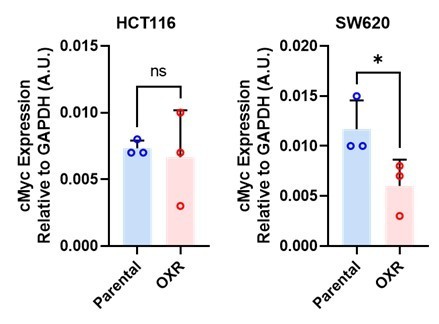
Oxaliplatin resistance in colorectal cancer enhances TRAIL sensitivity via death receptor 4 upregulation and lipid raft localization
Figures
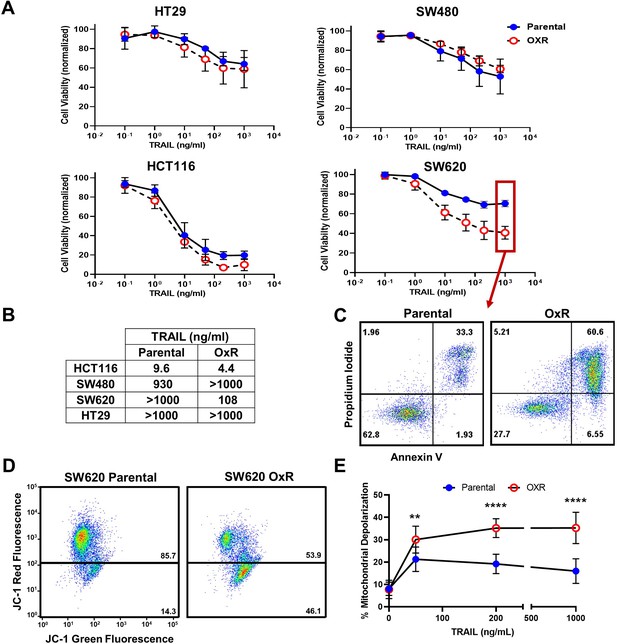
Oxaliplatin-resistant (OxR) colorectal cancer (CRC) cell lines exhibit enhanced sensitization to TRAIL-mediated apoptosis via the intrinsic pathway and mitochondrial permeabilization.
(A) Oxaliplatin-resistant SW620, SW480, HCT116, and HT29 colon cancer cell lines demonstrate similar or enhanced sensitivity to TRAIL compared to their parental counterparts after 24 hr of treatment. N = 3 (biological replicates); n = 9 (technical replicates). (B) IC50 values were calculated using a variable slope four-parameter nonlinear regression. (C) Representative Annexin-V/PI flow plots comparing SW620 parental and OxR cell viability after 24 hr of treatment with 1000 ng/ml TRAIL. The four quadrants represent viable cells (bottom left), early apoptosis (bottom right), necrosis (top left), and late apoptosis (top right). (D) Representative flow plots of JC-1 assay after treatment with 1000 ng/ml of TRAIL. Mitochondrial depolarization is evidenced by decreased red fluorescence and increased green fluorescence. (E) Mitochondrial depolarization as a function of TRAIL concentration for SW620 parental and OxR cell lines. N = 3 (n = 9). For all graphs, data are presented as mean ± SD. **p<0.01; ****p<0.0001 (unpaired two-tailed t-test).
-
Figure 1—source data 1
Raw viable cell counts from Annexin-V/PI assays and percent depolarized mitochondria in SW620 cells (panels A and E).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig1-data1-v1.xlsx
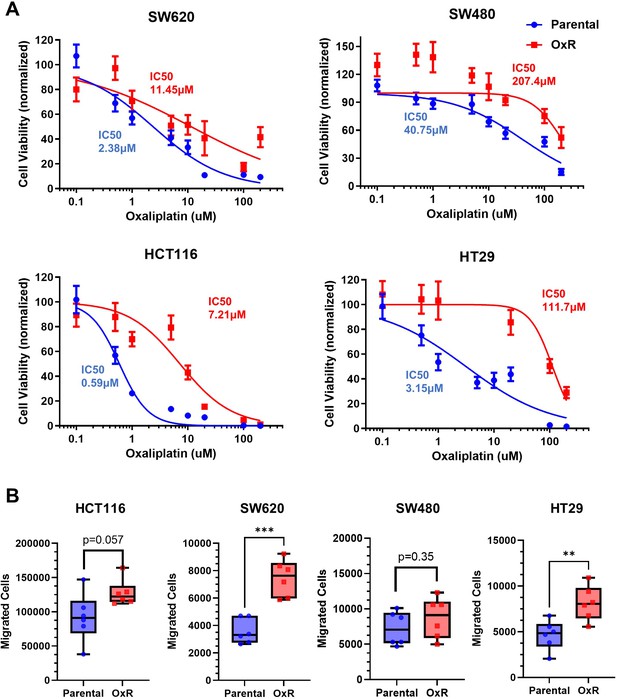
Oxaliplatin-resistant colorectal cancer (CRC) cells retain their increasingly chemoresistant and invasive phenotypes in culture.
(A) SW620, SW480, HCT116, and HT29 cells were treated with various concentrations of oxaliplatin for 72 hr and cell viability was measured using an MTT assay. IC50 values were calculated using a variable slope four-parameter nonlinear regression. Data are presented as mean ± SEM. N = 2 (n = 12). (B) Counts of successfully invasive cells after a 4-day Transwell assay with an initial seeding of 200,000 cells. N = 2 (n = 6). * p<0.05; **p<0.01 (unpaired two-tailed t-test).
-
Figure 1—figure supplement 1—source data 1
Raw data from MTT cell viability assays after oxaliplatin treatment (panel A).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig1-figsupp1-data1-v1.xlsx
-
Figure 1—figure supplement 1—source data 2
Invasive cell counts from Transwell assays (panel B).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig1-figsupp1-data2-v1.xlsx
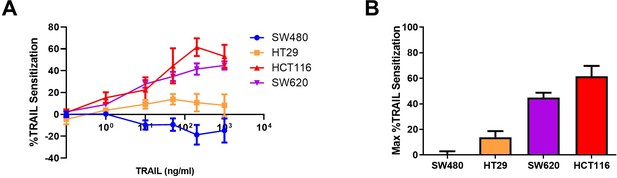
Sensitization of oxaliplatin-resistant colorectal cancer (CRC) cell lines to TRAIL.
(A) Sensitization of oxaliplatin-resistant SW620, HCT116, HT29, and SW480 cell lines compared to their parental counterparts as a function of TRAIL concentration. (B) Maximum TRAIL sensitization for each cell line between the tested concentrations of 0.1–1000 ng/ml. Data are presented as mean ± SEM.
-
Figure 1—figure supplement 2—source data 1
TRAIL sensitization calculations.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig1-figsupp2-data1-v1.xlsx
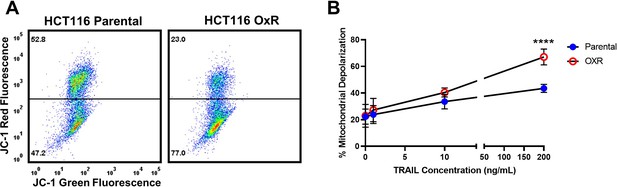
HCT116 oxaliplatin-resistant (OxR) cells have increased mitochondrial depolarization and activation of the intrinsic apoptotic pathway when treated with TRAIL.
(A) Representative flow plots of JC-1 assay after treatment with 200 ng/ml of TRAIL. Mitochondrial depolarization is evidenced by decreased red fluorescence and increased green fluorescence. (B) Mitochondrial depolarization as a function of TRAIL concentration for HCT116 parental and OxR cell lines. Data are presented as mean ± SD. N = 3 (n = 9). ****p<0.0001 (unpaired two-tailed t-test).
-
Figure 1—figure supplement 3—source data 1
Percent depolarized mitochondria in HCT116 cells measured from JC-1 assays.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig1-figsupp3-data1-v1.xlsx
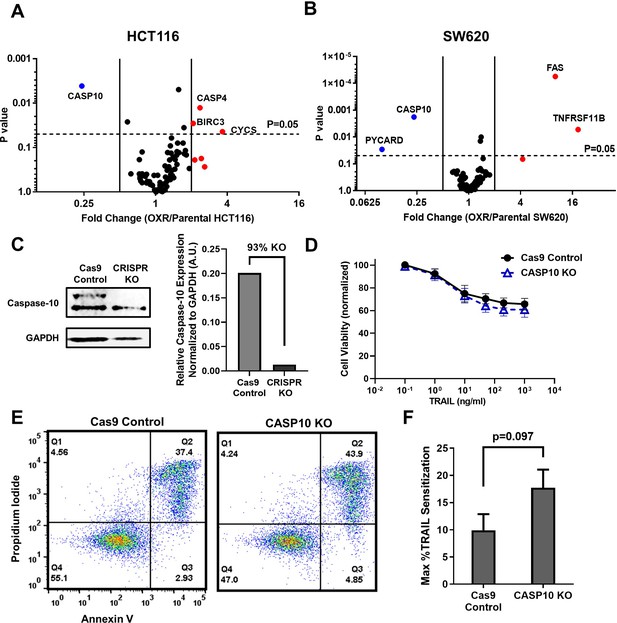
Microarray profiles show that parental and oxaliplatin-resistant (OxR) colorectal cancer (CRC) cell lines have similar expression of apoptotic transcripts while OxR derivatives have significantly downregulated CASP10.
