Design principles of the ESCRT-III Vps24-Vps2 module
Figures
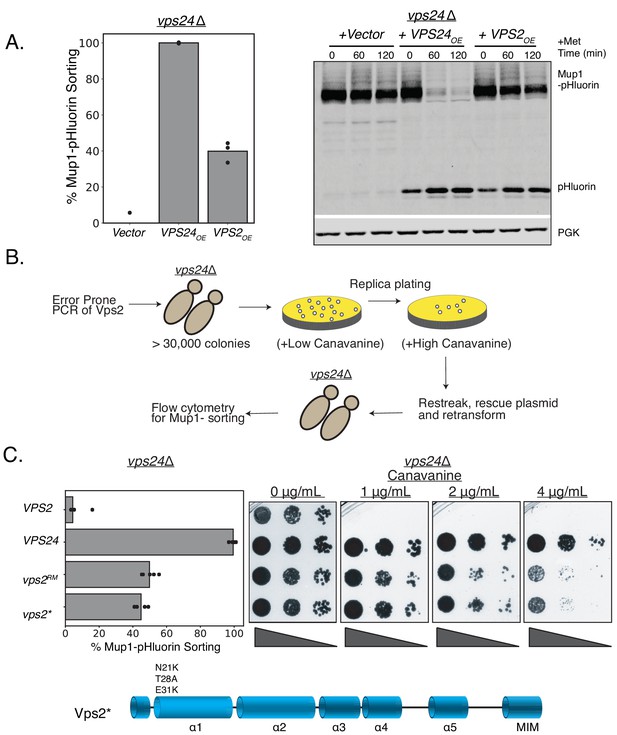
Minor modifications in Vps2 can replace the function of Vps24.
(A) Overexpression of Vps2 can rescue the defect of vps24∆ for Mup1 sorting. Image on the left represents Mup-pHluorin sorting through a flow cytometry assay and the image on the right represents an immunoblot for pHluorin upon methionine addition. Overexpression (OE) was achieved through a CMV promoter and Tet operator containing plasmid. (B) Flowchart of the random mutagenesis approach. (C) Top figure shows the flow cytometry and canavanine sensitivity assays with the mutants of Vps2 that can rescue the sorting defects of vps24∆. Bottom figure shows the domains of Vps2 highlighting the mutations in Vps2* .
-
Figure 1—source data 1
Mup1-pHluorin sorting data associated with data in Figure 1A, Figure 1C, Figure 1—figure supplement 1C, Figure 1—figure supplement 1D, Figure 1—figure supplement 3A, Figure 1—figure supplement 3B.
In Supplement 1C, two datapoints are from fluorescence measurements with flow cytometry and the third data set is from an immunoblot of Mup1-pHluorin cleavage. In Supplement 1D, source data are for different expression levels of Vps2 with the doxycycline-inducible operator at different concentrations of doxycycline. Also included is the sequence of the promoter region of Vps2* (from random mutagenesis), associated with Figure 1—figure supplement 3D.
- https://cdn.elifesciences.org/articles/67709/elife-67709-fig1-data1-v1.xls
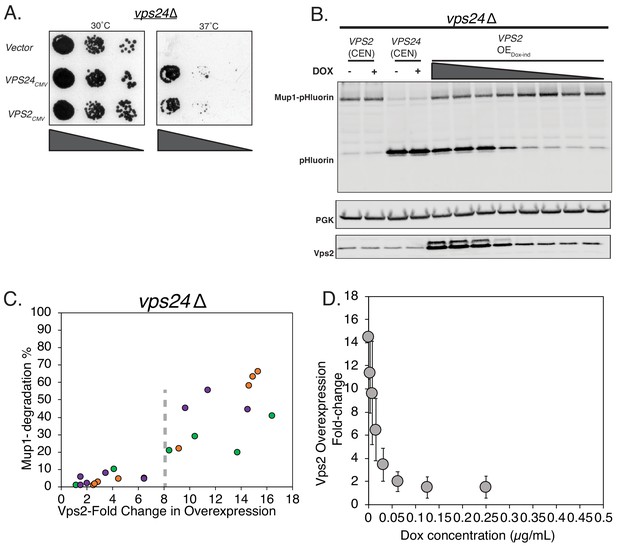
Overexpression of Vps2 can rescue the defect of vps24∆.
(A) Vps2 overexpression with a CMV promoter/Tet operator rescues the temperature sensitivity defect of vps24∆. (B) Immunoblot of pHluorin showing the cleavage of Mup1-pHluorin after 90 min of methionine addition. Vps2 was expressed either in a single-copy centromeric (CEN) plasmid or under a doxycycline-inducible CMV promoter. Expression of Vps2 was controlled by titrating the concentration of doxycycline. (C) Mup1-sorting characterization with changes in Vps2 expression level. The different colors represent different set of titration experiments. (D) Plot showing the control of Vps2 expression levels with doxycycline titration.
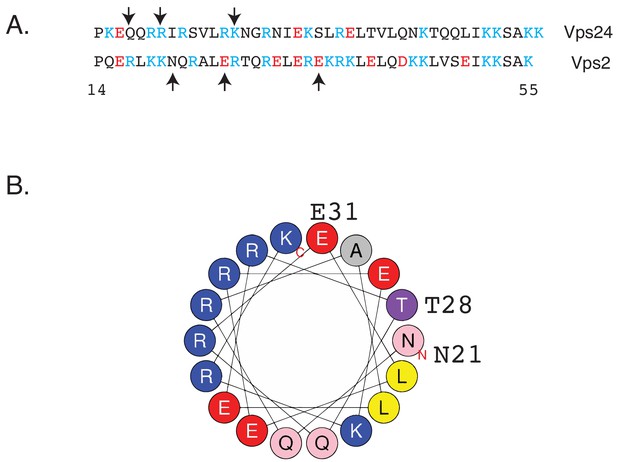
Helix-1 region of Vps2 is important for binding to Snf7.
