Functional interdependence of the actin nucleator Cobl and Cobl-like in dendritic arbor development
Figures
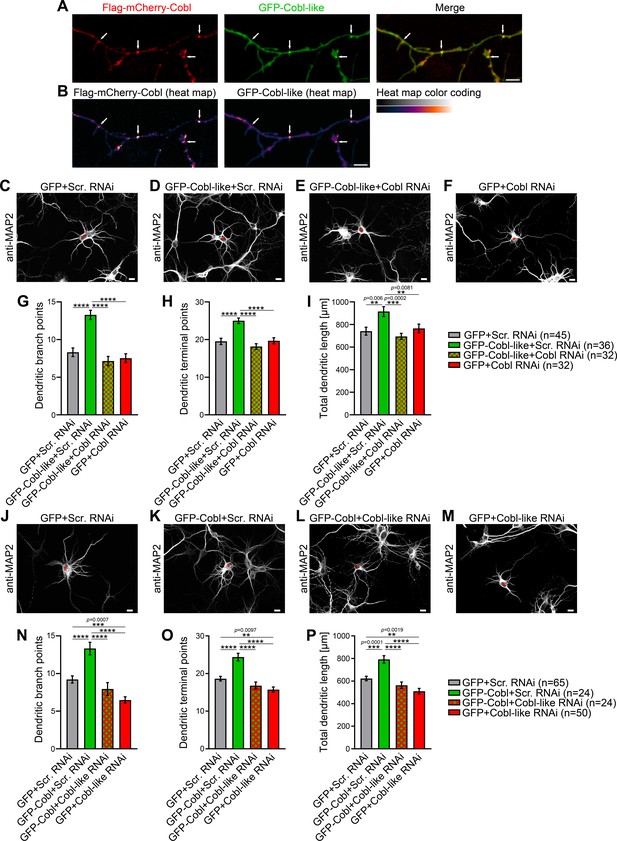
Cobl-like and the actin nucleator Cobl work at the same dendritic branching sites and show functional interdependence in dendritic arbor formation.
(A,B) Maximum intensity projections (MIPs) of GFP-Cobl-like and Flag-mCherry-Cobl in dendrites of developing hippocampal neurons (DIV6) in standard colors (A) and as heat map representation (B), respectively. Arrows, examples of putative, nascent dendritic branch induction sites with accumulations of both Cobl and Cobl-like. Bar, 5 µm. (C–F) MIPs of neurons showing the suppression of the Cobl-like gain-of-function phenotype (cotransfection at DIV4; fixation 34 hr thereafter) (D; GFP-Cobl-like+Scr. RNAi) by mCherryF-reported RNAi plasmids directed against Cobl (E; GFP-Cobl-like+Cobl RNAi) in comparison to control neurons (C; GFP+Scr. RNAi) and Cobl RNAi neurons (F; GFP+Cobl RNAi). (G–I) Quantitative determinations of indicated dendritic arborization parameters unveiling a full suppression of all Cobl-like functions in dendritic arbor formation by a lack of Cobl. (J–P) Related images (J–M) and quantitative data (N–P) of experiments revealing a functional dependence of Cobl on Cobl-like. Asterisks, transfected neurons. Bars, 10 µm. Data, mean ± SEM. One-way ANOVA+Tukey. Also see Figure 1—source data 1 and 2.
-
Figure 1—source data 1
Raw data and numerical data graphically presented in Figure 1G–I.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig1-data1-v1.xlsx
-
Figure 1—source data 2
Raw data and numerical data graphically presented in Figure 1N–P.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig1-data2-v1.xlsx

Comparison of domain structures and alignment of Cobl-like with the actin nucleator Cobl.
(A) Scheme of Cobl-like with its domains in comparison to the actin nucleator Cobl with its domains. Abbreviations: PRD, proline-rich domain (PRD next to the Cobl Homology domain is an Abp1- and PRMT2-binding site and therefore colored in light green reminiscent of the RD, that is, the Abp1-binding repeat domain in Cobl-like); WH2, WH2 domain. (B) Amino acid sequence alignment of murine Cobl-like (gi:74201419) and Cobl (gi:162135965) using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). Marked are the Cobl Homology domains of both proteins (boxed in light blue) and the ‘KRAP’ motifs (boxed in red) of both proteins. Furthermore, the single C terminal WH2 domain of Cobl-like and the three WH2 domains of Cobl are marked by orange letters. The CaM-binding region of Cobl in the Cobl Homology domain and in the C terminal CaM-binding regions in Cobl and Cobl-like are boxed in yellow, the Abp1- and PRMT2-binding PRD of Cobl is boxed in light green. The Abp1-binding repeat domain of Cobl-like is boxed in green and the proline-rich region located N terminal of the Abp1-binding area is boxed in purple. Note that although Cobl-like is considered as evolutionary ancestor of Cobl, the sequence conservation between both proteins in general is very low (*, identity; ‘:’, high similarity; ‘.’, moderate similarity). Even in the most-conserved part, the so-called Cobl Homology domain, the similarity is limited to the central core of this proposed domain and to the ‘KRAP’ motifs.
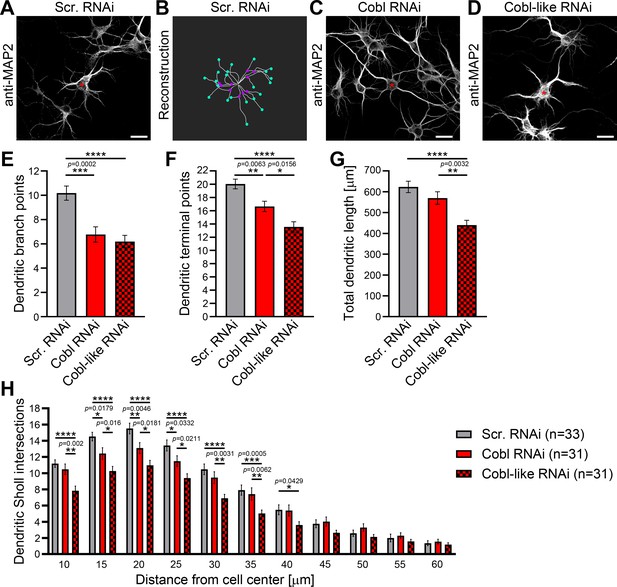
Cobl-like and the actin nucleator Cobl work at the same dendritic branching sites and largely phenocopy each other in their critical role in dendritic arborization.
(A–D) Maximum intensity projections (MIPs) of anti-MAP2 immunostained developing primary hippocampal neurons transfected as indicated at DIV4 and fixed 46 hr after transfection (A,C,D) and 2D representative of the 3D reconstruction of the morphology of the example neuron in A using IMARIS software (B). For detailed software parameters, see the Materials and methods section. Shown is the filament representing dendritic tracts (white lines) with branch points (magenta dots placed on the filament) and terminal points (turquoise dots placed on the filament) marked. Asterisks, transfected neurons. Bars, 20 µm. (E–H) Quantitative comparative Cobl-like and Cobl loss-of-function analyses of indicated dendritic parameters. Data, mean ± SEM. One-way ANOVA+Tukey (E–G) and two-way ANOVA+Bonferroni (H).