(A, B) Volcano plots of RT-PCR Apoptosis Profiler arrays demonstrate downregulation of CASP10 in OxR phenotypes. N = 3. (C) CRISPR/Cas9 knockout of caspase-10 in SW620 parental cells was confirmed via western blot. sgRNA/Cas9 ribonucleoprotein complexes reduced caspase-10 expression by 93% compared to cells treated with Cas9 alone. (D) CASP10 knock-out (KO) cells demonstrate slight decreases in viability when treated with TRAIL compared to Cas9 control. Data are presented as mean ± SD. N = 3 (n = 9). (E) Representative Annexin-V/PI flow plots comparing SW620 parental (Cas9 only) and CASP10 KO cell viability after 24 hr of treatment with 1000 ng/ml TRAIL. (F) Depletion of caspase-10 did not have a significant effect on TRAIL sensitization (unpaired two-tailed t-test). Data are presented as mean + SEM. N = 3 (n = 9).
-
Figure 2—source data 1
Apoptosis microarray data in HCT116 cells with fold regulation calculations generated using the GeneGlobe Data Analysis Center (panel A).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Apoptosis microarray data in SW620 cells with fold regulation calculations generated using the GeneGlobe Data Analysis Center (panel B).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig2-data2-v1.xlsx
-
Figure 2—source data 3
Western blot images (raw and annotated) confirming CASP10 KO (panel C).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig2-data3-v1.zip
-
Figure 2—source data 4
Quantification of CASP10 KO from western blots (panel C).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig2-data4-v1.xlsx
-
Figure 2—source data 5
Cell viability and TRAIL sensitization calculations in CASP10 KO cells (panels D and F).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig2-data5-v1.xlsx
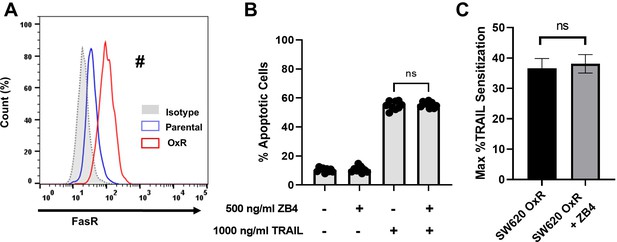
Upregulated FasR in SW620 oxaliplatin-resistant (OxR) cells has no effect on TRAIL sensitivity.
(A) Flow cytometry staining confirms increased surface expression of Fas receptor. #Significant according to a chi-squared test (see Supplementary file 1). (B) Percentage of apoptotic SW620 OxR cells after treating with 1000 ng/ml TRAIL and 500 ng/ml of the anti-FasR neutralizing antibody ZB4 (sum of early and late-stage apoptotic cells from Annexin/PI staining). Data are presented as mean + SD. N = 3 (n = 9). Significance was measured using an ordinary one-way ANOVA–Tukey’s multiple comparison test. (C) Neutralizing FasR has no effect on TRAIL sensitization. Data are presented as mean + SEM. N = 3 (n = 9). Significance was measured using an unpaired two-tailed t-test.
-
Figure 2—figure supplement 1—source data 1
Cell apoptosis and TRAIL sensitization calculations after ZB4 treatment with TRAIL.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig2-figsupp1-data1-v1.xlsx
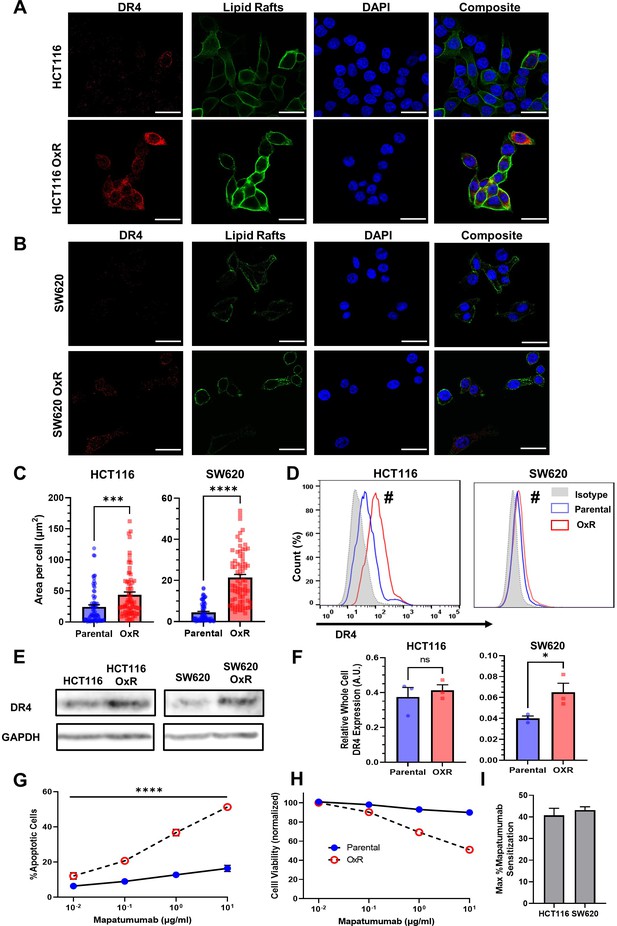
Oxaliplatin-resistant (OxR) colon cancer cell lines have upregulated DR4 expression.
(A, B) Confocal micrographs of HCT116 and SW620 cells, respectively. Red channel represents DR4, green is lipid rafts, and blue is DAPI (nuclei). Scale bar = 30 μm. (C) Quantification of DR4 area per cell in HCT116 and SW620 cells. For each cell line, N = 75 cells were analyzed. Data are presented as mean + SEM from N = 3 independent experiments. ***p<0.001; ****p<0.0001 (unpaired two-tailed t-test). (D) OxR cells had increased surface expression of DR4 in non-permeabilized cells analyzed via flow cytometry. #Significant according to a chi-squared test (see Supplementary file 1). (E) Western blots for DR4 in whole cell lysates of parental and OxR cells. (F) Quantification of western blots from three independent experiments (N = 3). Data are presented as mean + SEM. *p<0.05 (unpaired two-tailed t-test). (G) Percentage of apoptotic SW620 cells after treatment with 0.01–10 µg/ml mapatumumab (sum of early and late-stage apoptotic cells from Annexin/PI staining). Data are presented as mean ± SD. N = 3 (n = 6). ****p<0.0001 (multiple unpaired two-tailed t-tests). (H) Cell viability of SW620 cells after mapatumumab treatment, determined by Annexin-V/PI staining. Data are presented as mean ± SD. N = 3 (n = 6). (I) Maximum mapatumumab sensitization within OxR cell lines compared to their parental counterparts. Data are presented as mean + SEM.
-
Figure 3—source data 1
Quantification of DR4 area per cell in HCT116 and SW620 cells (panel C).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Western blot images (raw and annotated) for DR4 (panel E).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig3-data2-v1.zip
-
Figure 3—source data 3
Quantification of DR4 from western blots (panel F).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig3-data3-v1.xlsx
-
Figure 3—source data 4
Cell viabilty and percent apoptosis in SW620 cells after mapatumumab treatment (panels G-I).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig3-data4-v1.xlsx
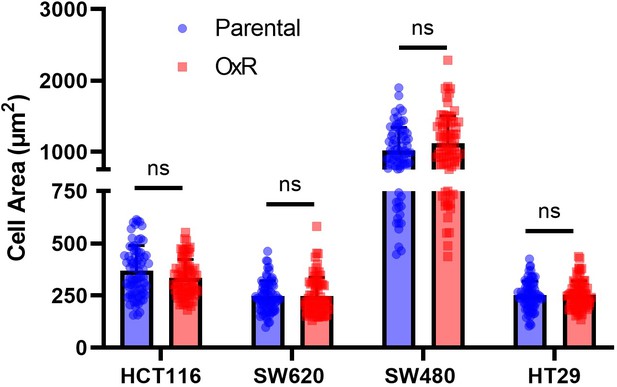
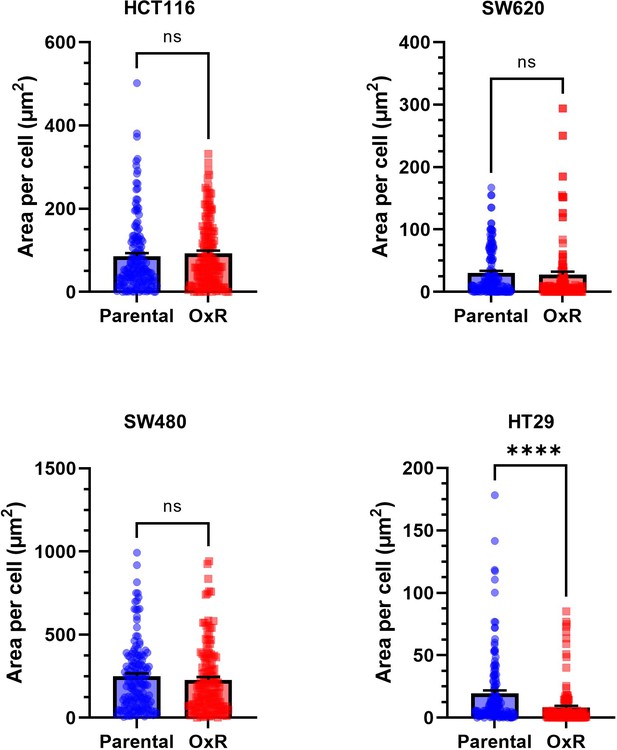
Parental and oxaliplatin-resistant (OxR) cell lines have no significant changes in cell area, analyzed from confocal microscopy images.