(A) Sequence alignment of helix 1 of Vps24 and Vps2. Cyan-colored residues are basic amino acids, and red colors represent acidic amino acids. Arrows in Vps24 sequence point to location of charge-inversion mutations that rescue the defect of the snf7D131K allele (Banjade et al., 2019a). Arrows in Vps2 sequence represent the location of the mutations that rescue vps24∆. (B) Helical wheel representation of part of the helix-1 region of Vps2 (Heliquest).
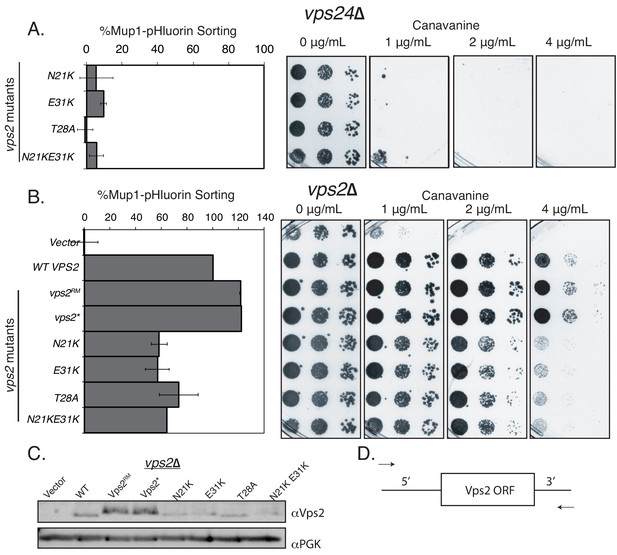
Vps2 N-terminal mutations can rescue the defect of vps24∆.
(A) Flow cytometry for Mup1-pHluorin sorting and canvanine sensitivity assay in vps24∆ for the N-terminal helix-1 mutations in Vps2 (compare with Figure 1C). In these constructs, the promoters were endogenous, wild-type (WT) promoters. (B) In a vps2∆ background, the suppressors vps2RM and vps2* also have higher sorting capabilities in both Mup1-sorting assay and canavanine sensitivity assay. The promoter regions of vps2RM and vps2* contain mutations, but other constructs used here are with WT promoters. (C) Immunoblots of various Vps2 mutants, the same constructs as used in Figure 1C, and A and B. (D) Design of the randomly mutagenized VPS2 plasmid – primers bind to the 5’ and 3’ UTR regions of VPS2.
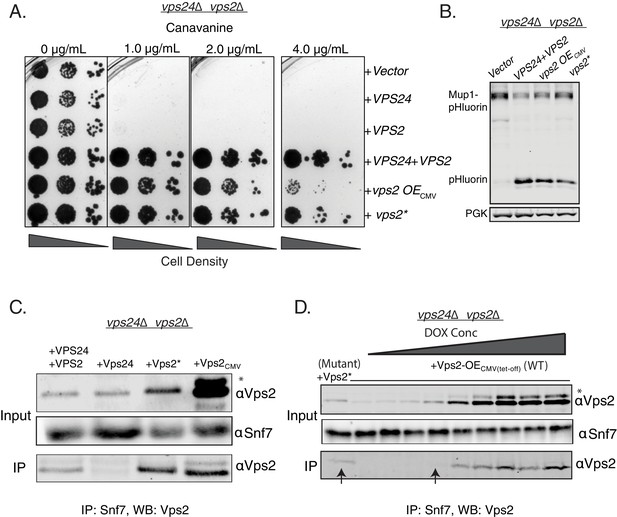
Properties of both Vps2 and Vps24 in a single Vps2 construct.
(A) Canavanine sensitivity data in vps24∆vps2∆ with an overexpression of Vps2 (CMV-Tet system) or with Vps2*. (B) Immunoblot for Mup1-pHluorin sorting upon overexpression of Vps2 (CMV) or with Vps2*. (C) Co-immunoprecipitation of Snf7 with Vps2 (CMV) and Vps2* in vps24∆vps2∆. (D) Co-immunoprecipation experiments of Snf7 with Vps2 at various expression levels of Vps2 after titration of the Tet-off operator with doxycycline. Arrows point to the relative binding to Snf at similar expression levels of Vps2 and Vps2*. In the gels, * refers to an unknown modified form of Vps2.
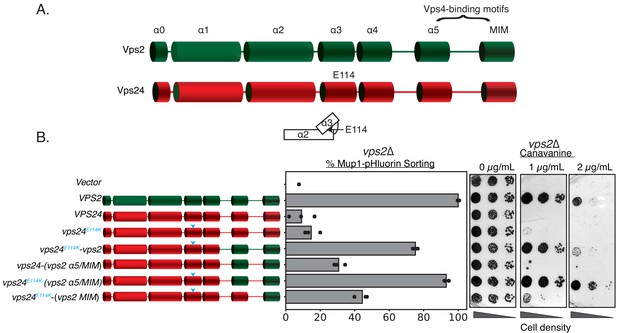
Simple modifications in Vps24 can be made to mimic Vps2.
(A) The domain organization of Vps2, highlighting the C-terminal region important for Vps4 binding. (B) Left panel denotes the chimeras made to replace regions of Vps2 onto Vps24. Cyan arrows in the helices are positions of the E114K mutation. Right panel represents Mup1-pHluorin sorting and canavanine sensitivity assays. In this assay, the constructs were overexpressed under a CMV promoter-Tet-off operator system.
-
Figure 3—source data 1
Mup1-pHluorin sorting data associated with figure in Figure 3B, Figure 3—figure supplement 1A, Figure 3—figure supplement 1B, Figure 3—figure supplement 3A, Figure 3—figure supplement 3B.