-
Figure 1—figure supplement 2—source data 1
Raw data and numerical data graphically presented in Figure 1—figure supplement 2 A-D.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig1-figsupp2-data1-v1.xlsx
-
Figure 1—figure supplement 2—source data 2
Raw data and numerical data graphically presented in Figure 1—figure supplement 2E-G.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig1-figsupp2-data2-v1.xlsx
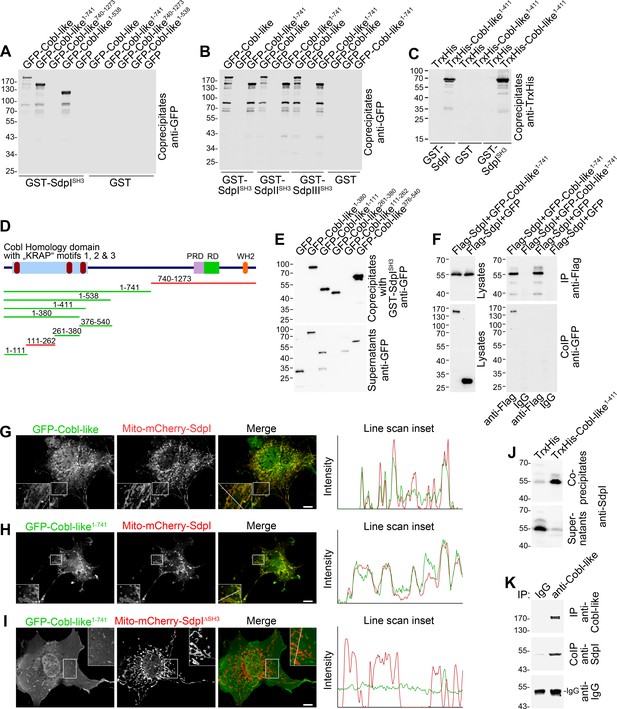
Cobl-like associates with syndapins.
(A) Coprecipitation analyses of GFP-tagged Cobl-like and deletion mutants thereof with immobilized syndapin I SH3 domain (GST-SdpISH3). (B) Related coprecipitation studies with the SH3 domains of syndapin I, syndapin II (SdpIISH3), and syndapin III (SdpIIISH3), respectively. (C) Reconstitution of the association of TrxHis-Cobl-like1-411 with purified GST-syndapin I and GST-syndapin I SH3 domain but not with GST. (D) Scheme of Cobl-like with its domains (PRD, proline-rich domain; RD, Abp1-binding repeat domain; WH2, WH2 domain) and deletion mutants used (red, not binding syndapins; green, binding). Red in the Cobl Homology domain, ‘KRAP’ motifs (not drawn to scale). (E) Coprecipitation assays with Cobl-like deletion mutants mapping Cobl-like’s syndapin binding sites. (F) Coimmunoprecipitations unveiling a specific association of GFP-Cobl-like1-741 with Flag-syndapin I. (G–I) Reconstitution and visualization of Cobl-like/syndapin I complexes in COS-7 cells using mitochondrially targeted syndapin I (G,H) as well as a mutant lacking the SH3 domain (Mito-mCherry-SdpI∆SH3) (I) with GFP-Cobl-like (G) and GFP-Cobl-like1-741 (H,I). Boxed areas are shown at higher magnification as insets. Line scans of fluorescence intensities of both channels are along the respective line indicated in the merged insets in G–I. Bars, 10 µm. (J) Coprecipitation of endogenous syndapin I from mouse brain lysates by TrxHis-Cobl-like1-411. (K) Endogenous coimmunoprecipitation of Cobl-like and syndapin I from mouse brain lysates.
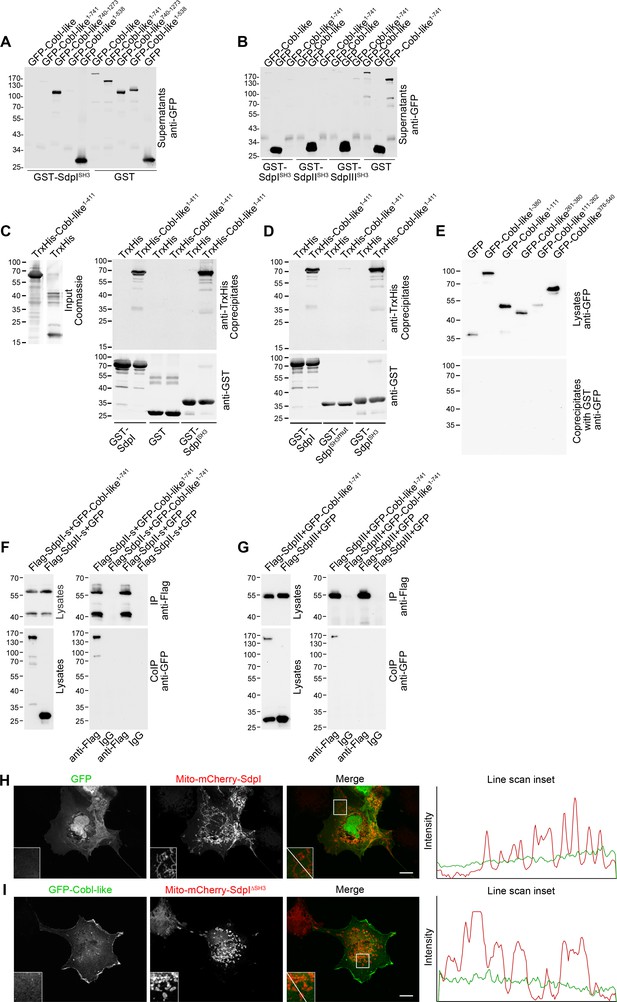
Cobl-like associates with syndapins.
(A,B) Anti-GFP immunoblotting analyses of supernatants of coprecipitation analyses of GFP-tagged Cobl-like and deletion mutants thereof with immobilized syndapin I SH3 domain (GST-SdpISH3) and GST (control) (A) and of related coprecipitation studies with the SH3 domains of syndapin I, syndapin II (SdpIISH3), and syndapin III (SdpIIISH3), respectively (B). For eluate analyses, see Figure 2A,B. (C) More complete immunoblotting analyses of the reconstitution of the association of TrxHis-Cobl-like1-411 with purified GST-syndapin I and GST-syndapin I SH3 domain but not with GST shown in Figure 2C containing a characterization of the input and both the anti-TrxHis and anti-GST immunoblotting analyses of the eluates. (D) Immunoblot analyses of a reconstitution of the association of TrxHis-Cobl-like1-411 with recombinant, purified GST-syndapin I and GST-syndapin I SH3 domain but not with a mutated syndapin I SH3 domain (P434L; SdpISH3mut). (E) Anti-GFP immunoblotting of the cell lysates and of the GST controls of the syndapin I coprecipitation assays with Cobl-like deletion mutants shown in Figure 2E. (F,G) Immunoblotting analyses of specific coimmunoprecipitations of GFP-Cobl-like1-741 but not GFP with Flag-tagged syndapin II-s (short splice variant, SdpII-s) and syndapin III (SdpIII). (H,I) Maximum intensity projections (MIPs) showing control experiments accompanying the protein complex reconstitutions between GFP-Cobl-like and GFP-Cobl-like1-741 and mitochondrially targeted syndapin I (Mito-mCherry-SdpI) shown in Figure 2G–I. (H) GFP is not recruited to Mito-mCherry-SdpI-enriched sites. (I) GFP-Cobl-like is not recruited to mitochondria when a Mito-mCherry-SdpI lacking the SH3 domain (Mito-mCherry-SdpI∆SH3) is coexpressed. Boxed areas are shown at higher magnification as insets. Line scans of fluorescence intensities of both channels are along the respective line indicated in the merged insets of H and I. Bars, 10 µm.