Data are presented as mean + SD. For each cell line, N = 75 cells were analyzed. An unpaired two-tailed t-test was used to measure significance between groups.
-
Figure 3—figure supplement 1—source data 1
Cell area measurements for all cell lines.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig3-figsupp1-data1-v1.xlsx
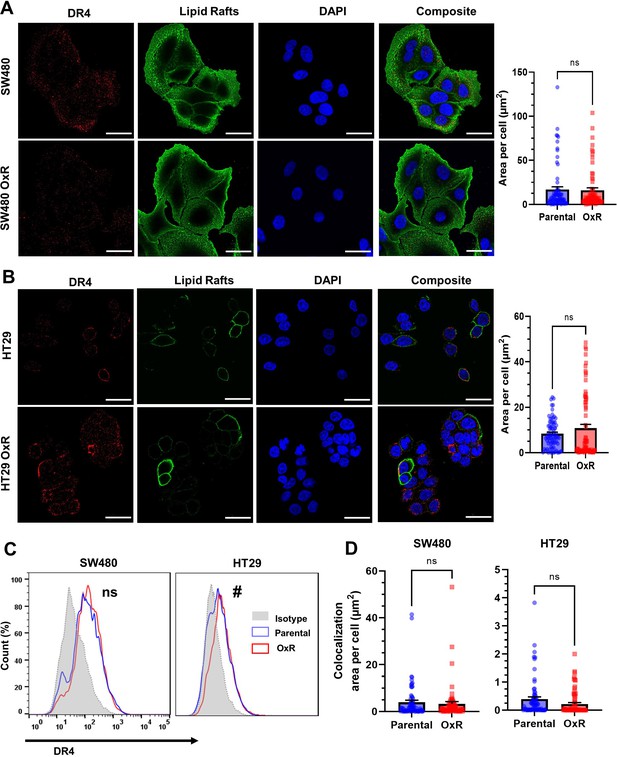
Unsensitized SW480 and HT29 oxaliplatin-resistant (OxR) cell lines show no significant changes in DR4 expression or lipid raft colocalization relative to their parental counterparts.
(A, B) Confocal micrographs and DR4 quantification of SW480 and HT29 cells, respectively. Red channel is DR4, green is lipid rafts, and blue is DAPI (nuclei). Scale bar = 30 μm. (C) Surface expression of DR4 in non-permeabilized cells analyzed via flow cytometry. #Significant according to a chi-squared test (see Supplementary file 1). (D) DR4/LR colocalization area per cell for SW480 and HT29 was found to be insignificant between parental and OxR phenotypes. Data are presented as mean + SEM. An unpaired two-tailed t-test was performed for panels (A), (B), and (D). For each cell line, N = 75 cells were analyzed.
-
Figure 3—figure supplement 2—source data 1
Quantification of DR4 and lipid raft colocalized DR4 area per cell in SW480 and HT29 cells.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig3-figsupp2-data1-v1.xlsx
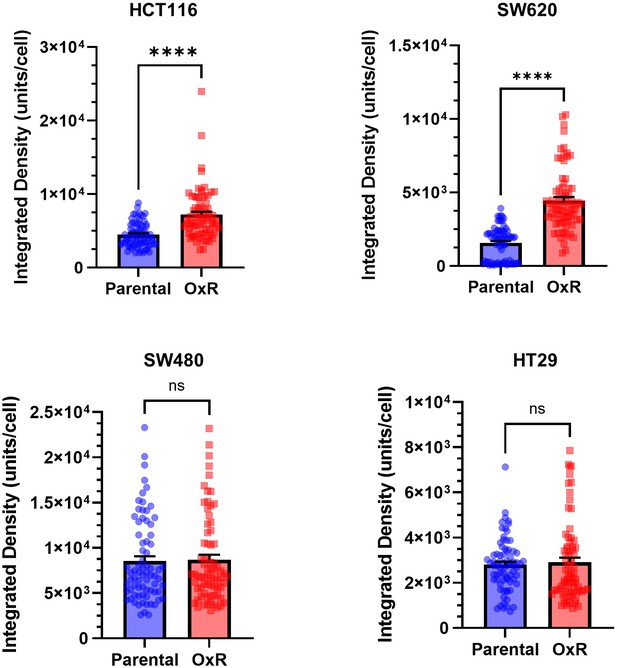
Sensitized oxaliplatin-resistant cell lines have significantly increased DR4 integrated density per cell.
Data are presented as mean + SEM. For each cell line, N = 75 cells were analyzed. ****p<0.0001 (unpaired two-tailed t-test).
-
Figure 3—figure supplement 3—source data 1
Integrated density measurements of DR4.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig3-figsupp3-data1-v1.xlsx
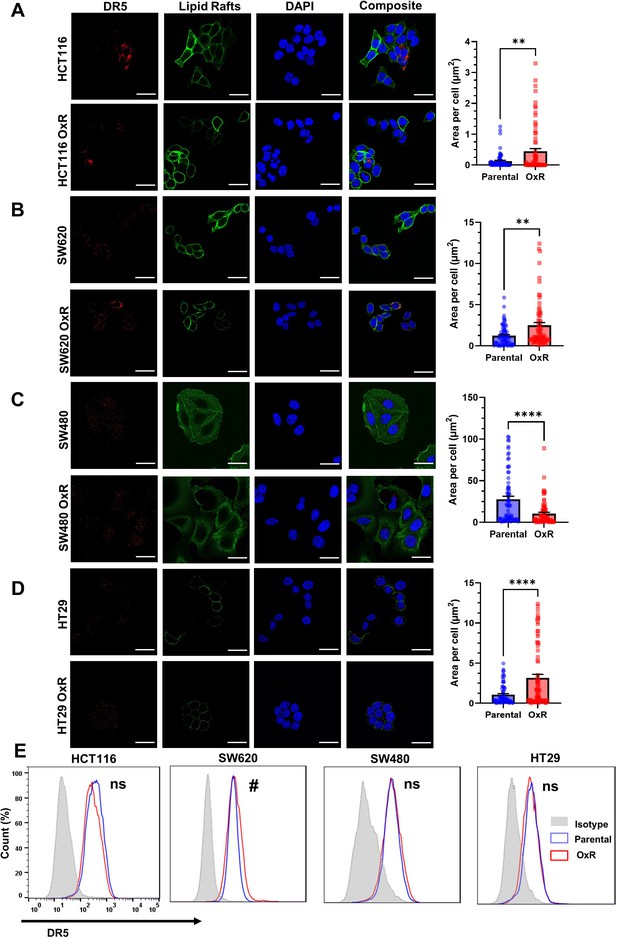
Chemoresistant HCT116, SW620, and HT29 cells have upregulated DR5 while in chemoresistant SW480 cells, DR5 is decreased.
(A–D) Confocal micrographs and DR5 quantification of HCT116, SW620, SW480, and HT29 cells, respectively. Red channel is death receptor 5, green is lipid rafts, and blue is DAPI (nuclei). Scale bar = 30 μm. ** p<0.01; ****p<0.0001 (unpaired two-tailed t-test). Data are presented as mean + SEM. For each cell line, N = 75 cells were analyzed. (E) Oxaliplatin-resistant (OxR) cells only demonstrate increased surface expression of DR5 in non-permeabilized SW620 cells. #Significant according to a chi-squared test (see Supplementary file 1).
-
Figure 3—figure supplement 4—source data 1
Quantification of DR5 area per cell for all cell lines.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig3-figsupp4-data1-v1.xlsx
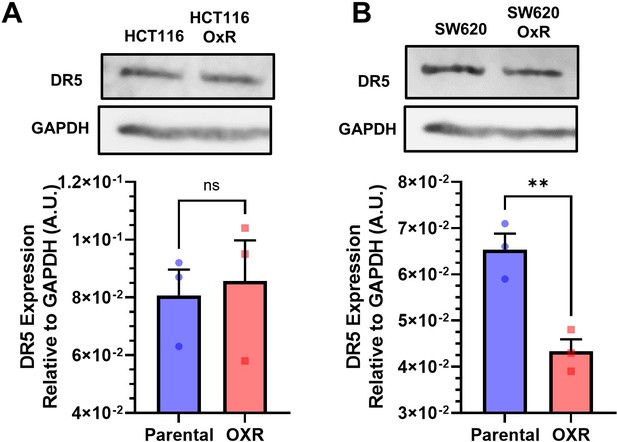
Western blots show TRAIL-sensitized HCT116 (A) and SW620 (B) oxaliplatin-resistant (OxR) cells have no increases in DR5.
