An adhesion G protein-coupled receptor is required in cartilaginous and dense connective tissues to maintain spine alignment
Figures
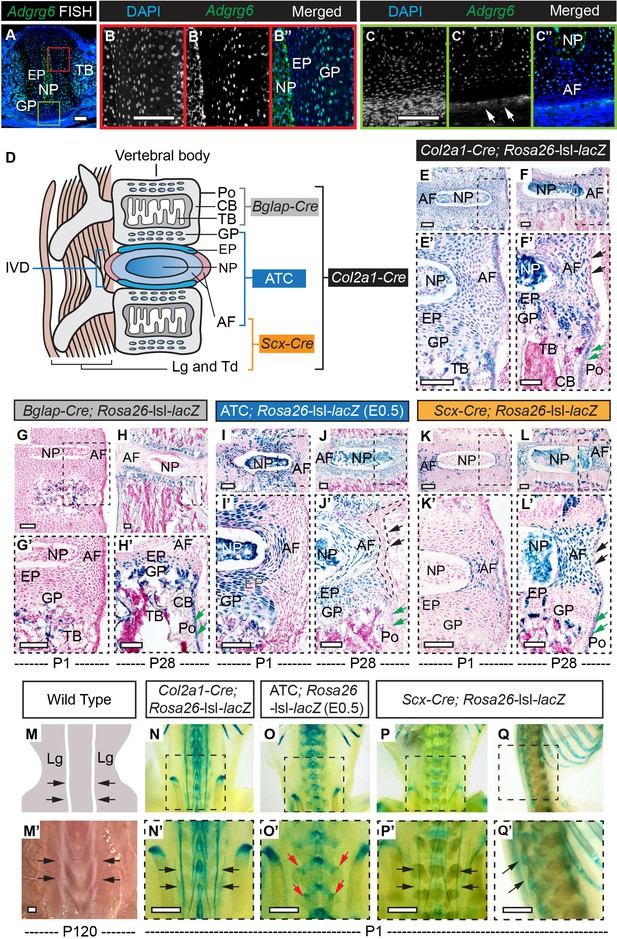
β-Galactosidase staining of Rosa26-lsl-lacZ reporter mice recombined with different Cre strains.
(A–C”) Fluorescent in situ hybridization (FISH) analysis of Adgrg6 on thoracic spine sections of wild-type mice at P1. Adgrg6 signal is detected in the GP, NP, and EP (B’, B”). Adgrg6 is also expressed in TB, AF (A, C’, C’’), and the outmost AF (white arrows, C'); n = 3 mice for each group. (D) Schematic of Cre targeting outlined in this study. Generally, Bglap-Cre targets bony tissues, ATC targets cartilaginous tissues, Scx-Cre targets dense connective tissues, and Col2a1-Cre targets all these tissues. (E–F’) β-galactosidase staining of Col2a1-Cre; Rosa26-lsl-lacZ spine sections at P1 (E, E’) and P28 (F, F’). Col2a1-Cre targets NP, AF, EP, and GP, as well as some cells in the bony tissues at both P1 and P28. Note that Col2a1-Cre also targets the outmost AF (black arrows, F’) and the periosteum (Po) (green arrows, F’) at P28. (G–H’) β-Galactosidase staining of Bglap-Cre; Rosa26-lsl-lacZ spine sections at P1 (G, G’) and P28 (H, H’). Bglap-Cre targets TB at P1 (G, G’), but targets TB (H’), CB (H’), and Po (green arrows, H’) at P28. Bglap-Cre also targets hypertrophic cells within the EP and GP (H’). (I–J’) β-Galactosidase staining of ATC; Rosa26-lsl-lacZ (Dox-induced from E0.5–P20) spine sections at P1 (I, I’) and P28 (J, J’). ATC targets most cells in NP, AF, EP, and GP at both time points. Note that ATC did not target the outmost AF (black dash line and arrows, J’), nor the Po (green arrows, J’). (K–L’) β-Galactosidase staining of Scx-Cre; Rosa26-lsl-lacZ spine sections at P1 (K, K’) and P28 (L, L’). Scx-Cre targets AF at P1 (K’) and P28 (L’), and also recombines in several cells of NP, EP, and GP (L’) at P28. Note that Scx-Cre also targets some cells of the outmost AF (black arrows, L’) and the Po (green arrows, L’) at P28. An illustration (M) and a bright-field image (M’) of the dorsal side of a wild-type mouse show the supraspinous ligaments (black arrows, M, M’). (N–Q’) Whole-mount β-galactosidase staining of Rosa26-lsl-lacZ reporter mice at P1. Dorsal view of Col2a1-Cre; Rosa26-lsl-lacZ mouse is shown in (N, N’). Supraspinous ligaments are indicated with black arrows (N’). Dorsal view of ATC; Rosa26-lsl-lacZ mouse (Dox-induced from E0.5) is shown in (O, O’). Facet joints are indicated with red arrows (O’). Dorsal and sagittal views of the whole-mount Scx-Cre; Rosa26-lsl-lacZ mice are shown in (P, P’) and (Q, Q’), respectively. Supraspinous ligaments are indicated with black arrows (P’, Q’). n = 3 mice in each group. Scale bars: 100 μm in (A–C) and (E–L’); 1 mm in (M’–Q’). AF: annulus fibrosis; EP: endplate; GP: growth plate; NP: nucleus pulposus; TB: trabecular bone; CB: cortical bone; Po: periosteum; Lg: ligament; Td: tendon.