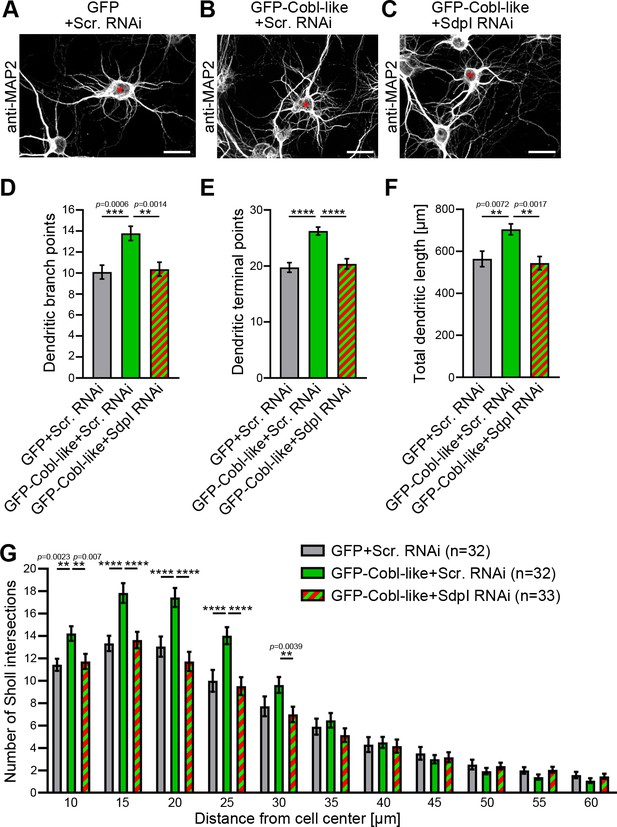
Cobl-like functions in dendritic arbor formation rely on syndapin I.
(A–C) Maximum intensity projections (MIPs) of DIV5.5 neurons transfected as indicated. Asterisks, transfected neurons. Bars, 20 µm. (D–G) Quantitative determinations of key dendritic arborization aspects promoted by Cobl-like for their dependence on syndapin I (cotransfection at DIV4; fixation 34 hr thereafter). Data, mean ± SEM. One-way ANOVA+Tukey (D–F) and two-way ANOVA+Bonferroni (G). Also see Figure 3—source data 1 and 2.
-
Figure 3—source data 1
Raw data and numerical data graphically presented in Figure 3D–F.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Raw data and numerical data graphically presented in Figure 3G.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig3-data2-v1.xlsx
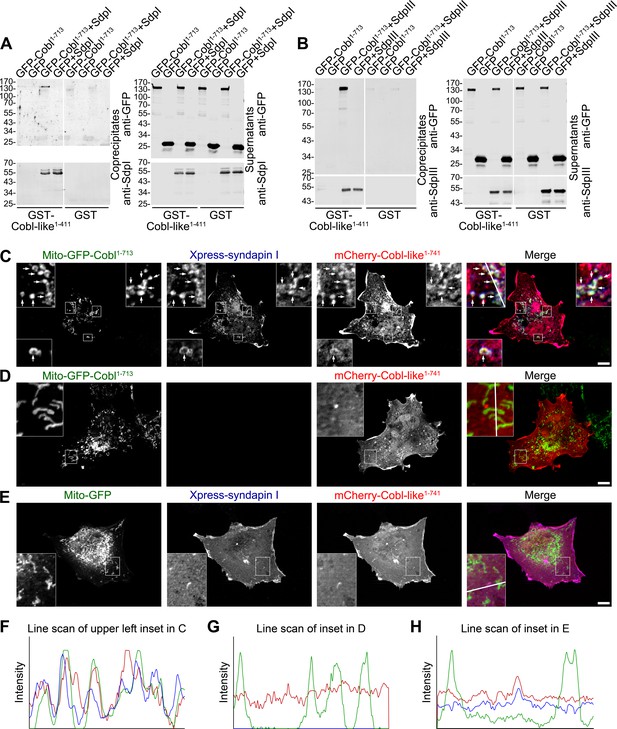
Cobl-like is physically linked to Cobl via syndapin I acting as a bridging component.
(A,B) Coprecipitation analyses unveiling specific and syndapin-dependent formation of complexes composed of immobilized GST-Cobl-like, syndapin I (A) and syndapin III (B), respectively, as well as GFP-Cobl1-713. White lines indicate omitted blot lanes. (C–E) Reconstitution and visualization of Cobl-like/syndapin I/Cobl protein complexes in COS-7 cells. Mito-GFP-Cobl1-713 (C,D) but not Mito-GFP (E) recruited mCherry-Cobl-like1-741 in the presence of Xpress-syndapin I (C) but not in its absence (D). Boxes in C–E areas presented as magnified insets (C,D, fourfold; E, threefold). Arrows, examples of colocalization of all three channels. Boxed areas are shown at higher magnification as insets. (F–H) Line scans of fluorescence intensities of all three channels are along the respective line indicated in the insets of C–E. Bars, 10 µm.
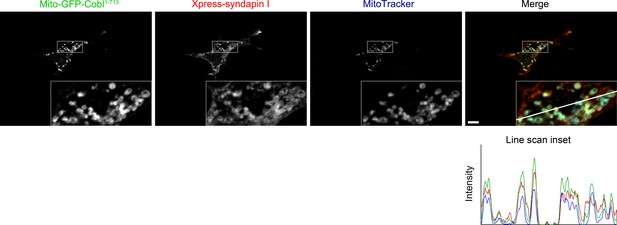
Recruitment of syndapin I to mitochondrial surfaces in intact cells by Mito-GFP-Cobl1-713.
Reconstitution and visualization of Cobl/syndapin I protein complexes inside of intact COS-7 cells. Mitochondrially targeted Cobl1-713 (green in merge) recruits Xpress-tagged syndapin I (red in merge; anti-Xpress immunolabeling) to mitochondria (blue in merge; visualized by MitoTracker). Boxes mark the area presented as magnified inset (fourfold enlargement). Colocalization of all three channels appears in whitish colors (white, beige, turquoise, rosé) in the merge. The line scan of fluorescence intensities of all three channels is along the line indicated in the merged inset. Bar, 10 µm.