Data are presented as mean + SEM. N = 3. **p<0.01 (unpaired two-tailed t-test).
-
Figure 3—figure supplement 5—source data 1
Quantification of DR5 from western blots.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig3-figsupp5-data1-v1.xlsx
-
Figure 3—figure supplement 5—source data 2
Western blot images (raw and annotated) for DR5.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig3-figsupp5-data2-v1.zip
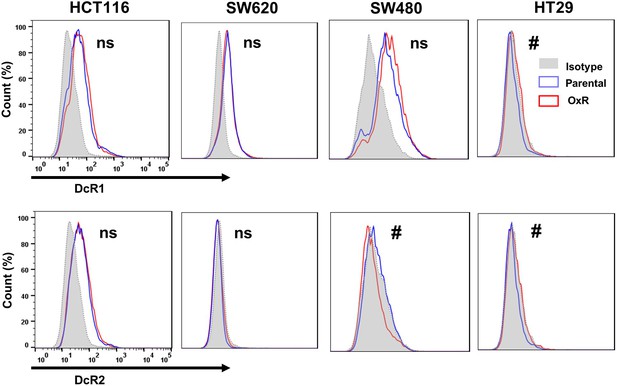
Flow cytometry analysis of the surface expression of decoy death receptors 1 (DcR1) and 2 (DcR2) on nonpermeabilized parental and oxaliplatin-resistant (OxR) cell lines.
#Significant according to a chi-squared test (see Supplementary file 1).
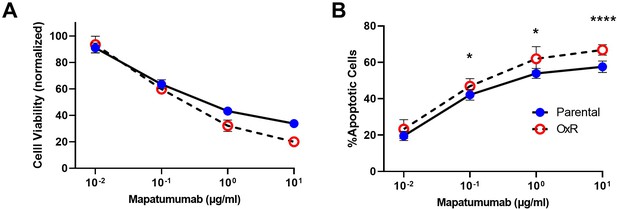
HCT116 oxaliplatin-resistant (OxR) cells are increasingly sensitive to DR4 agonist antibody treatment.
(A) Cell viability of HCT116 cells after 0.01–10 µg/ml mapatumumab treatment, determined by Annexin-V/PI staining. (B) Percentage of apoptotic SW620 cells after mapatumumab treatment (sum of early and late-stage apoptotic cells from Annexin/PI staining). For all graphs, data are presented as mean + SD. N = 3 (n = 6). *p<0.05; ****p<0.0001 (multiple unpaired two-tailed t-tests).
-
Figure 3—figure supplement 7—source data 1
Cell viabilty and percent apoptosis in HCT116 cells after mapatumumab treatment.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig3-figsupp7-data1-v1.xlsx
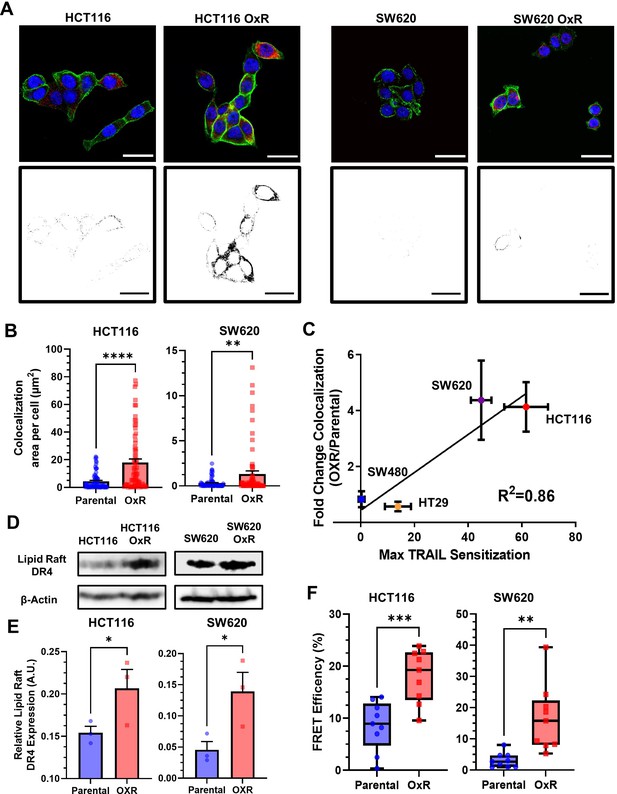
Oxaliplatin-resistant (OxR) colon cancer cell lines have enhanced colocalization of DR4 into lipid rafts.
(A) Composite images and binary projections of DR4/LR colocalization areas in HCT116 and SW620 cell lines. Lipid raft and DR4 binary images were generated for a specified threshold, then multiplied by one another to generate images with positive pixels in double-positive areas. Red is DR4, green is lipid rafts, and blue is DAPI. Scale bar = 30 μm. (B) Quantification of DR4 and lipid raft colocalization area per cell in HCT116 and SW620 cells. For each cell line, N = 75 cells were analyzed. **p<0.01; ****p<0.0001 (unpaired two-tailed t-test). (C) Correlation between the fold change in DR4/LR colocalization (OxR phenotype/parental) and maximum TRAIL sensitization observed by the OxR phenotype for each of the four cell lines (simple linear regression analysis). (D) Lipid raft fractions were isolated and analyzed for DR4 via western blot in parental and OxR cells. (E) Quantification of lipid raft DR4 blots in (D). *p<0.05 (unpaired two-tailed t-test). For all graphs, data are presented as mean + SEM. (F) Förster resonance energy transfer (FRET) efficiencies of FITC-labeled DR4 (donor) and Alexa 555-labeled lipid rafts (acceptor) in parental and OxR cells analyzed via flow cytometry. **p<0.01; ***p<0.001 (unpaired two-tailed t-test).
-
Figure 4—source data 1
Quantification of LR-colocalized DR4 area per cell in HCT116 and SW620 cells (panel B).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig4-data1-v1.xlsx
-
Figure 4—source data 2
Correlation analysis of LR-colocalized DR4 area per cell with TRAIL sensitization (panel C).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig4-data2-v1.xlsx
-
Figure 4—source data 3
Western blot images (raw and annotated) for DR4 from LR isolates (panel D).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig4-data3-v1.zip
-
Figure 4—source data 4
Quantification of LR DR4 from western blots (panel E).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig4-data4-v1.xlsx
-
Figure 4—source data 5
FRET efficency calculations (panel F).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig4-data5-v1.xlsx
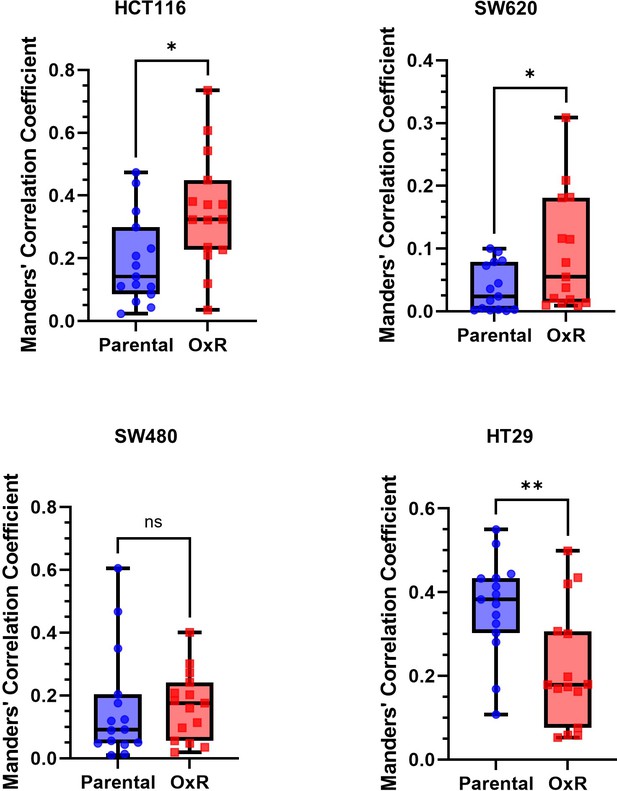
TRAIL-sensitized oxaliplatin-resistant (OxR) cells have significantly increased colocalization of DR4 with lipid rafts according to Manders’ Correlation Coefficient.
N = 15 analyzed micrographs across three independent trials. *p<0.05; *p<0.001 (unpaired two-tailed t-test).