- https://cdn.elifesciences.org/articles/67709/elife-67709-fig3-data1-v1.xls
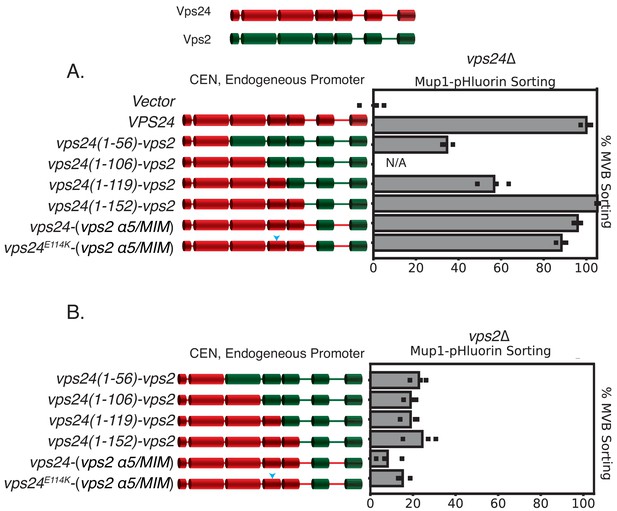
Chimeras of Vps24-Vps2 are functional proteins.
Top figure depicts the domain organization of Vps2 and Vps24. (A) Mup-pHluoring sorting assay with several chimeras of Vps24-Vps2, showing that the replacement of the C-terminal regions of Vps2 onto Vps24 keeps the constructs functional. (B) The same constructs as in (A) do not suppress vps2∆, as they are under endogeneous promoters. ‘CEN’ represents denotation for centromeric plasmid.
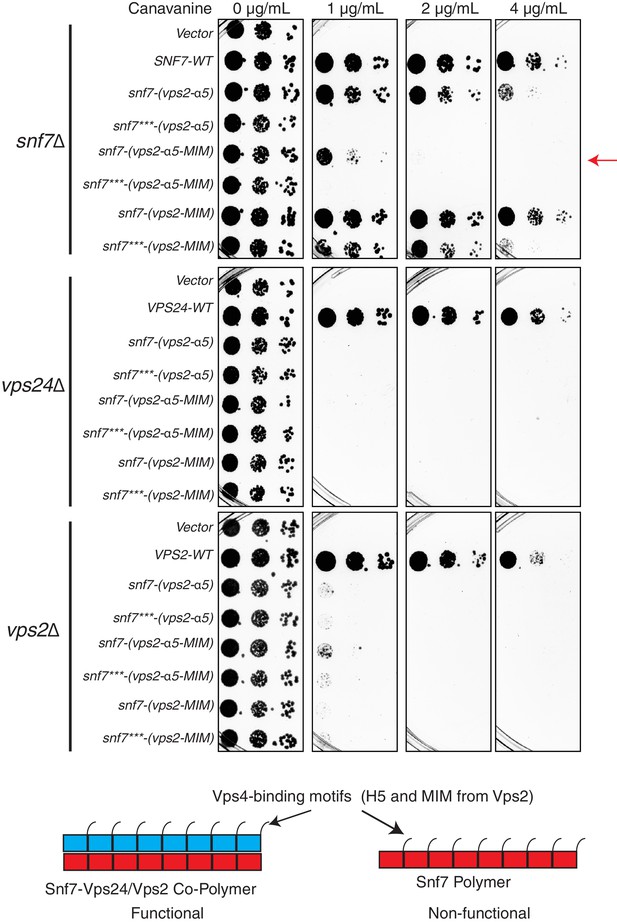
Simply adding Vps4 binding sites to Snf7 do not replace the functions of Vps24-Vps2.
Canavanine sensitivity assays of Snf7 constructs with the helix-5 and MIM regions replaced from Vps2 onto Snf7. Similar replacements were also done with Snf7***, which consists of the triple mutation R52EQ90LN100I. Bottom models illustrate the idea that Vps24-Vps2 copolymer and recruitment of Vps4 through the copolymer may be required to obtain the functional heteropolymer.
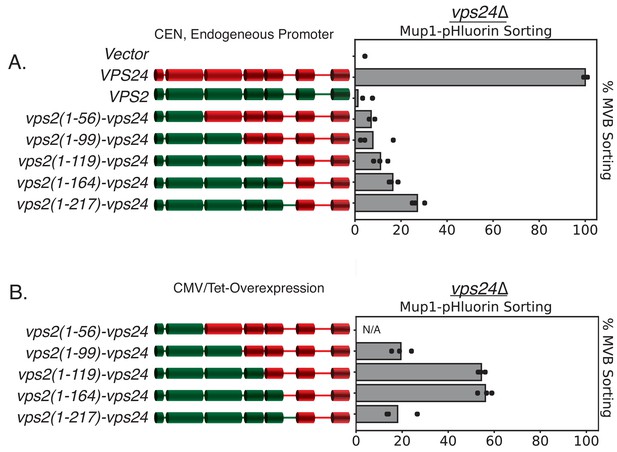
Simple modifications in Vps24 can be made to mimic Vps2.
(A) Under the endogenous promoter and centromeric plasmid (CEN), various chimeras of Vps24-Vps2 do not support the sorting of Mup1-pHluorin, but some of the same constructs when overexpressed can rescue vps24∆ (B). Note that the N-terminus of Vps2 needs to be intact to mimic Vps24. Overexpression was achieved with a CMV promoter, Tet operator system.
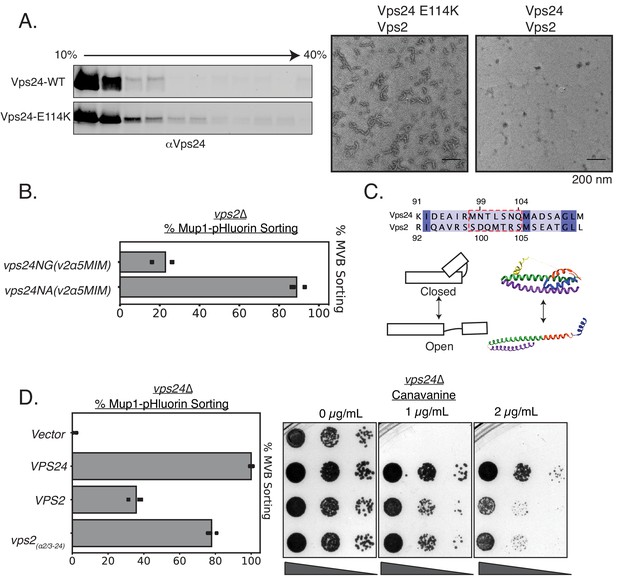
Vps24 and Vps2 may exhibit different conformations.