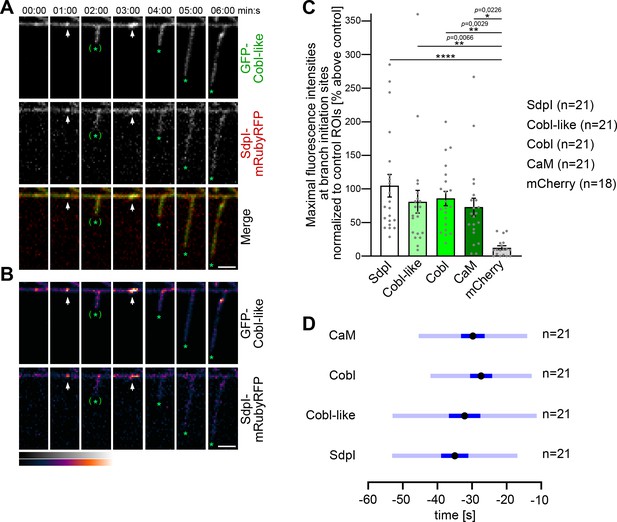
Cobl-like and syndapin I coincide at nascent dendritic branch sites.
(A) Maximum intensity projections (MIPs) of individual frames of a 3D time-lapse recording of a dendrite segment of a DIV7 rat hippocampal neuron coexpressing GFP-Cobl-like and syndapin I (SdpI)-mRubyRFP. Arrows, GFP-Cobl-like and syndapin I enrichments prior to protrusion initiation from these dendritic sites; *, tips of growing dendritic protrusions; (*), abandoned protrusions. (B) Heat map representations. Bars, 2.5 µm. (C) Quantitation of maximal intensities of fluorescent syndapin I, Cobl-like, Cobl and CaM fusion proteins as well as of mCherry as control at dendritic branch induction sites prior to protrusion formation (time frame: the six 10 s frames prior to protrusion start) in relation to a control ROI at the same dendrite at a position neighboring the branch induction site (shown as % above this control ROI). Data, mean ± SEM. Bar/dot plot overlay of individual data points averaged. (D) Temporal analyses of the maximal fluorescence intensities of syndapin I, Cobl-like, Cobl, and CaM occurring at dendritic branch induction sites prior to protrusion formation. Data, mean (black dot) ± SD (light blue) and ± SEM (dark blue). One-way-ANOVA (C). Also see Figure 5—source data 1.
-
Figure 5—source data 1
Raw data and numerical data graphically presented in Figure 5C,D.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig5-data1-v1.xlsx
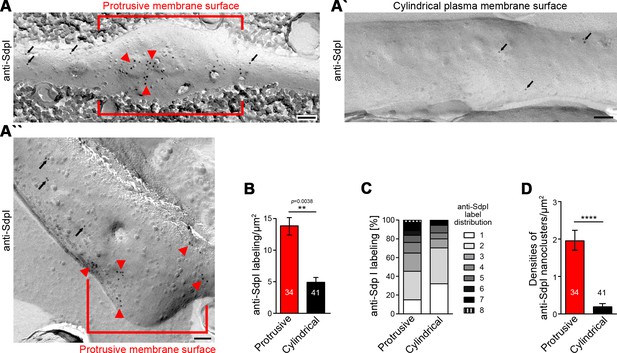
Syndapin I nanoclusters are enriched at sites of dendritic protrusion.
(A,A’,A’’) Transmission electron microscopy (TEM) images of anti-syndapin I immunogold-labeled freeze-fracture replica of developing neurons (DIV7). Red lines highlight membrane topologies protruding from regular cylindrical topology. Arrowheads, abundant and clustered anti-syndapin I immunogold labeling (10 nm) at protrusive sites. Arrows, sparse and rarely clustered anti-syndapin immunogold labeling at regular, cylindrical membrane structures. Bars, 200 nm. (B) Quantitative evaluations of anti-syndapin I labeling densities at protrusive and cylindrical membrane topologies. (C) Quantitative analysis of the relative abundance of differently clustered syndapin I labels (ROIs, 35 nm radius). In total, 335 (protrusive) and 130 (cylindrical) labels were evaluated. (D) Quantitative analysis of the density of anti-syndapin I nanoclusters (≥3 anti-syndapin I immunogold labels/ROI) at regular cylindrical membrane surfaces and at those with protrusive topology. Data (B,D), mean ± SEM. One-way ANOVA (B); two-tailed Student’s t-test (D). Also see Figure 6—source data 1.
-
Figure 6—source data 1
Raw data and numerical data graphically presented in Figure 6B–D.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig6-data1-v1.xlsx
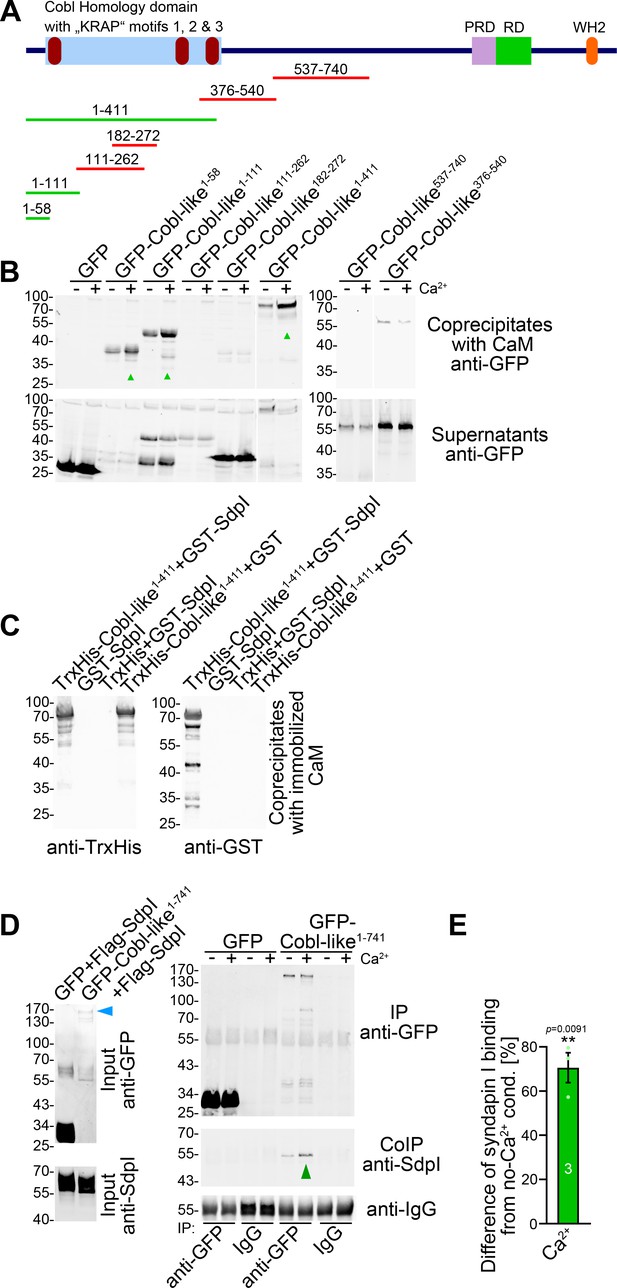
Ca2+/CaM associates with the N terminus of Cobl-like and positively regulates Cobl-like’s syndapin I association.