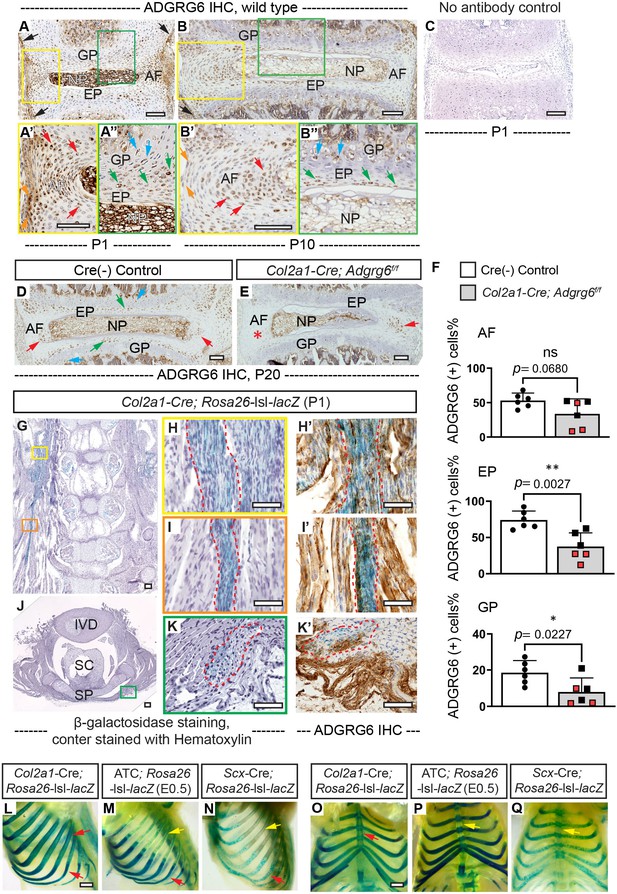
Immunohistochemistry (IHC) analyses of ADGRG6 and β-galactosidase staining of Rosa26-lsl-lacZ reporter mice recombined with different Cre strains.
(A–C) IHC analyses of ADGRG6 on thoracic spine sections of wild-type mice at P1 and P10. ADGRG6 is mainly expressed in AF (red arrows, A’, B’), NP (A”, B’’), EP (green arrows, A”, B’’), and GP (blue arrows, A”, B’’). Note that ADGRG6 is also expressed in the outermost AF (orange arrows, A', B’) and the periosteum (black arrows, A). No antibody control staining is shown in (C). n = 3 mice. (D–F) IHC analyses of ADGRG6 on thoracic spine sections of Cre (-) control and scoliotic Col2a1-Cre; Adgrg6f/f mice at P20. ADGRG6 expressed is reduced in EP and GP of the mutants (E) compared with that in the controls (green arrow and blue arrows, D). ADGRG6 expression in the AF of the convex side of the Col2a1-Cre; Adgrg6f/f mice (red asterisk, E) is also reduced compared with that in the controls (red arrows, D), though ADGRG6 expression in the AF of the concave side is relatively normal (red arrow, E). The percentile of ADGRG6 (+) cells were quantified in (F). Cre (-) control mice, n = 6 mice; Col2a1-Cre; Adgrg6f/f mutant mice, n = 6 mice (three scoliotic mice, three non-scoliotic mice). Bars are plotted with mean and 95% CI. Each dot represents one mouse analyzed. Scoliotic Col2a1-Cre; Adgrg6f/f mutant mice are marked in red in (F). The statistical difference is evaluated by a two-tailed Student's t-test. The p-value for each comparison is shown. ns: not significant. (H–L’) Coronal (G–I’) and transverse (J–K’) sections of the Col2a1-Cre; Rosa26-lsl-lacZ mouse spine as shown in Figure 1N. Blue signal represents recombination within the ligamentous tissues, which is also outlined with red dot lines in (H–K’). IHC analyses of ADGRG6 performed on adjacent sections of (H), (I), and (K) are shown in (H’), (I'), and (K’). Most cells that are targeted by Col2a1-Cre (blue signal) also express ADGRG6 (brown signal). n = 3 mice in each group. (L–Q) Whole-mount β-galactosidase staining of Rosa26-lsl-lacZ reporter mice at P1. Sagittal and ventral views of Col2a1-Cre; Rosa26-lsl-lacZ mouse (L, O) and ATC; Rosa26-lsl-lacZ mouse (M, P, induced from E0.5) show robust recombination within cartilaginous tissues of the rib cage (red arrows, L, M). Sagittal