-
Figure 4—figure supplement 1—source data 1
MCC calculations for all cell lines.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig4-figsupp1-data1-v1.xlsx
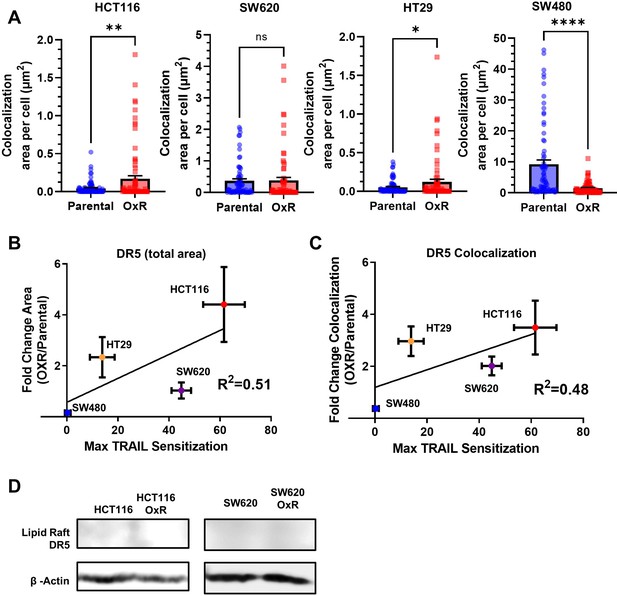
Sensitization to TRAIL in oxaliplatin-resistant (OxR) cell lines poorly correlates with DR5 expression while lipid raft fractions have no detectable DR5.
(A) Quantification of DR5/LR colocalization in HCT116, SW620, SW480, and HT29 cells. *p<0.05; **p<0.01; ****p<0.0001 (unpaired two-tailed t-test). For each cell line, N = 75 cells were analyzed. (B) Correlation of total DR5 area per cell and (C) DR5/LR colocalization with maximum TRAIL sensitization observed in OxR cells (linear regression analysis). For all graphs, data are presented as mean + SEM. (D) Western blots show DR5 was undetectable in lipid raft isolated fractions.
-
Figure 4—figure supplement 2—source data 1
Quantification of LR-colocalized DR5 area per cell (panel A).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig4-figsupp2-data1-v1.xlsx
-
Figure 4—figure supplement 2—source data 2
Correlation analysis of DR5 and LR-colocalized DR5 area per cell with TRAIL sensitization (panels B and C).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig4-figsupp2-data2-v1.xlsx
-
Figure 4—figure supplement 2—source data 3
Western blot images (raw and annotated) for DR5 from LR isolates (panel D).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig4-figsupp2-data3-v1.zip
Quantification of lipid raft area per cell.
For each cell line, N = 150 cells were analyzed. Data are presented as mean + SEM. ****p<0.0001 (unpaired two-tailed t-test).
-
Figure 4—figure supplement 3—source data 1
Quantification of LR area per cell.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig4-figsupp3-data1-v1.xlsx
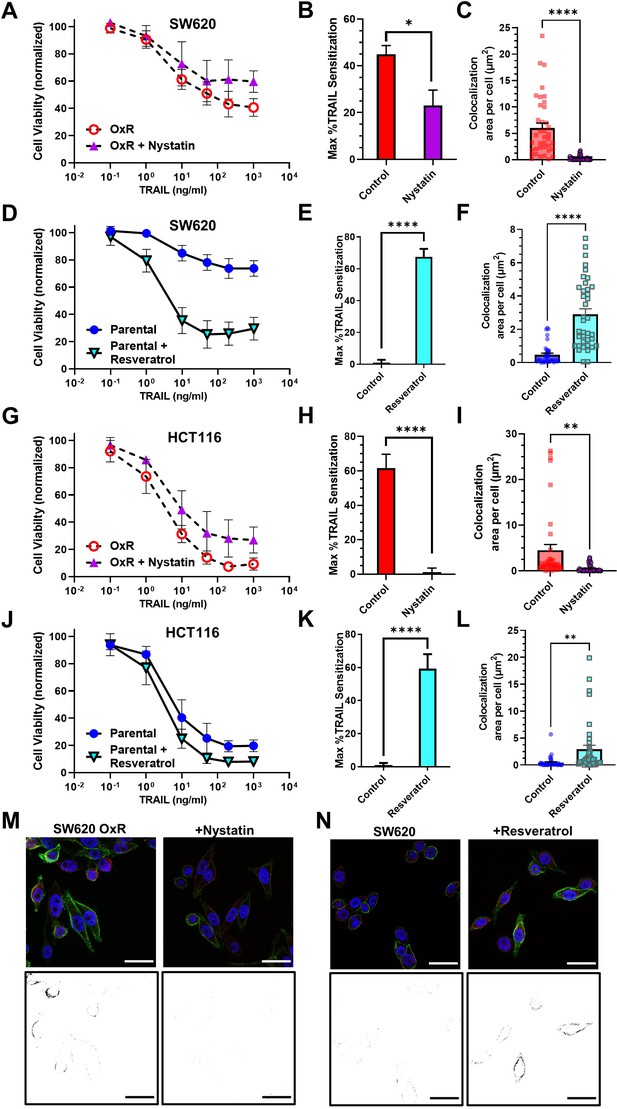
Pharmacological perturbation of DR4 localization in lipid rafts significantly alters cellular apoptosis in response to TRAIL.
(A, G) SW620 oxaliplatin-resistant (OxR) and HCT116 OxR cells, respectively, treated for 24 hr with a combination of TRAIL and 5 µM nystatin. (B, H) SW620 OxR and HCT116 OxR cells, respectively, showed a significant decrease in TRAIL sensitization when treated in combination with nystatin. N = 3 (n = 9). (C, I) Treatment with 5 µM nystatin significantly decreased DR4/LR colocalization area in SW620 OxR and HCT116 OxR cells, respectively. For each cell line, N = 40 cells were analyzed. (D, J) SW620 Par and HCT116 Par cells, respectively, treated for 24 hr with a combination of TRAIL and 70 µM resveratrol. N = 3 (n = 9). (E, K) SW620 Par and HCT116 Par cells, respectively, showed a significant increase in TRAIL sensitization when treated in combination with resveratrol. N = 3 (n = 9). (F, L) Treatment with 70 µM nystatin significantly increased DR4/LR colocalization area in SW620 Par and HCT116 Par cells, respectively. For each cell line, N = 40 cells were analyzed. (M) Representative composite images and binary projections of DR4/LR colocalization in SW620 OxR cells before and after nystatin treatment. (N) Representative composite images and binary projections of DR4/LR colocalization in parental SW620 cells before and after resveratrol treatment. Red represents DR4, green is lipid rafts, and blue is DAPI. Scale bar = 30 μm. **p<0.01; ****p<0.0001 (unpaired two-tailed t-test for all graphs). (A, D, G, J) Data are presented as mean ± SD. (B, C, E, F, H, I, K, L) Data are presented as mean + SEM.
-
Figure 5—source data 1
Cell viability after TRAIL combination treatments with resveratrol and nystatin (panels A, D, G, J).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig5-data1-v1.xlsx
-
Figure 5—source data 2
TRAIL sensitization calculations after resveratrol and nystatin (panels B, E, H, K).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig5-data2-v1.xlsx
-
Figure 5—source data 3
Quantification of LR-colocalized DR4 after resveratrol and nystatin treatment (panels C, F, I, L).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig5-data3-v1.xlsx
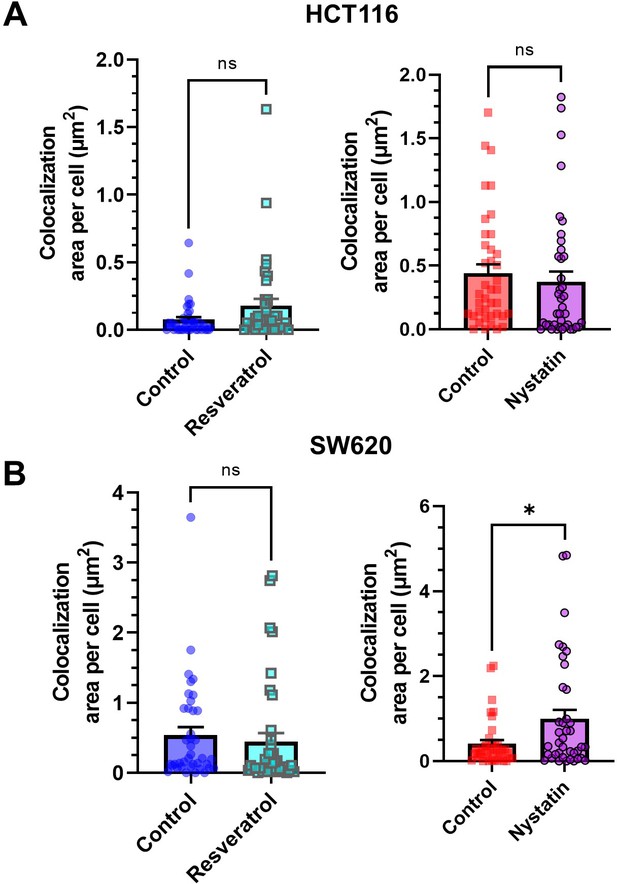
Quantification of the effects of resveratrol and nystatin on DR5 colocalization with lipid rafts in HCT116 (A) and SW620 cells (B).