(A) Scheme of Cobl-like and deletion mutants used for CaM-binding studies (B) (red, no Ca2+-dependent binding; green, Ca2+-dependent binding). (B) Coprecipitations with immobilized CaM in presence (500 µM) and absence of Ca2+ and different Cobl-like deletion mutants. Green arrowheads, increased CaM interactions in the presence of Ca2+. White lines, lanes omitted from blots. (C) Coprecipitation analyses with immobilized CaM and purified TrxHis-Cobl-like1-411 and GST-syndapin I (GST-SdpI) showing direct and simultaneous interactions of Cobl-like1-411 with both CaM and syndapin I. (D,E) Quantitative coimmunoprecipitation analyses demonstrating that Ca2+/CaM signaling leads to increased syndapin I coimmunoprecipitation with Cobl-like1-741. Blue arrowhead, position of the only faintly detected GFP-Cobl-like1-741 in the lysates (D). Green arrowhead, increase of coimmunoprecipitated syndapin I (D). (E) Anti-syndapin I signal per immunoprecipitated Cobl-like (expressed as change from conditions without Ca2+). Data, bar/dot plot overlays with mean ± SEM. Unpaired Student’s t-test. Also see Figure 7—source data 1.
-
Figure 7—source data 1
Raw data and numerical data graphically presented in Figure 7E.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig7-data1-v1.xlsx
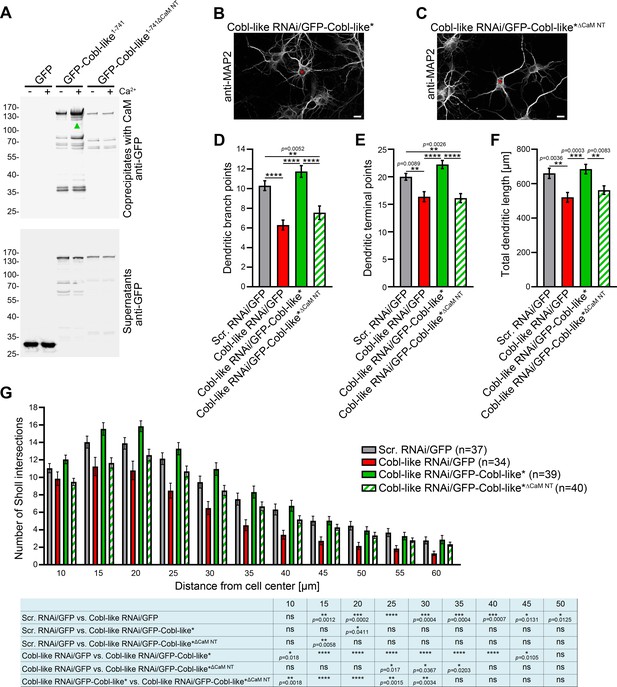
The N terminal CaM-binding site of Cobl-like is indispensable for all critical functions of Cobl-like in dendritic arbor formation.
(A) Coprecipitation analyses of Cobl-like1-741, Cobl-like1-741∆CaM NT (∆11–45), and GFP with immobilized CaM in Ca2+ presence and absence. Arrowhead, increased CaM interaction of Cobl-like1-741 upon Ca2+ (disrupted in Cobl-like1-741∆CaM NT). (B,C) Functional analyses in primary hippocampal neurons (transfection at DIV4; fixation 37 hr thereafter) unveiling that an RNAi-insensitive (*) Cobl-like mutant lacking the N terminal CaM-binding site (GFP-Cobl-like*∆CaM NT) failed to rescue the Cobl-like loss-of-function phenotypes. Red asterisks, transfected neurons (transfection, DIV4; analyses, DIV5.5). Bars, 10 µm. (D–G) Quantitative evaluations of indicated dendritic parameters. Data, mean ± SEM. One-way ANOVA+Tukey (D–F); two-way ANOVA+Bonferroni (G). Also see Figure 8—source data 1 and 2.
-
Figure 8—source data 1
Raw data and numerical data graphically presented in Figure 8D–F.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig8-data1-v1.xlsx
-
Figure 8—source data 2
Raw data and numerical data graphically presented in Figure 8G.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig8-data2-v1.xlsx
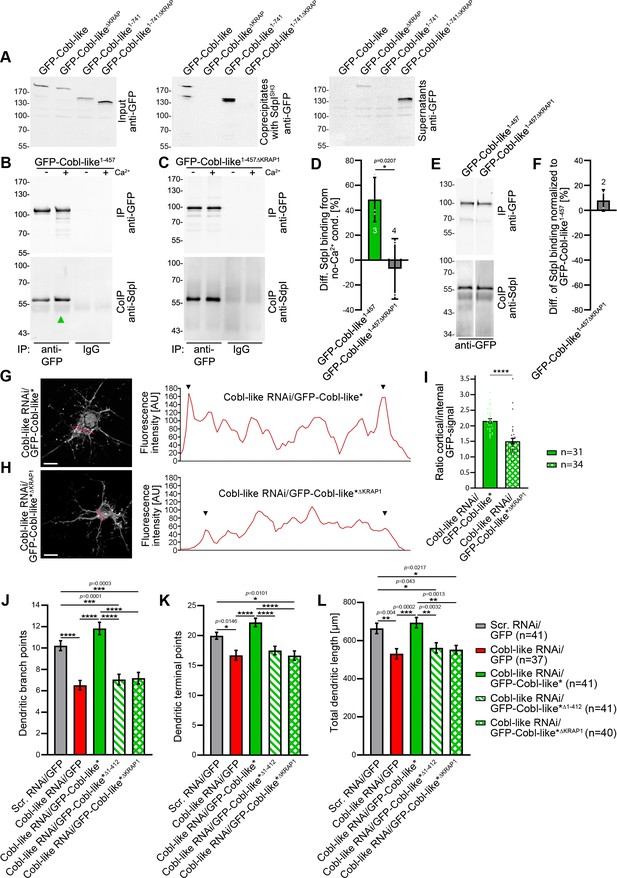
Ca2+/CaM signaling exclusively promotes the syndapin I association with the first of the three ‘KRAP’ motifs and this single Ca2+/CaM-regulated motif is crucial for Cobl-like’s functions.