(A) Left: Glycerol-gradient experiments with Vps24 and Vps24E114K suggest that the mutant can form higher molecular-weight species. Right: Negative stain electron microscopy of Vps24 E114K or wild-type (WT) Vps24 at 1 µM each of the proteins in the presence of Vps2. (B) Mup1-pHluorin assays with Vps24 mutations in the asparagines (N99 and N103) ɑ2/ɑ3 hinge region to Ala or Gly residues in constructs that have the Vps4 binding sites H5 (helix 5) and MIM from Vps2 (V2). These constructs are expressed with the CMV promoter, Tetoff system. See Figure 3 for direct comparison with other Vps24 mutants and chimeras. (C) Top: Sequences of the ɑ2/ɑ3 hinge region of Vps24 and Vps2. Bottom left: Model showing the two conformations of ESCRT-III proteins. Structural model on the right is that of CHMP3 (closed) (Bajorek et al., 2009) and of Snf7 (open) (Tang et al., 2015). (D) Mup1-pHluorin sorting and canavanine sensitivity assays with overexpression of Vps2 (CMV-Tet) and with a mutant replacing the ɑ2/ɑ3 hinge region of Vps2 with that of Vps24 (also CMV-Tet system).
-
Figure 4—source data 1
Mup1-pHluorin sorting data associated with figure in Figure 4B and Figure 4C.
- https://cdn.elifesciences.org/articles/67709/elife-67709-fig4-data1-v1.csv
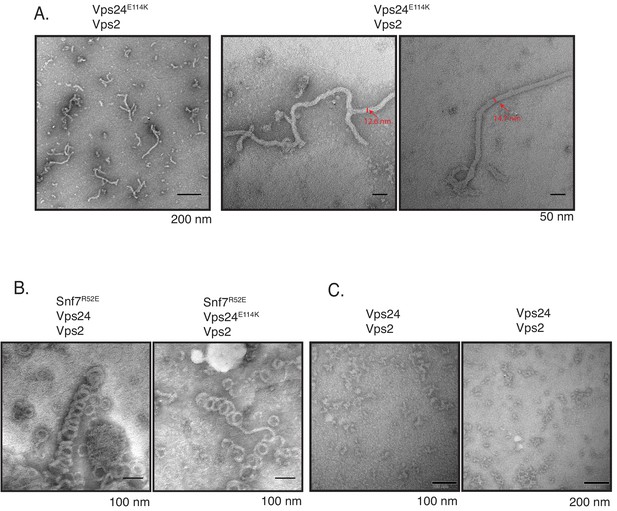
Vps24 (E114K) associates with Vps2.
(A) Electron microscopy images of Vps24E114K assembled with Vps2 at concentrations of 5 µM each. Two images on the right are zoomed-in images of the same polymers. Vps24 E114K alone or Vps24 with Vps2 do not form such polymers. (B) Vps24E114K mutant with Vps2 and Snf7-R52E still form 3D helices. Snf7-R52E is a mutant that has a lower critical concentration for polymerization as a higher fraction of this protein is in an open conformation (Henne et al., 2012). (C) Vps24-Vps2 at higher concentrations (15 µM each) form amorphous structures.
Structures of the autoinhibited CHMP3, and the filament forming conformations of Vps24 and Snf7.
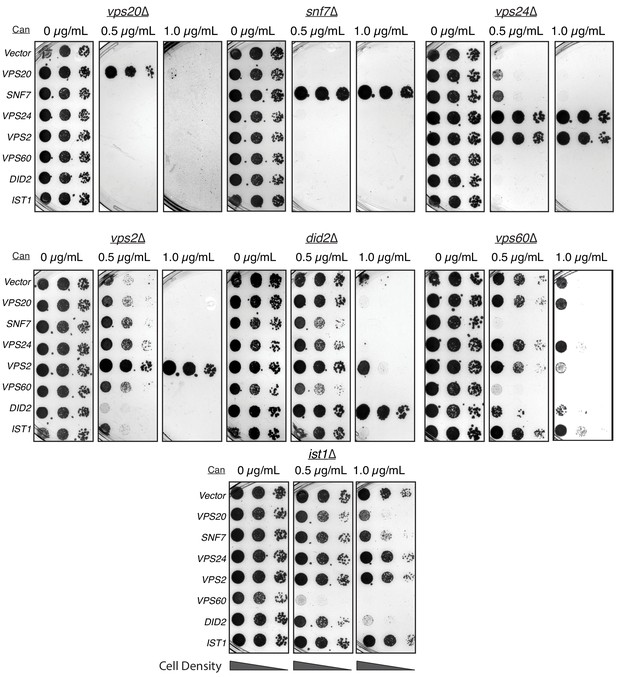
Overexpressing ESCRT-III proteins in the background of other ESCRT-III mutants show selective rescue phenotypes.
In the annotated mutants, ESCRT-III proteins were expressed with a CMV promoter/Tet operator system and plated in canavanine-containing plates. Vps2 overexpression can rescue the defect of vps24∆. Vps2 overexpression in a did2∆ partially rescues canavanine sensitivity. Vps60 overexpression appears to be dominant negative.