and ventral views of Scx-Cre; Rosa26-lsl-lacZ mouse (N, Q) only show some recombination signals within cartilaginous portions of the rib cage (red arrow, N). Note that Col2a1-Cre targets both bony and cartilaginous portions of the ribs (red arrows, L), while ATC and Scx-Cre do not target the bony ribs (yellow arrows, M, N). In addition, the sternum is only targeted by Col2a1-Cre (red arrow, O), but not ATC or Scx-Cre (yellow arrows, P, Q). n = 3 mice in each group. Scale bars: 100 μm in (A–E) and (G–K’); 1 mm in (L, O). AF: annulus fibrosis; EP: endplate; GP: growth plate; NP: nucleus pulposus; IVD: intervertebral disc; SC: spinal cord; SP: spinous process.
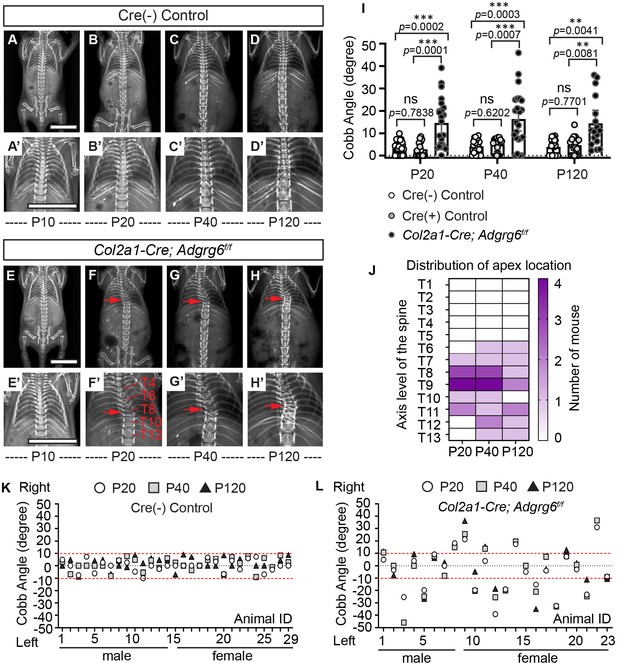
Ablation of Adgrg6 in osteochondral progenitor cells models progressive adolescent idiopathic scoliosis (AIS) in mouse.
(A–D’) Longitudinal X-ray analysis of a representative Cre (-) control mouse at P10 (A, A’), P20 (B, B’), P40 (C, C’), and P120 (D, D’). (E–H’) Longitudinal X-ray analysis of a representative Col2a1-Cre; Adgrg6f/f mutant mouse at P10 (E, E’), P20 (F, F’), P40 (G, G’), and P120 (H, H’), showing adolescent-onset (F) and progressive (F–H) thoracic scoliosis, with the apex of scoliosis indicated (red arrows; F–H’). Thoracic (T) vertebrae are labeled in (F’). (I) Longitudinal Cobb angle analysis; Cre (-) control mice, n = 29 mice; Cre (+) control mice, n = 18, 17, and 18 mice at P20, P40, and P120, respectively; and Col2a1-Cre; Adgrg6f/f mutant mice, n = 23, 23, and 17 mice at P20, P40, and P120, respectively. Dots are plotted with mean ± 95% CI. The statistical difference is evaluated by two-way ANOVA followed by Tukey's multiple comparison test. The p-value for each comparison is shown. (J) Heat map of the apex distribution of the scoliotic Col2a1-Cre; Adgrg6f/f mice at P20, P40, and P120. The heat map is plotted with the axis level of the thoracic (T) spine (T1–T13, left axis) and the number of mice with scoliosis (right axis) with apex observed at each level. The apex of scoliosis is distributed along the middle to lower thoracic spine (T6–T13), with hotspots at T8 and T9. (K, L) Cobb angle values for all the Cre (-) control mice (K) and Col2a1-Cre; Adgrg6f/f mice (L) showed in (I). Thresholds of scoliosis (Cobb angle >10°) are indicated with two red dot lines. Scale bars: 10 mm.