For each cell line, N = 40 cells were analyzed. Data are presented as mean + SEM. *p<0.05 (unpaired two-tailed t-test).
-
Figure 5—figure supplement 1—source data 1
Quantification of the effects of resveratrol and nystatin on DR5 colocalization with LRs in HCT116 and SW620 cells.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig5-figsupp1-data1-v1.xlsx
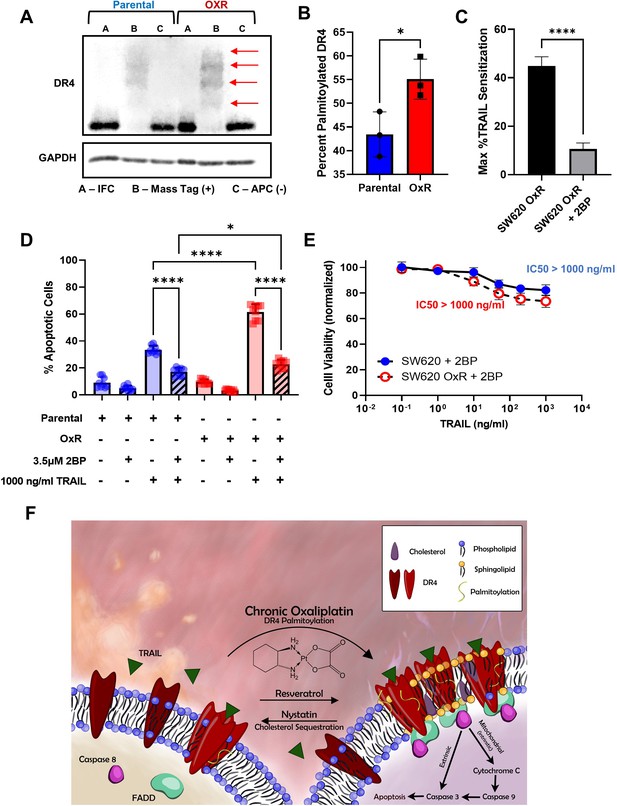
Oxaliplatin resistance enhances palmitoylation of DR4, selectively.
(A) Death receptor palmitoylation was determined by protein precipitation, thioester cleavage, and conjugation of a mass tag to enumerate and quantify the degree of S-palmitoylation between cellular phenotypes. Samples with a mass tag ‘B’ have distinct bands of equivalent increasing mass, with each mass shift indicating a palmitoylated site. Input fraction control (IFC) samples ‘A’ were collected before thioester cleavage, while the acyl preservation negative control (APC) samples were incubated with an acyl-preservation reagent to block free thiols in place of the mass tag reagent. Arrows show palmitoylation bands. (B) Quantification of the percentage of palmitoylated DR4, calculated by dividing the total palmitoylated mass shift intensity by the average intensity of IFC and APC for each sample. Data are presented as mean ± SD (N = 3). *p<0.05 (unpaired two-tailed t-test). (C) Treatment with the irreversible palmitoylation inhibitor 2BP in combination with TRAIL significantly reduced TRAIL sensitization in SW620 OxR cells. Data are presented as mean + SEM. N = 3 (n = 9). *p<0.0001 (unpaired two-tailed t-test). (D) Percentage of apoptotic SW620 parental and OxR cells after treating with 1000 ng/ml TRAIL and 3.5 μM 2BP in combination (sum of early and late-stage apoptotic cells from Annexin/PI staining). Data are presented as mean + SD. N = 3 (n = 9). *p<0.05; ****p<0.0001 (ordinary one-way ANOVA–Tukey’s multiple comparison test). (E) Cell viability determined by Annexin-V/PI staining for cells treated with 0.1–1000 ng/ml TRAIL and 3.5 μM 2BP. IC50 values were calculated using a variable slope four-parameter nonlinear regression. Data are presented as mean ± SD. N = 3 (n = 9). (F) Proposed mechanism of enhanced TRAIL-mediated apoptosis in oxaliplatin-resistant cells.
-
Figure 6—source data 1
Quantification of palmitoylated DR4 from western blots (panel B).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig6-data1-v1.xlsx
-
Figure 6—source data 2
Cell viability and percent apoptosis in SW620 cells after 2BP and TRAIL combination treatment (panels C-E).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig6-data2-v1.xlsx
-
Figure 6—source data 3
Western blot images (raw and annotated) for palmitoylated DR4.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig6-data3-v1.zip
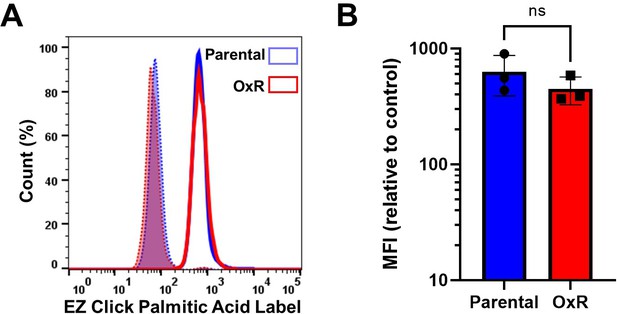
Total palmitoylation remains unchanged between SW620 parental and oxaliplatin-resistant (OxR) cells.
(A) Parental and OxR SW620 cells were labeled with EZClick Palmitic Acid/Fluorescent Azide staining kit and analyzed via flow cytometry to determine total protein palmitoylation between cell lines. Blue histograms represent parental SW620 cells, and red histograms are SW620 OxR cells. Shaded histograms are background controls for each cell line (Palmitic Acid [-]/Fluorescent Azide [+]). (B) Quantification of median fluorescence intensity (MFI) shows no significant change in total palmitoylation between cellular phenotypes (unpaired two-tailed t-test). Data are presented as mean ± SD. N = 3. Significance was measured using an unpaired two-tailed t-test.
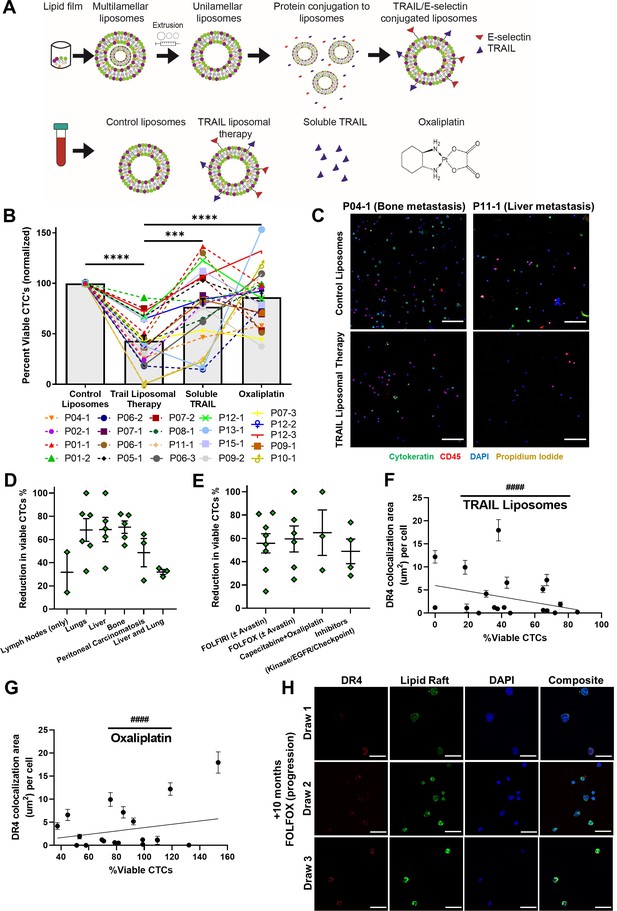
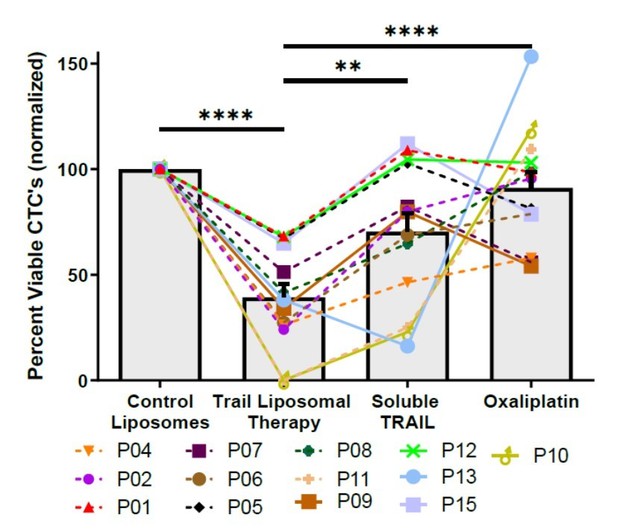
TRAIL-conjugated liposomes neutralize circulating tumor cells (CTCs) from the blood of patients with metastatic, oxaliplatin-resistant colorectal cancer.