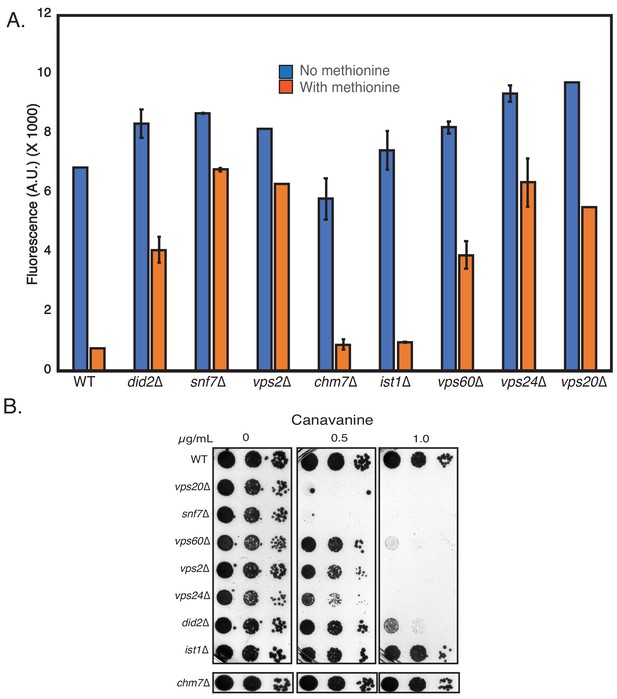
Relative effects of all ESCRT-III mutants for defects in cargo sorting.
(A) Cargo sorting analysis with ESCRT-III mutants using the Mup1-pHluorin assay. Fluorescence of 100,000 cells were measured after 90 min of adding of 20 µg methionine. (B) Canavanine sensitivity assays of the ESCRT-III mutants.
-
Figure 5—figure supplement 1—source data 1
Mup1-pHluorin sorting data associated with figure in Figure 5—figure supplement 1A.
- https://cdn.elifesciences.org/articles/67709/elife-67709-fig5-figsupp1-data1-v1.csv
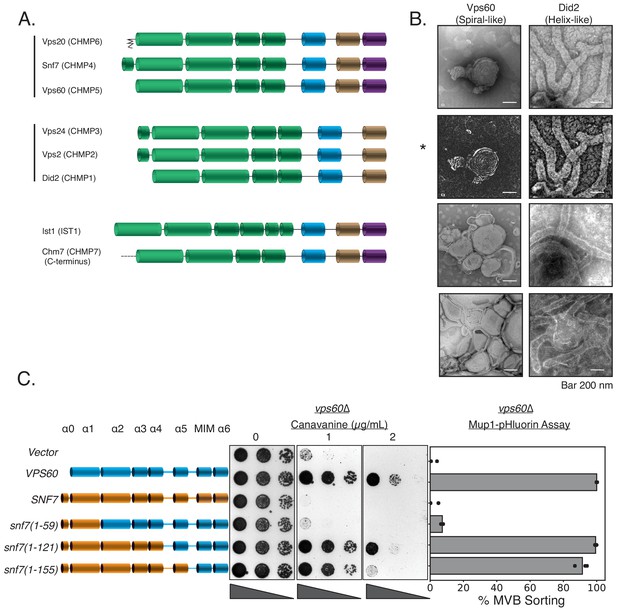
Vps60 possesses features of Snf7.
(A) Domain subunits of the eight ESCRT-III proteins in yeast. The mammalian names are in parentheses. (B) Electron microscopy images of 1 µM Vps60 or 1 µM Did2 on lipid monolayers, incubated for 1 hr. Bar is 200 nm each. Top two images (highlighted by an asterisk, *) are different constrast-adjusted depictions of the same image. (C) Domain swaps from Snf7 onto Vps60 can rescue the defects of canavanine sensitivity and Mup1-pHluorin sorting in a vps60∆ strain.
-
Figure 6—source data 1
Zip file contains Mup1-pHluorin sorting data associated with figure in Figure 6—figure supplement 1C and the raw images used to make Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/67709/elife-67709-fig6-data1-v1.zip
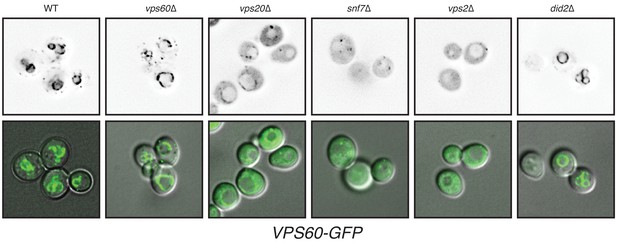
Localization of VPS60-GFP in different ESCRT-III mutants.
Top panel represents grayscaled GFP fluorescence and bottom panel represents a merge of GFP and DIC channels. While in wild-type (WT) strains VPS60-GFP primarily localizes to punctae (membranes), in various mutants the cytoplasmic signal is increased.
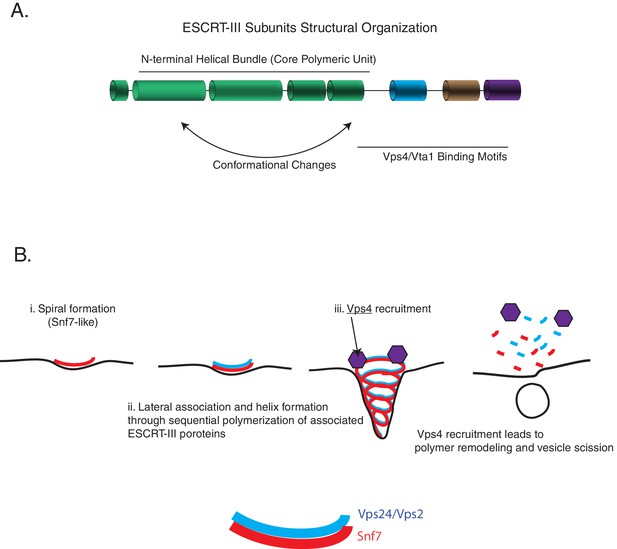
ESCRT-III assembly principles.