(A) Coprecipitations with immobilized syndapin I SH3 domain (SdpISH3) and Cobl-like and ∆KRAP mutants thereof. (B,C) Quantitative coimmunoprecipitation analyses with GFP-Cobl-like1-457 (B) in comparison to a corresponding mutant solely lacking the first ‘KRAP’ motif (GFP-Cobl-like1-457∆KRAP1) in the presence and absence of Ca2+, respectively (C). Arrowhead, increase of coimmunoprecipitated Flag-syndapin I with GFP-Cobl-like1-457 upon Ca2+. (D) Quantitation of anti-syndapin I coimmunoprecipitation upon Ca2+ presence normalized to immunoprecipitated GFP-Cobl-like1-457 and GFP-Cobl-like1-457∆KRAP1, respectively (as deviation from conditions without Ca2+). (E,F) Side-by-side comparison of syndapin I coimmunoprecipitations with GFP-Cobl-like1-457 and GFP-Cobl-like1-457∆KRAP1 (E) and quantitative analysis thereof (F). White line, lanes omitted from blot. (G,H) Images of Apotome sections showing the cortical localization of GFP-Cobl-like expressed in Cobl-like-deficient background (Cobl-like RNAi/GFP-Cobl-like*) in primary hippocampal neurons transfected at DIV4 and imaged 37 hr later (G) compared to the subcellular distribution of GFP-Cobl-like*∆KRAP1 in the same background (H). Lines indicate positions of line scans shown. Bars, 10 µm. (I) Quantitative assessment of cortical GFP intensities (marked with arrowheads in the line scans) normalized to the GFP intensity of an internal ROI in the same cell. (J–K) Functional analyses of the importance of Cobl-like’s CaM-regulated syndapin I binding site (KRAP1) by loss-of-function rescue experiments evaluating the indicated dendritic arbor parameters of developing neurons (transfection, DIV4; analysis, DIV5.5). Note that neither a Cobl-like mutant lacking the entire N terminal part (GFP-Cobl-like*∆1-412) nor GFP-Cobl-like*∆KRAP1 was able to rescue Cobl-like’s loss-of-function phenotypes. Data, bar/dot plot overlays with mean ± SEM (D,I) and mean ± absolute error (F) as well as bar plots with mean ± SEM (J–L). Unpaired Student’s t-test (D,F,I); one-way ANOVA+Tukey (J–L). Also see Figure 9—source data 1–4.
-
Figure 9—source data 1
Raw data and numerical data graphically presented in Figure 9D.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig9-data1-v1.xlsx
-
Figure 9—source data 2
Raw data and numerical data graphically presented in Figure 9F.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig9-data2-v1.xlsx
-
Figure 9—source data 3
Raw data and numerical data graphically presented in Figure 9I.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig9-data3-v1.xlsx
-
Figure 9—source data 4
Raw data and numerical data graphically presented in Figure 9J–L.
- https://cdn.elifesciences.org/articles/67718/elife-67718-fig9-data4-v1.xlsx
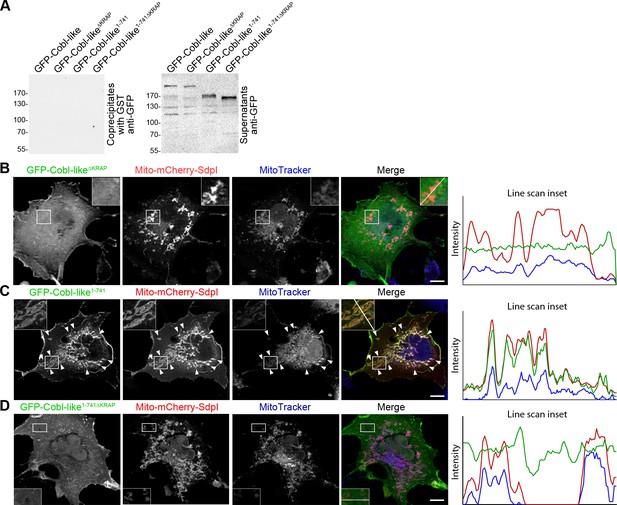
The three ‘KRAP’ motifs are critical for Cobl-like’s association with syndapin I.
(A) Anti-GFP immunoblotting of control experiments of the coprecipitation analyses shown in Figure 10A demonstrating that neither Cobl-like nor any of the Cobl-like mutants associated with GST. (B–D) Maximum intensity projections (MIPs) of COS-7 cells transfected with Mito-mCherry-syndapin I and GFP-Cobl-like∆KRAP (B), Mito-mCherry-syndapin I and GFP-Cobl-like1-741 (C), and a GFP-Cobl-like mutant lacking the regions with the three ‘KRAP’ motifs (GFP-Cobl-like1-741∆KRAP) (D), respectively. Note the successful recruitment of GFP-Cobl-like1-741 to syndapin I-decorated mitochondria (C; colocalization of all three channels shown appears white in merge; examples are marked by arrowheads), whereas GFP-Cobl-like∆KRAP and GFP-Cobl-like1-741∆KRAP were not recruited to mitochondria (colocalization of only Mito-mCherry-syndapin I [red in merge] and MitoTracker [blue in merge] appears purple in merge; B,D). Boxed areas are shown at higher magnification as insets. Line scans of fluorescence intensities of all three channels are shown along the respective line indicated in the merged insets of B–D. Bars, 10 µm.
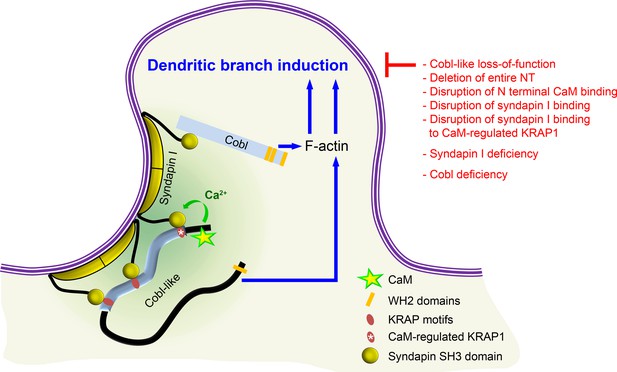
Model depicting how Cobl and Cobl-like functions in dendritic branch initiation are joined, coordinated, and controlled.