(A) Liposomes were synthesized using a thin-film hydration method, followed by extrusion and his-tag conjugation of TRAIL and E-selectin protein. Patient blood samples were treated in a cone-and-plate viscometer under circulatory shear conditions with either control liposomes, TRAIL liposomes, soluble TRAIL, or oxaliplatin. (B) Effects of TRAIL liposomes and control treatments on the number of viable CTCs, normalized to control liposome treatment. Bars represent the average of all patients and time points (N = 21). **p<0.001; ****p<0.0001 (ordinary one-way ANOVA–Tukey’s multiple comparison test). (C) Representative micrographs of two patients showing neutralization of CTCs in TRAIL liposomes compared to control liposomes, stained for cytokeratin (green), DAPI (blue), CD45 (red), and propidium iodide (yellow). Scale bar = 100 µm. (D, E) Reduction in viable CTCs categorized by location of metastasis and treatment administered at the time of blood draw, respectively. (F) DR4/LR colocalization area of patient CTCs plotted against the percentage of viable CTCs following TRAIL liposome treatments. Each point corresponds with one patient draw. ####p<0.0001 (simple linear regression to confirm significant deviation from zero). (G) DR4/LR colocalization area of patient CTCs plotted against the normalized percentage of viable CTCs following oxaliplatin treatment. Each point corresponds with one patient draw. ####p<0.0001 (simple linear regression to confirm significant deviation from zero). (H) CTCs of patient 7, stained for DR4 (red) and lipid rafts (green), demonstrating increased DR4/LR colocalization over the course of 10 months of FOLFOX treatment (with progressive disease despite treatment). Scale bar = 30 µm. For all graphs, data are presented as mean ± SEM.
-
Figure 7—source data 1
Viability of patient CTCs following treatment.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig7-data1-v1.xlsx
-
Figure 7—source data 2
Correlation analysis of LR-colocalized DR4 area with percent viable CTCs after treatment.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig7-data2-v1.xlsx
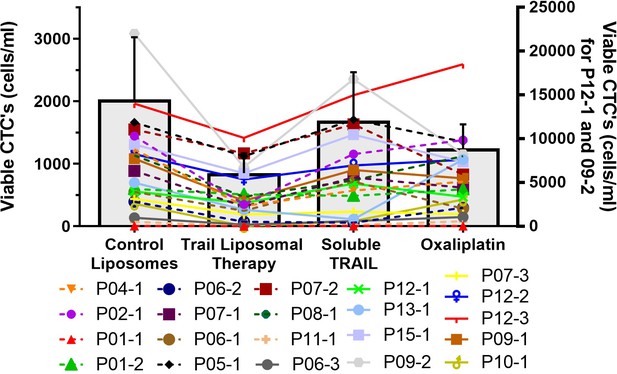
Absolute numbers of viable circulating tumor cells (CTCs) per ml of blood following TRAIL liposomal therapy and control treatments.
Bars represent mean of all patient samples, and error bars represent SEM. Samples 12-1 and 09-2 showed very large CTC concentrations and were plotted using the alternative scale shown on the right. All other samples (and average) were plotted using the scale shown on the left.
-
Figure 7—figure supplement 1—source data 1
Absolute numbers of viable circulating tumor cells (CTCs) per ml of blood following TRAIL liposomal therapy and control treatments.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig7-figsupp1-data1-v1.xlsx
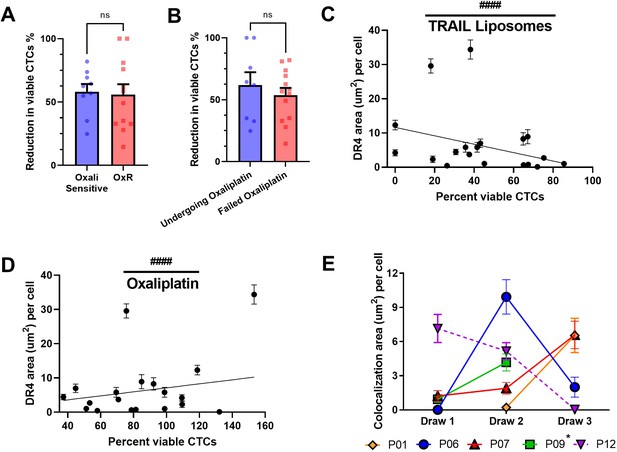
TRAIL liposomes are effective in oxaliplatin-sensitive and -refractory patients.
(A) Patients were categorized as either oxaliplatin-sensitive (viability <80%, N = 9) or oxaliplatin-resistant (viability >80%, N = 12) to compare changes in the reduction of viable circulating tumor cells (CTCs) (unpaired two-tailed t-test). (B) Patients undergoing oxaliplatin chemotherapy at the time of blood draw (N = 8) showed insignificant changes in viable CTC reduction compared to patients that have previously failed oxaliplatin (N = 13) (unpaired two-tailed t-test). (C) DR4 area of patient CTCs plotted against the percentage of viable CTCs following TRAIL liposome treatments. Each point corresponds with one patient draw. ####p<0.0001 (simple linear regression to confirm significant deviation from zero). (D) DR4 area of patient CTCs plotted against the normalized percentage of viable CTCs following oxaliplatin treatment. Each point corresponds with one patient draw. ####p<0.0001 (simple linear regression to confirm significant deviation from zero). (E) Lipid raft/DR4 analysis of repeat patients, analyzing the changes in DR4 colocalization over the course of therapy. *Patient 9 died after draw 2, precluding further blood collection. For all graphs, data are presented as mean ± SEM.
-
Figure 7—figure supplement 2—source data 1
Treatment efficacy in oxaliplatin resistant and sensitive patients (panels A and B).
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig7-figsupp2-data1-v1.xlsx
-
Figure 7—figure supplement 2—source data 2
Correlation analysis of DR4 area in CTCs with percent viable CTCs after treatment.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig7-figsupp2-data2-v1.xlsx
-
Figure 7—figure supplement 2—source data 3
Analysis of LR-colocalized DR4 in patient CTCs over time.
- https://cdn.elifesciences.org/articles/67750/elife-67750-fig7-figsupp2-data3-v1.xlsx
Tables
Demographic and clinical information of metastatic colorectal cancer (CRC) patients enrolled in this study.
*Denotes missing treatment analysis for this sample. †Denotes missing DR4/LR analysis for this sample.