(A) The domain organization of ESCRT-III subunits and the various functional parts of the structures/sequence. (B) The minimal features of ESCRT-III assembly may involve spiral formation, lateral association between copolymers that induce helicity, and recruitment of a disassembly factor such as Vps4. Schematic on the bottom shows how spirals of Snf7 and Vps24/Vps2 may laterally associate.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Saccharomyces cerevisiae, Matα) | WT | PMID:3062374 | SEY6210 | (Background strain) MATα ura3-52 his3-200 leu2-3,112 trp1-901 lys2-801 suc2-9 |
| Strain, strain background (S. cerevisiae, Mata) | WT | PMID:3062374 | SEY6210.1 | (Background strain) MATa leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 |
| Strain, strain background (S. cerevisiae, Mata) | WT; Mup-pHluorin | PMID:24139821 | NBY40 | (SEY6210.1); MUP1-pHLUORIN::KANMX |
| Strain, strain background (S. cerevisiae, Mata) | snf7Δ | PMID:23063125 | NBY44 | (SEY6210.1); snf7Δ::HIS3; MUP1-PHLUORIN::KAN |
| Strain, strain background (S. cerevisiae, Mata) | vps24Δ | PMID:24139821 | NBY47 | (SEY6210.1); vps24Δ::HIS3; MUP1-pHLUORIN::KANMX |
| Strain, strain background (S. cerevisiae, Mata) | vps2Δ | PMID:24139821 | NBY69 | (SEY6210.1); vps2Δ::HIS3 MUP1-pHLUORIN::KANMX |
| Strain, strain background (S. cerevisiae, Mata) | did2∆ | This study | STY35 | (SEY6210.1); did2Δ::TRP1; Mup1-pHluorin::KAN |
| Strain, strain background (S. cerevisiae, Mata) | chm7∆ | This study | STY25 | (SEY6210.1); yjl049wΔ::TRP1; Mup1-pHluorin::KAN |
| Strain, strain background (S. cerevisiae, Matα) | ist1∆ | This study | STY34 | (SEY6210); ist1Δ::TRP1; Mup1-pHluorin::NAT |
| Strain, strain background (S. cerevisiae, Mata) | vps60∆ | This study | SBY465 | (SEY6210.1); vps60∆::hph; Mup1-pHluorin::Kan |
| Strain, strain background (S. cerevisiae, Mata) | vps20∆ | PMID:24139821 | NBY42 | (SEY6210.1); vps20::HIS3 MUP1-pHLUORIN::KANMX |
| Strain, strain background (S. cerevisiae, Mata) | vps24Δ vps2Δ | This study | SBY05 | (SEY6210.1); vps24Δ::HIS3; vps2Δ::Hph; MUP1-pHLUORIN::KANMX |
| Strain, strain background (S. cerevisiae, Mata) | vps60∆ | PMID:18032584 | SMY24 | (SEY6210.1); vps60∆::TRP1 |
| Strain, strain background (S. cerevisiae, Mata) | vps20∆ | PMID:12194857 | MBY25 | (SEY6210.1); vps20∆::HIS3 |
| Strain, strain background (S. cerevisiae, Mata) | snf7Δ | PMID:12194857 | MBY24 | (SEY6210.1); snf7∆::HIS3 |
| Strain, strain background (S. cerevisiae, Mata) | vps2Δ | PMID:12194857 | MBY29 | (SEY6210.1); vps2∆::HIS3 |
| Strain, strain background (S. cerevisiae, Mata) | did2∆ | PMID:18032584 | SMY61 | (SEY6210.1); did2∆::TRP1 |
| Strain, strain background (S. cerevisiae, Matα) | vps2Δ | PMID:12194857 | MBY28 | (SEY6210); vps2∆::HIS3 |
| Strain, strain background (S. cerevisiae, Matα) | did2∆ | PMID:18032584 | SMY60 | (SEY6210); did2∆::TRP1 |
| Recombinant DNA reagent | Vector | PMID:2659436 | pRS416 | |
| Recombinant DNA reagent | Vector | PMID:2659436 | pRS414 | |
| Recombinant DNA reagent | Vector | PMID:2659436 | pRS415 | |
| Recombinant DNA reagent | pCM189 | PMID:9234672 | pCM189 | CMV promoter controlling Tet-off operator |
| Recombinant DNA reagent | pCM189 Vps24 (OE) | PMID:31246173 | pCM189 Vps24 | Progenitor: pRS 414 Vps24 (PMID:23063125), Vector: pCM189 (PMID:9234672), S. cerevisiae sequence |
| Recombinant DNA reagent | pCM189 Vps2 (OE) | PMID:31246173 | pCM189 Vps2 | Progenitor: pRS 415 Vps2 (PMID:23063125), Vector: pCM189 (PMID:9234672), S. cerevisiae sequence |
| Recombinant DNA reagent | Vps24 | PMID:31246173 | pRS 416 Vps24 | Progenitor: pRS 414 Vps24 (PMID:23063125), Vector: pRS416 (PMID:2659436), S. cerevisiae sequence |
| Recombinant DNA reagent | Vps2 | PMID:31246173 | pRS 416 Vps2 | Progenitor: pRS 415 Vps2 (PMID:23063125), Vector: pRS416 (PMID:2659436), S. cerevisiae sequence |
| Recombinant DNA reagent | Snf7 R52E | PMID:31246173 | pET28aH6SUMOSnf7R52E | Progenitor: pRS416 Snf7R52E (PMID:2659436), Vector: pET28aH6SUMO (PMID:26670543), S. cerevisiae sequence |
| Recombinant DNA reagent | Vps24 | PMID:31246173 | pET28aH6SUMOVps24 | Progenitor: pOPTVps24 (PMID:18786397), Vector: pET28aH6SUMO (PMID:26670543), S. cerevisiae protein sequence, DNA seq optimized for Escherichia coli |