Cobl and Cobl-like functions are not only both critical for dendritic branch formation but both factors promoting the formation of actin filaments were found to act in an interdependent manner. The underlying mechanisms of coordination and control are depicted and include physical linkage of Cobl and Cobl-like by syndapin I forming dimers and multimeric clusters at the convexly bent membrane areas at the base of nascent branch sites. The newly identified interaction of syndapin I with Cobl-like is mediated by three independent KRAP motifs (red), the most N terminal of which (marked by white asterisk) is regulated by a newly discovered CaM association to Cobl-like’s N terminus. All mechanistic aspects unveiled in this study are depicted in detail and the corresponding functional evaluations conducted are listed in brief. The WH2 domains of Cobl and Cobl-like are shown to indicate the C terminal domains of both proteins and their cytoskeletal functions.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Cobl-like (Cobll1) | Izadi et al., 2018 | AK144943.1, GI: 74201418 | |
| Gene (Mus musculus) | Cordon-Bleu (Cobl) | Ahuja et al., 2007 | NM_172496.3, GI: 162135965 | The common abbreviation of Cordon-Bleu is Cobl |
| Gene (Rattus norvegicus) | Syndapin I (Pacsin1) | Braun et al., 2005 | AF104402.1, GI: 4324451 | Syndapin is used as gene name in most model organisms, such as rat, worms, flies |
| Gene (Rattus norvegicus) | Calmodulin (Calm1) | SenGupta et al., 1987 | M19312.1, GI: 203255 | The common abbreviation of calmodulin is CaM |
| Strain, background (Escherichia coli) | E. coli commercial strain BL21-CodonPlus(DE3)-RIPL | Agilent | Cat#230280 | |
| Strain, background (Escherichia coli) | E. coli commercial strain XL10-Gold | Agilent | Cat#200314 | |
| Cell line (African green monkey) | COS-7 | Cell Lines Services GmbH | RRID:CVCL_0224 | |
| Cell line (human) | HEK293 | Cell Lines Services GmbH | RRID:CVCL_0045 | |
| Biological sample (Rattus norvegicus) | Primary hippocampal neurons (Wistar rat; Crl:WI; mixed sex) | Charles River | RRID:RGD_68115 | Primary neurons isolated from E18 rat embryos (sex undetermined) |
| Biological sample (Mus musculus) | Isolated brains (Mouse strain C57BL/6J, female) | Jackson Labs | RRID:IMSR_JAX:000664 | Brain material processed for protein biochemical examinations |
| Antibody | Anti-Cobl-like (Rabbit polyclonal) | Izadi et al., 2018 | N/A | WB (1:1000) |
| Antibody | Anti-syndapin I (Guinea pig polyclonal) | Braun et al., 2005 | N/A | WB (1:500) EM (1:50) |
| Antibody | Anti-syndapin III (Guinea pig polyclonal) | Koch et al., 2011 | N/A | WB (1:500) |
| Antibody | Anti-TrxHis (Rabbit polyclonal) | This paper | N/A | WB (1:1000) |
| Antibody | Anti-GST (Rabbit polyclonal) | Qualmann and Kelly, 2000 | N/A | WB (1:1000) |
| Antibody | Anti-TrxHis (Guinea pig polyclonal) | Schwintzer et al., 2011 | N/A | WB (1:2000) |
| Antibody | Anti-GST (Guinea pig polyclonal) | Braun et al., 2005 | N/A | WB (1:1000) |
| Antibody | Anti-GFP (ab290) (Rabbit polyclonal) | Abcam | Cat#ab290 RRID:AB_303395 | WB (1:2000) |
| Antibody | Anti-GFP (JL-8) (Mouse monoclonal) | Clontech | Cat#632380 RRID:AB_10013427 | WB (1:4000) |
| Antibody | Anti-Flag antibody (M2) (Mouse monoclonal) | Sigma-Aldrich | Cat#F3165 RRID:AB_259529 | WB (1:500) |
| Antibody | Anti-MAP2 (HM-2) (Mouse monoclonal) | Sigma-Aldrich | Cat#M4403 RRID:AB_477193 | IF (1:500) |
| Antibody | Anti-Flag antibody (Rabbit polyclonal) | Sigma-Aldrich | Cat#F7425 RRID:AB_439687 | WB (1:1000) |
| Antibody | Anti-Xpress antibody (Mouse monoclonal) | Invitrogen | Cat#R910-25; RRID:AB_2556552 | IF (1:500) |
| Antibody | Alexa Fluor488-labeled goat anti-guinea pig (Goat polyclonal) | Molecular Probes | Cat#A-11073 RRID:AB_142018 | IF (1:1000) |
| Antibody | Alexa Fluor568-labeled goat anti-guinea pig (Goat polyclonal) | Molecular Probes | Cat#A-11075 RRID:AB_141954 | IF (1:1000) |
| Antibody | Alexa Fluor488-labeled donkey anti-mouse (Donkey polyclonal) | Molecular Probes | Cat#A-21202 RRID:AB_141607 | IF (1:1000) |
| Antibody | Alexa Fluor568-labeled donkey anti-mouse (Donkey polyclonal) | Molecular Probes | Cat#A10037 RRID:AB_2534013 | IF (1:1000) |
| Antibody | Alexa Fluor647-labeled goat anti-mouse (Goat polyclonal) | Molecular Probes | Cat#A-21236 RRID:AB_141725 | IF (1:1000) |
| Antibody | Alexa Fluor488-labeled donkey anti-rabbit (Donkey polyclonal) | Molecular Probes | Cat#A-21206 RRID:AB_141708 | IF (1:1000) |
| Antibody | Alexa Fluor568-labeled goat anti-rabbit (Goat polyclonal) | Molecular Probes | Cat#A-11036 RRID:AB_143011 | IF (1:1000) |
| Antibody | Alexa Fluor647-labeled goat anti-rabbit (Goat polyclonal) | Molecular Probes | Cat#A-21245 RRID:AB_141775 | IF (1:1000) |
| Antibody | Alexa Fluor680-labeled goat-anti-rabbit (Goat polyclonal) | Molecular Probes | Cat#A-21109 RRID:AB_2535758 | WB (1:10000) |
| Antibody | Alexa Fluor680-labeled goat-anti-mouse (Goat polyclonal) | Molecular Probes | Cat#35519 RRID:AB_1965956 | WB (1:10000) |
| Antibody | DyLight800-conjugated goat anti-rabbit (Goat polyclonal) | Thermo Fisher Scientific | Cat#SA5-35571 RRID:AB_2556775 | WB (1:10000) |
| Antibody | DyLight800-conjugated goat anti-mouse (Goat polyclonal) | Thermo Fisher Scientific | Cat#SA5-35521 RRID:AB_2556774 | WB (1:10000) |
| Antibody | IRDye680-conjugated donkey anti-guinea pig (Donkey polyclonal) | LI-COR Bioscience | Cat#926–68077 RRID:AB_10956079 | WB (1:10000) |
| Antibody | IRDye800-conjugated donkey anti-guinea pig (Donkey polyclonal) | LI-COR Bioscience | Cat#926–32411 RRID:AB_1850024 | WB (1:10000) |
| Antibody | Peroxidase-AffiniPure donkey anti-rabbit antibody (Donkey polyclonal) | Jackson ImmunoResearch Labs | Cat#711-035-152 RRID:AB_10015282 | WB (1:5000) |
| Antibody | Peroxidase-AffiniPure goat anti-guinea pig antibody (Goat polyclonal) | Jackson ImmunoResearch Labs | Cat#106-036-003 RRID:AB_2337405 | WB (1:5000) |
| Antibody | Peroxidase-goat F(ab')2 anti-mouse (Goat polyclonal) | Dianova | Cat#115-036-003 RRID:AB_2617176 | WB (1:5000) |
| Antibody | Goat anti-guinea pig IgG 10 nm gold (Goat polyclonal) | BBI Solutions | Cat#EM.GAG10 RRID:AB_2892072 | EM (1:50) |
| Recombinant DNA reagent | Flag-mCherry Cobl (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like (Plasmid) | Izadi et al., 2018 | N/A | |
| Recombinant DNA reagent | Scr. RNAi in pRNAT-H1.1 (Plasmid) | Pinyol et al., 2007 | N/A | |
| Recombinant DNA reagent | Scr. RNAi in pRNAT-mCherryF (Plasmid) | Schneider et al., 2014 | N/A | |
| Recombinant DNA reagent | Cobl-RNAi in pRNAT-mCherryF (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | Cobl-like RNAi (#1) in pRNAT-H1.1 (Plasmid) | Izadi et al., 2018 | N/A | |
