory between one extreme Control male and one extreme LS1 male, selected from non-overlapping regions of morphospace (see Figure 1). The skull configurations are scaled to the pooled mean centroid size of the total sample (n = 327). Bottom: as above, but the two skulls have been scaled to those individuals’ centroid sizes, revealing substantial differences in centroid size between these two individuals as well.
Tables
Longshanks adult (F01, F09, F20) and neonate (F32) sample composition.
Target sample sizes of n = 40/group represent a compromise between sampling effort (e.g. scanning capacity) and the ability to detect small-to-moderate effect sizes at a power of 0.8 in the case of univariate analyses (Cohen’s d = 0.25). For multivariate analyses (e.g. principal components analyses), target sample sizes of n = 40 with 50–68 landmarks produce highly repeatable covariance matrices (average RV coefficient of a sample’s Procrustes-adjusted covariance matrix with 1000 covariance matrices derived from bootstrapped data > 0.99).
| Longshanks samples (n) | |||
|---|---|---|---|
| CTL | LS1 | LS2 | |
| Generation 1 (F01) | 24 | 40 | 40 |
| Generation 9 (F09) | 23 | 40 | 40 |
| Generation 20 (F20) | 40 | 40 | 40 |
| Generation 32 (F32) | 32 | 36 | 36 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Mouse (Mus musculus) | Longshanks 1 | Campbell Rolian | LS1 | Stock: HSD:ICR |
| Mouse (Mus musculus) | Longshanks 2 | Campbell Rolian | LS2 | Stock: HSD:ICR |
| Mouse (Mus musculus) | Control | Campbell Rolian | CTL | Stock: HSD:ICR |
| Software, algorithm | R, R packages geomorph, Morpho, RRPP | R Development Core Team, 2020, Schlager, 2020, Collyer and Adams, 2018, Adams et al., 2020 | n/a | |
| Software, algorithm | Amira | Visage Imaging, Berlin, Germany | Version 5.4.2 | |
| Software, algorithm | Python and Bash (Medical Imaging NetCDF library) | Percival et al., 2019, Devine et al., 2020 | Python 3.6 Bash-5.1 MINC 2.0 | https://github.com/BIC-MNI/minc-toolkit-v2 https://github.com/jaydevine/Landmarking |
| Other | Haematoxylin (for Weigert’s) | Sigma | H3136 | Histological stain |
| Other | Iron (III) chloride (for Weigert’s) | Sigma | 157740 | Histological stain |
| Other | Fast Green (FCF) | Sigma | F7252 | Histological stain |
| Other | Safranin-O | Sigma | S2255 | Histological stain |
| Other | Gill’s Haematoxylin | Sigma | GHS332 | Histological stain |
| Other | Eosin Y | Sigma | 588X | Histological stain |
Additional files
-
Supplementary file 1
Morphometric data for adult mice among lines and generations.
Body mass data represent means and SEM, whereas centroid size and tibia length are least squared means and SEM. Superscripts denote significant differences in means (p<0.05) between a given group and: Controls CTL, Longshanks Line 1 LS1, Longshanks Line 2 LS2, from either: Generation 1 F01, Generation 9 F09, or Generation 20 F20, as determined using Tukey’s HSD tests. Differences in body mass were determined by ANOVA, whereas tibia length and centroid size differences were assessed by ANCOVA with body mass as a covariate. Bold and italic superscripts indicate significant intergenerational differences and intragenerational differences, respectively.
- https://cdn.elifesciences.org/articles/67612/elife-67612-supp1-v2.docx
-
Supplementary file 2
Mean Euclidean distances.
Euclidean distances between the multivariate mean PC scores of each group, based on Procrustes shape data adjusted for sex only (above diagonal), or sex and cranial centroid size (below diagonal). The only non-significant Euclidean distance, based on a post-hoc Procrustes ANCOVA, is indicated in bold.
- https://cdn.elifesciences.org/articles/67612/elife-67612-supp2-v2.docx
-
Supplementary file 3
Morphometric data for neonate mice among lines and generations.
Tibia and centroid size data represent least squared means (SEM) and litter sizes are means (SEM). Differences in litter size were determined by ANOVA, whereas tibia length and centroid size differences were assessed by ANCOVA with litter size as a covariate. Superscripts denote significant differences in means (p<0.05) between a given group and: Controls CTL, Longshanks Line 1 LS1, Longshanks Line 2 LS2.
- https://cdn.elifesciences.org/articles/67612/elife-67612-supp3-v2.docx
-
Supplementary file 4
Adult cranium landmarks and their anatomical definitions.
- https://cdn.elifesciences.org/articles/67612/elife-67612-supp4-v2.docx
-
Supplementary file 5
Neonate cranium landmarks and their anatomical definitions.
- https://cdn.elifesciences.org/articles/67612/elife-67612-supp5-v2.docx
-
Supplementary file 6
Neonate cranial base landmarks and their anatomical definitions.
- https://cdn.elifesciences.org/articles/67612/elife-67612-supp6-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67612/elife-67612-transrepform-v2.pdf