| Recombinant DNA reagent | Vps2 | PMID:31246173 | pET28aH6SUMOVps2 | Progenitor: pOPTVps2 (PMID:18786397), Vector: pET28aH6SUMO (PMID:26670543) S. cerevisiae protein sequence, DNA seq optimized for E. coli |
| Recombinant DNA reagent | Vps60 | This study | pET23d + Vps60 | His6 tagged, S. cerevisiae sequence |
| Recombinant DNA reagent | Did2 | This study | pET23d + Did2 | His6 tagged, S. cerevisiae sequence |
| Recombinant DNA reagent | Vps2RM | This study | pRS 416 Vps2RM | Progenitor: obtained from random mutagenesis of pRS416 Vps2, transformed into NBY47 strain and selected for canvavanine resistance |
| Recombinant DNA reagent | Vps2* | This study | pRS 416 Vps2* | Vps2 N21K T28A E31K, Progenitor: pRS416 Vps2RM(promoter is the same as in the pRS416 Vps2RM plasmid) |
| Recombinant DNA reagent | pCM189 Vps20 | This study | pCM189 Vps20 | CMV promoter controlling Tet-off operator |
| Recombinant DNA reagent | pCM189 Vps60 | This study | pCM189 Vps60 | CMV promoter controlling Tet-off operator |
| Recombinant DNA reagent | pCM189 Ist1 | This study | pCM189 Ist1 | CMV promoter controlling Tet-off operator |
| Recombinant DNA reagent | pCM189 Did2 | This study | pCM189 Did2 | CMV promoter controlling Tet-off operator |
| Recombinant DNA reagent | Vps2 N21K | This study | pRS 416 Vps2 N21K | WT promoter |
| Recombinant DNA reagent | Vps2 E31K | This study | pRS 416 Vps2 E31K | WT promoter |
| Recombinant DNA reagent | Vps2 T28A | This study | pRS 416 Vps2 T28A | WT promoter |
| Recombinant DNA reagent | Vps2 N21K E31K | This study | pRS 416 Vps2 N21K E31K | WT promoter |
| Recombinant DNA reagent | Vps2 N21K T28A E31K | This study | pRS 416 Vps2 N21K T28A E31K | WT promoter |
| Recombinant DNA reagent | Vps24WT | PMID:23063125 | pRS 414 Vps24 | S. cerevisiae sequence |
| Recombinant DNA reagent | Vps24E114K | This study | pCM189 Vps24 E114K | Figure 3 |
| Recombinant DNA reagent | Vps24E114K - Vps2 | This study | pCM189 Vps24 E114K - Vps2 | Figure 3 |
| Recombinant DNA reagent | Vps24-(Vps2 α5/MIM) | This study | pCM189 Vps24 -(Vps2 α5/MIM) | Figure 3 |
| Recombinant DNA reagent | Vps24E114K-(Vps2 α5/MIM) | This study | pCM189 Vps24-E114K-(Vps2 α5/MIM) | Figure 3 |
| Recombinant DNA reagent | Vps24E114K-(Vps2 MIM) | This study | pCM189 Vps24-E114K-(Vps2 MIM) | Figure 3 |
| Recombinant DNA reagent | vps24(1-56)-vps2 | This study | pRS414 vps24(1-56)-vps2 | Figure 3—figure supplement 1 |
| Recombinant DNA reagent | vps24(1-106)-vps2 | This study | pRS414 vps24(1-106)-vps2 | Figure 3—figure supplement 1 |
| Recombinant DNA reagent | vps24(1-119)-vps2 | This study | pRS414 vps24(1-119)-vps2 | Figure 3—figure supplement 1 |
| Recombinant DNA reagent | vps24(1-152)-vps2 | This study | pRS414 vps24(1-152)-vps2 | Figure 3—figure supplement 1 |
| Recombinant DNA reagent | vps24-(vps2 α5/MIM) | This study | pRS414 vps24-(vps2 α5/MIM) | Figure 3—figure supplement 1 |
| Recombinant DNA reagent | vps24E114K-(vps2 α5/MIM) | This study | pRS414 vps24E114K-(vps2 α5/MIM) | Figure 3—figure supplement 1 |
| Recombinant DNA reagent | Vps2(1-56)-Vps24 | This study | 416-Vps2(1-56)-Vps24(57-224) | Figure 3—figure supplement 3 |
| Recombinant DNA reagent | Vps2(1-99)-Vps24 | This study | 416-Vps2(1-99)-Vps24(101-224) | Figure 3—figure supplement 3 |
| Recombinant DNA reagent | Vps2(1-119)-Vps24 | This study | 416-Vps2(1-119)-Vps24(120-224) | Figure 3—figure supplement 3 |
| Recombinant DNA reagent | Vps2(1-164)-Vps24 | This study | 416-Vps2(1-164)-Vps24(164-224) | Figure 3—figure supplement 3 |
| Recombinant DNA reagent | Vps2(1-217)-Vps24 | This study | 416-Vps2(1-217)-Vps24(213-224) | Figure 3—figure supplement 3 |
| Recombinant DNA reagent | Vps2(1-56)-Vps24 | This study | pCM189 Vps2(1-56)-Vps24(57–224) | Figure 3—figure supplement 3 |
| Recombinant DNA reagent | Vps2(1-99)-Vps24 | This study | pCM189 Vps2(1-99)-Vps24(101-224) | Figure 3—figure supplement 3 |
| Recombinant DNA reagent | Vps2(1-119)-Vps24 | This study | pCM189 Vps2(1-119)-Vps24(120-224) | Figure 3—figure supplement 3 |
| Recombinant DNA reagent | Vps2(1-164)-Vps24 | This study | pCM189 Vps2(1-164)-Vps24(164-224) | Figure 3—figure supplement 3 |