| Patient | Age | Sex | Cancer | Metastatic location | Treatment history at draw 1 | Draw 2 | Draw 3 |
|---|---|---|---|---|---|---|---|
| P01 | 59 | F | Colon | Paraaortic lymph nodes | †FOLFOX (2016), FOLFIRI, 5-FU + Avastin | +2 months 5-FU + Avastin | * +7 months 5-FU + Avastin |
| P02 | 83 | F | Colon | Liver | †FOLFOX, FOLFIRI, FOLFOX + Avastin | ||
| P04 | 53 | F | Rectal | Pelvis, mesenteric lymph nodes | FOLFOX + Avastin, capecitabine +radiation, regorafenib + nivolumab | ||
| P05 | 68 | M | Rectal | Pulmonary | Capecitabine + oxaliplatin | ||
| P06 | 68 | F | Rectal | Lung, bone | FOLFOX, FOLFIRI | +6 months FOLFIRI | +7 months FOLFIRI |
| P07 | 64 | M | Cecum | Peritoneal carcinomatosis | FOLFOX (1st cycle) | +7 months FOLFOX (progression) | +10 months FOLFOX (progression) Started FOLFIRI |
| P08 | 69 | M | Colon | Lung, abdomen | FOLFOX + Avastin (progression) capecitabine + Avastin, 5-FU + cetuximab + panitumumab | ||
| P09 | 73 | M | Sigmoid | Liver, mesentery | FOLFOX, capecitabine, FOLFIRI, Lonsurf | +7 months FOLFIRI | N/A (patient deceased) |
| P10 | 52 | M | Rectal | Lung | Radiation + capecitabine capecitabine + oxaliplatin | ||
| P11 | 70 | M | Colon | Liver | FOLFOX | ||
| P12 | 59 | M | Colon | Liver, lungs, R adrenal | FOLFOX + Avastin, FOLFIRI + Avastin | Cetuximab + encorafenib (progression) | +3 months cetuximab + encorafenib (progression) |
| P13 | 63 | F | Colon | Adnexa pelvis | FOLFOX, irinotecan + panitumumab, capecitabine + oxaliplatin | ||
| P15 | 79 | M | Colon | Liver | FOLFOX (1st cycle) |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | SW620 adenocarcinoma,colorectal, Dukes' type C | ATCC | #CCL-227 | RRID:CVCL_0547 L15 Media |
| Cell line (Homo sapiens) | SW480 adenocarcinoma,colorectal, Dukes' type B | ATCC | #CCL-228 | RRID:CVCL_0546 L15 Media |
| Cell line (Homo sapiens) | HT29 adenocarcinoma, colorectal | ATCC | #HTB-38 | RRID:CVCL_0320 McCoy’s 5A Media |
| Cell line (Homo sapiens) | HCT116 carcinoma, colorectal | ATCC | #CCL-247 | RRID:CVCL_0291 McCoy’s 5A Media |
| Cell line (Homo sapiens) | SW620 OxRadenocarcinoma,colorectal, Dukes' type C | Kobe Pharmaceutical University | #CCL-227 | RRID:CVCL_4V77 L15 Media |
| Cell line (Homo sapiens) | SW480 OxRadenocarcinoma, colorectal, Dukes' type B | MD Anderson Cancer Center Characterized Cell Line Core | #CCL-228 | RRID:CVCL_AU18 L15 Media |
| Cell line (Homo sapiens) | HT29 OxR adenocarcinoma, colorectal | MD Anderson Cancer Center Characterized Cell Line Core | #HTB-38 | RRID:CVCL_ 5949 McCoy’s 5A Media |
| Cell line (Homo sapiens) | HCT116 OxR carcinoma, colorectal | MD Anderson Cancer Center Characterized Cell Line Core | #CCL-247 | RRID:CVCL_4V73 McCoy’s 5A Media |
| Chemical compound, drug | Oxaliplatin | MedChemExpress | Cat# HY-17371 | CAS No: 61825-94-3 |
| Commercial assay or kit | MTT Assay Kit | Abcam | Cat# ab211091 | |
| Peptide, recombinant protein | Recombinant Human sTRAIL/Apo2L | PeproTech | Cat# 310-04 | |
| Antibody | Mouse Anti-TNFRSF10A Recombinant Antibody (clone mAY4) | Creative Biolabs | Cat# HPAB-1616-FY | Mapatumumab (0.01–10 μg/ml) |
| Commercial assay or kit | FITC Annexin-V Apoptosis Detection Kit I | BD Pharmingen | Cat# 556547 | Includes propidium iodide |
| Software, algorithm | FlowJo v10.7.1 | FlowJo | RRID:SCR_008520 | |
| Commercial assay or kit | JC1 – Mitochondrial Membrane Potential Assay Kit | Abcam | Cat# ab113850 | |
| Commercial assay or kit | RNeasy Plus Mini Kit | Qiagen | Cat# 74134 | |
| Commercial assay or kit | RT2 First Strand Kit | Qiagen | Cat# 330404 | |
| Commercial assay or kit | RT2 Profiler PCR Human Apoptosis Array | Qiagen | Cat# PAHS-012Z | |
| Software, algorithm | CFX Maestro Software | Bio-Rad | RRID:SCR_018064 | |
| Software, algorithm | GeneGlobe Data Analysis Center | Qiagen | RRID:SCR_021211 | |
| Commercial assay or kit | Gene Knockout Kit v2 – Human CASP10 with Cas9 2NLS Nuclease | Synthego | sgRNA:Cas9 (90 pmol:10 pmol) | |
| Commercial assay or kit | Vybrant Alexa Fluor 488 Lipid Raft Labeling Kit | Invitrogen | Cat# V34403 | |
| Commercial assay or kit | Vybrant Alexa Fluor 555 Lipid Raft Labeling Kit | Invitrogen | Cat# V34404 | |
| Antibody | Mouse anti-human CD261 (DR4) Monoclonal Antibody (clone DJR1) | Invitrogen | Cat# 14-6644-82 | RRID:AB_468188 (1:50 IF) |
| Antibody | Mouse anti-human CD262 (DR5) Monoclonal Antibody (clone DJR2-4) | Invitrogen | Cat# 14-9908-82 | RRID:AB_468592 (1:50 IF) |
| Antibody | Alexa Fluor 555 goat anti-mouse IgG (H+L) | Invitrogen | Cat# A28180 | RRID:AB_2536164 (1:1000 IF) |
| Other | DAPI | Invitrogen | Cat# D1306 | RRID:AB_2629482 (1 µg/ml) |
| Software, algorithm | Fiji – ImageJ | FIJI | RRID:SCR_002285 JaCOP plugin | |
| Antibody | Human TruStain FcX | BioLegend | Cat# 422301 | RRID:AB_2818986 |
| Antibody | PE mouse anti-human CD261 (DR4) (clone DJR1) | BioLegend | Cat# 307206 | RRID:AB_2287472 (5 µl per sample, FC) |
| Antibody | PE mouse anti-human CD262 (DR5) (clone DJR2-4) | BioLegend | Cat# 307406 | RRID:AB_2204926 (5 µl per sample, FC) |
| Antibody | PE mouse anti-human TRAILR3 (DcR1) (clone DJR3) | BioLegend | Cat# 307006 | RRID:AB_2205089 (5 µl per sample, FC) |
| Antibody | PE mouse anti-human TRAILR4 (DcR2) (clone 104918) | BioLegend | Cat# FAB633P | RRID:AB_2205217 (5 µl per sample, FC) |
| Antibody | PE Mouse IgG1 κ Isotype Control (clone MOPC-21) | BioLegend | Cat# 400114 | RRID:AB_326435 (5 µl per sample, FC) |
| Antibody | FITC mouse anti-human DR4 (clone DR-4-02) | Thermo Fisher Scientific | Cat# MA1-19757 | RRID:AB_1955203 (5 µl per sample, FC) |
| Chemical compound, drug | Resveratrol | Sigma-Aldrich | Cat# R5010-100MG | RRID:AB_309682 CAS: 501-36-0 |
| Chemical compound, drug | Nystatin | Thermo Fisher Scientific | Cat# BP29495 | CAS: 1400-61-9 |
| Chemical compound, drug | 2-Bromopalmitate | Sigma-Aldrich | Cat# 21604-1G | CAS: 18263-25-7 |
| Antibody | Mouse Anti-Fas Antibody (human, neutralizing) (clone ZB4) | Sigma-Aldrich | Cat# 05-338 | RRID:AB_309682 500 ng/ml (neutralization) |
| Commercial assay or kit | Minute Plasma Membrane-Derived Lipid Raft Isolation Kit | Invent Biotechnologies | Cat# LR-042 | |
| Antibody | DR4 Rabbit monoclonal antibody (clone D9S1R) | Cell Signalling Technologies | Cat# 42533 | RRID:AB_2799223 (1:500 WB) |
| Antibody | DR5 Rabbit polyclonal antibody | Thermo Fisher Scientific | Cat# PA1-957 | RRID:AB_2303474 (1:500 WB) |
| Antibody | Caspase-10 Rabbit polyclonal antibody | Thermo Fisher Scientific | Cat# PA5-29649 | RRID:AB_2547124 (1:1000 WB) |
| Antibody | Mouse anti-human GAPDH (clone 6C5) | MilliporeSigma | Cat# MAB374 | RRID:AB_2107445 (1:2000 WB) |
| Antibody | Mouse Anti-β-Actin monoclonal antibody (clone C4) | Santa Cruz | Cat# sc-47778 | RRID:AB_2714189 (1:1000 WB) |
| Antibody | IRDye 800CW goat anti-rabbit secondary antibody | LICOR | Cat# 926-32211 | RRID:AB_621843 (2:15,000 WB) |
| Antibody | IRDye 800CW goat anti-mouse secondary antibody | LICOR | Cat# 926-32210 | RRID:AB_621842 (2:15,000 WB) |
| Software, algorithm | LICOR housekeeping protein normalization protocol | LICOR Odyssey Fc | RRID:SCR_013715 | |
| Commercial assay or kit | SiteCounter S-Palmitoylated Protein Kit | Badrilla | Cat# K010312 | |
| Commercial assay or kit | EZClick Palmitoylated Protein Assay Kit | BioVision | Cat# K416-100 | |
| Commercial assay or kit | CD45 magnetic beads (human) | Mylteni Biotech | Cat# 130-045-801 | |
| Antibody | Biotin mouse anti-human CD45 Antibody (clone HI30) | BioLegend | Cat# 304004 | RRID:AB_314392 (1:50 IF) |
| Antibody | Streptavidin-conjugated Alexa Fluor 647 | Thermo Fisher Scientific | Cat# S21374 | RRID:AB_2336066 (1:200 IF) |
| Antibody | FITC Mouse Anti-Human Cytokeratin (clone CAM5.2) | BD Pharmingen | Cat# 347653 | (20 µl per sample, IF) |
| Antibody | Goat anti-mouse Alexa Fluor 647 | Thermo Fisher Scientific | Cat# A21235 | RRID:AB_2535804 (1:200 IF) |
Additional files
-
Supplementary file 1
Statistical reporting of chi-squared T(x) values for comparing distribution differences in flow cytometry staining.
Significance was determined if chi-squared T(x) sample > chi-squared T(x) background.
- https://cdn.elifesciences.org/articles/67750/elife-67750-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67750/elife-67750-transrepform-v1.docx