| Recombinant DNA reagent | Cobl-like RNAi (#1) in pRNAT-mCherryF (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl (Plasmid) | Hou et al., 2015 | N/A | |
| Recombinant DNA reagent | GFP-Cobl1-713 (Plasmid) | Hou et al., 2015 | N/A | |
| Recombinant DNA reagent | Mito-GFP-Cobl1-713 (Plasmid) | This paper | N/A | |
| Recombinant DNA reagent | GFP-Cobl-like1-741 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like740-1273 (Plasmid) | Izadi et al., 2018 | N/A | |
| Recombinant DNA reagent | GFP-Cobl-like1-538 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like1-411 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like1-380 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like376-540 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like261-380 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like111-262 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like1-111 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like537-740 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like182-272 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like1-58 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | Cobl-like RNAi/GFP-Cobl-like* in pRNAT H1.1 (Plasmid) | Izadi et al., 2018 | N/A | |
| Recombinant DNA reagent | Cobl-like RNAi/GFP-Cobl-like*∆CaM NT in pRNAT H1.1 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | mCherry-Cobl-like1-711 | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like1-741∆CaM NT (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like∆KRAP (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like1-741∆KRAP (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like1-457∆KRAP1 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-Cobl-like1-457 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | Cobl-like RNAi/GFP-Cobl-like*∆KRAP1 in pRNAT H1.1 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | Cobl-like RNAi/GFP-Cobl-like*∆1-412 in pRNAT H1.1 (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | Flag-syndapin I (SdpI) (Plasmid) | Qualmann and Kelly, 2000 | N/A | |
| Recombinant DNA reagent | Flag-syndapin II-s (SdpII) (Plasmid) | Dharmalingam et al., 2009 | N/A | |
| Recombinant DNA reagent | Flag-syndapin III (SdpIII) (Plasmid) | Braun et al., 2005 | N/A | |
| Recombinant DNA reagent | Xpress-syndapin I (SdpI) (Plasmid) | Qualmann et al., 1999 | N/A | |
| Recombinant DNA reagent | Syndapin I–mRubyRFP (Plasmid) | This paper | N/A | See Materials and methods |
| Recombinant DNA reagent | GFP-syndapin I (Plasmid) | Kessels and Qualmann, 2006 | N/A | |
| Recombinant DNA reagent | Mito-mCherry-SdpI (Plasmid) | Kessels and Qualmann, 2002 | N/A | |
| Recombinant DNA reagent | Mito-mCherry-SdpI∆SH3 (Plasmid) | Braun et al., 2005 | N/A | |
| Recombinant DNA reagent | Mito-mCherry (Plasmid) | Dharmalingam et al., 2009 | N/A | |
| Recombinant DNA reagent | SdpI-RNAi in pRNAT-mCherryF (Plasmid) | Dharmalingam et al., 2009 Schneider et al., 2014 | N/A | |
| Recombinant DNA reagent | GFP-CaM (Plasmid) | This paper | N/A | See Materials and methods |
| Sequence-based reagent | Cobl-like aa1 fw | This paper | PCR primer | 5’-AATTAGATCTATGGACCGCAGCGTCCCCGATCC-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa261 fw | This paper | PCR primer | 5’-AAAGATCTGATATCAGCAGAGAG-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa537 fw | This paper | PCR primer | 5’-AAAGATCTAAGGATCCTGATTCAGC-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa740 fw | This paper | PCR primer | 5’-GCCTCAAGAGAATTCAGG-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa376 fw | This paper | PCR primer | fw: 5’-TTGAATTCTTAAACCATGATCGCTTC-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa182 fw | This paper | PCR primer | 5’- TTAGATCTCCTACACCTATAATC-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa457 rv | This paper | PCR primer | 5’- AACTCGAGCCCGGGACCAAGGGAGC-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa741 rv | This paper | PCR primer | 5’-TCCTGAATTCTCTTGAGG-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa540 rv | This paper | PCR primer | 5’-TTCTCGAGTTAATCAGGATCCTTCTC-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa411 rv | This paper | PCR primer | 5’-GCAAGCTTGGTTTTCGAAGGTGG-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa272 rv | This paper | PCR primer | 5’-AAGAATTCTCAGTTGTGTGATATTTG-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa380 rv | This paper | PCR primer | 5’-TTGAATTCGAAGCGATCATGGTG-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like∆CaM NT aa1-10+46–51 fw | This paper | PCR primer | 5’-AAAGATCTATGGACCGCAGCGT CCCGGATCCCGTACCCAAGAATCAC AAATTCCTG-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa413 fw | This paper | PCR primer | 5’-TTAAGCTTCTGGCTCAGAC TGATG-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa58 rv | This paper | PCR primer | 5’-TTAAGCTTGCTCTGACAAATATG-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa70 fw | This paper | PCR primer | 5’-TTAAGCTTGCCGAGACGAAGGGC-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa333 rv | This paper | PCR primer | 5’-TTAAGCTTTGCATCCGAGGGC-3’ (see Materials and methods) |
| Sequence-based reagent | Cobl-like aa411 rv Sal I | This paper | PCR primer | 5’-GGGTCGACGGTTTTCGAAGGTGG-3’ (see Materials and methods) |
| Recombinant protein | TrxHis | Hou et al., 2015 | N/A | |
| Recombinant protein | TrxHis-Cobl-like1-411 | This paper | N/A | |
| Recombinant protein | GST-Cobl-like1-411 | This paper | N/A | |
| Recombinant protein | GST-SdpISH3 | Qualmann et al., 1999 | N/A | |
| Recombinant protein | GST-SdpIISH3 | Qualmann and Kelly, 2000 | N/A | |
| Recombinant protein | GST-SdpIIISH3 | Seemann et al., 2017 | N/A | |
| Recombinant protein | GST-SdpI | Qualmann et al., 1999 | N/A | |
| Recombinant protein | GST-SdpISH3mut | Qualmann and Kelly, 2000 | N/A | |
| Commercial assay or kit | NucleoSpin Plasmid | Macherey-Nagel | Cat#740588.50 | |
| Commercial assay or kit | NucleoBond Xtra Midi | Macherey-Nagel | Cat#740410.50 | |
| Commercial assay or kit | Lipofectamine 2000 transfection reagent | Invitrogen | Cat#11668019 | |
| Commercial assay or kit | Turbofect transfection reagents | Thermo Fisher Scientific | Cat#R0532 | |
| Commercial assay or kit | Calmodulin Sepharose 4B | GE Healthcare | Cat#GE17-0529-01 | |
| Commercial assay or kit | PreScission protease | GE Healthcare | Cat#27-0843-01 | |
| Chemical compound, drug | MitoTracker Deep Red Alexa Fluor633 | Molecular Probes | Cat#M22426 | |
| Software, algorithm | ZEN2012 | Zeiss | RRID:SCR_013672 | |
| Software, algorithm | Prism5, Prism6 | GraphPad Prism | RRID:SCR_002798 | |
| Software, algorithm | ImageJ | Other | RRID:SCR_003070 | Open source software |
| Software, algorithm | IMARIS 7.6 | Bitplane | RRID:SCR_007370 | |
| Software, algorithm | Adobe Photoshop | Adobe | RRID:SCR_014199 |