| Recombinant DNA reagent | Vps2(1-217)-Vps24 | This study | pCM189 Vps2(1-217)-Vps24(213-224) | Figure 3—figure supplement 3 |
| Recombinant DNA reagent | Vps24 NG (V2 H5 MIM) | This study | pCM189 Vps24 N99G N103G (Vps2 H5 MIM) | Figure 4 |
| Recombinant DNA reagent | Vps24 NA (V2 H5 MIM) | This study | pCM189 Vps24 N99A N103A (Vps2 H5 MIM) | Figure 4 |
| Recombinant DNA reagent | Vps2 (α2/α3–24) | This study | pCM189 Vps2 (α2/α3–24) | Figure 4 |
| Recombinant DNA reagent | Vps60-GFP | PMID:18032584 | pSM11; pRS415-Vps60-GFP | |
| Recombinant DNA reagent | VPS60 | This study | pRS416-Vps60 | Figure 6 |
| Recombinant DNA reagent | snf7(1-59) | This study | pRS416-snf7(1-59)-Vps60 | Figure 6 |
| Recombinant DNA reagent | snf7(1-121) | This study | pRS416-snf7(1-121)-Vps60 | Figure 6 |
| Recombinant DNA reagent | snf7(1-155) | This study | pRS416-snf7(1-155)-Vps60 | Figure 6 |
| Recombinant DNA reagent | Snf7 R52E | PMID:31246173 | pET28aH6SUMOSnf7R52E | Progenitor: pRS416 Snf7R52E (PMID:2659436), Vector: pET28aH6SUMO (PMID:26670543), S. cerevisiae sequence |
| Recombinant DNA reagent | Snf7(vps2α5MIM) | This study | 416-Snf7(vps2α5MIM) | Figure 3—figure supplement 2 |
| Recombinant DNA reagent | Snf7***(vps2α5MIM) | This study | 416-Snf7***(vps2α5MIM) | Figure 3—figure supplement 2 |
| Recombinant DNA reagent | Snf7(vps2MIM) | This study | 416-Snf7(vps2MIM) | Figure 3—figure supplement 2 |
| Recombinant DNA reagent | Snf7***(vps2MIM) | This study | 416-Snf7***(vps2MIM) | Figure 3—figure supplement 2 |
| Recombinant DNA reagent | Snf7(Vps2α5) | This study | 416-Snf7(Vps2α5) | Figure 3—figure supplement 2 |
| Recombinant DNA reagent | Snf7***(Vps2α5) | This study | 416-Snf7***(Vps2α5) | Figure 3—figure supplement 2 |
| Antibody | anti-Vps2 (Rabbit polyclonal) | PMID:24711499 | anti-Vps2 Ab, S. cerevisiae Vps2 | (1:500) |
| Antibody | anti-GFP (Rabbit polyclonal) | Torrey Pines Biolabs | (Torrey Pines Biolabs Cat# TP401, RRID:AB_2313770) | (1:2500) |
| Antibody | anti-G6PDH (Rabbit polyclonal) | Sigma-Aldrich | (Sigma-Aldrich Cat# A9521, RRID:AB_258454) | (1: 10000) |
| Antibody | anti-PGK (Mouse monoclonal) | Thermo Fisher | (Thermo Fisher Scientific Cat# 459250, RRID:AB_2532235) | (1:4000) |
| Antibody | anti-Mouse IRdye 800/680 (Goat polyclonal) | LI-COR Biosciences | LI-COR Biosciences Cat# 926–32210, RRID:AB_621842 | (1:10,000) |
| Antibody | anti-Rabbit IRdye 800/680 (Rabbit polyclonal) | LI-COR Biosciences | LI-COR Biosciences Cat# 926–32211,RRID:AB_621843 | (1:10,000) |
| Antibody | anti-Snf7 (Rabbit polyclonal) | PMID:9606181 | anti-Snf7 Ab, S. cerevisiae Snf7 | (1:10000) |
| Antibody | anti-Vps24 (Rabbit polyclonal) | PMID:9606181 | anti-Vps24 Ab, S. cerevisiae Vps24 | (1:1000) |
| Software | Odyssey, Image Studio Lite | LI-COR Biosciences | (Image Studio Lite, RRID:SCR_013715) | |
| Software | Mafft | http://mafft.cbrc.jp/alignment/server/ | (MAFFT, RRID:SCR_011811) | |
| Software | Jalview | PMID:9151095 | (Jalview, RRID:SCR_006459) | |
| Software | Modeller | https://salilab.org/modeller/download_installation.html | (MODELLER, RRID:SCR_008395) | |
| Software | UCSF Chimera | PMID:15264254 | (UCSF Chimera, RRID:SCR_004097) | |
| Software | Fiji (ImageJ) | PMID:22743772 | (Fiji, RRID:SCR_002285) | |
| Other | Canavanine | Sigma-Aldrich | L-Canavanine sulfate, Cat.# G8772 | Stock solution 5–10 mg/mL, made in water |
| Other | Cobalt resin | Clontech | TALON Metal Affinity Resin, Cat.# 635502 | |
| Other | HiTrap Q column | GE Healthcare | HiTrap Q Sepharose FF, Cat.# 17-5053-01 | |
| Other | SD200increase | GE Healthcare | Superdex 200 Increase 10/300 GL, Cat.# 28990944 | |
| Other | EM grids | Electron Microscopy Sciences | Formvar Carbon Coated 200 Mesh Copper Grids, Cat.# FCF200-Cu | |
| Other | Ammonium molybdate | Sigma-Aldrich | Ammonium molybdate 99.98%, Cat.# 277908 | Made 2% (w/v) in water |
| Other | POPC | Avanti Polar Lipids | 16:0-18:1 PC (POPC), Cat.# 850457C | |
| Other | POPS | Avanti Polar Lipids | 16:0-18:1 PC (POPC), Cat.# 840034 | |
| Other | PI3P | Avanti Polar Lipids | 18:1 PI(3)P, Cat.# 850150 | Dissolved in 10% methanol, 90% chloroform, to make 0.25–1 mg/mL of solution |
| Other | Doxycycline | Sigma-Aldrich | Doxycycline hyclate, Cat.# D9891 | Stock solution 5 mg/mL, made in ethanol |