-
Figure 2—source data 1
Cobb angle measurements of mice with Adgrg6 ablation in osteochondral progenitor cells.
- https://cdn.elifesciences.org/articles/67781/elife-67781-fig2-data1-v3.xlsx
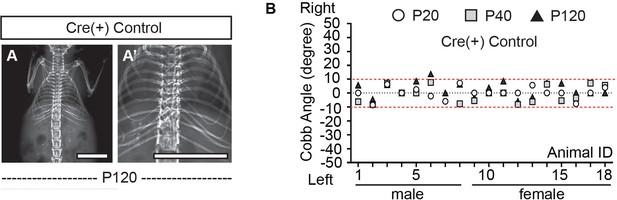
X-ray analysis and Cobb angle measurements of Cre (+) control mice.
(A, A’) X-ray analysis of a representative Cre (+) control mouse at P120. (B) Cobb angle values for all the Cre (+) control mice as shown in Figure 2I. Thresholds of scoliosis (Cobb angle >10°) are indicated with two red dot lines. Only one Cre (+) control mice exhibited scoliosis at P120 (1/18). Scale bars: 10 mm in (A, A’).
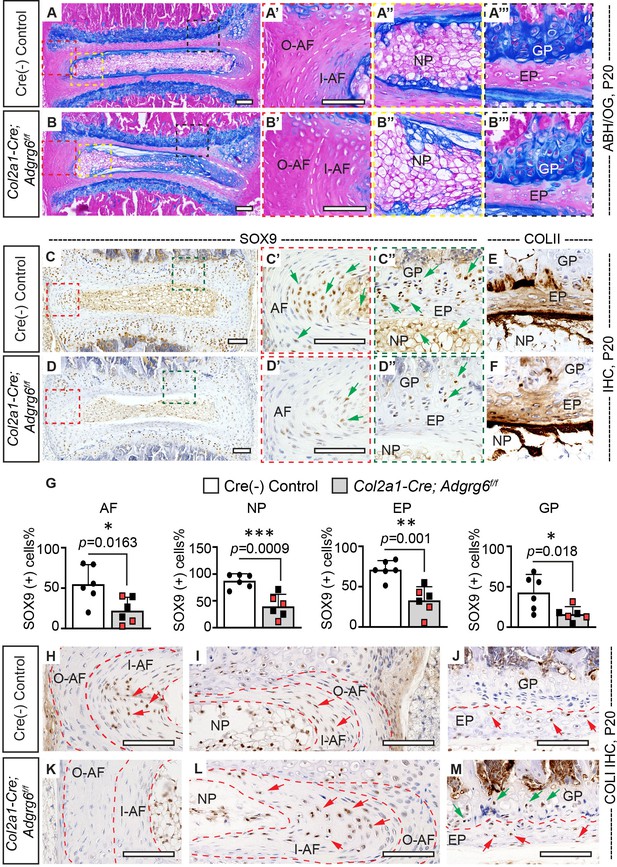
Col2a1-Cre; Adgrg6f/f mice display alterations in intervertebral discs (IVD).
(A–B’’’) Midline sectioned IVD sections from Cre (-) control (A–A’’’) and scoliotic Col2a1-Cre; Adgrg6f/f mutant mice (B–B’’’) stained with Alcian Blue Hematoxylin/Orange G (ABH/OG) at P20. The IVD close to the apex of the curve in the Col2a1-Cre; Adgrg6f/f mice is mildly wedged, associated with a shift in the position of the nucleus pulposus towards the convex side of the curve (B, B"). The inner AF (I–AF) and outer AF (O–AF) in the convex side of the Col2a1-Cre; Adgrg6f/f mice are composed of more vertical lamellae compared with that in Cre (-) control mice (white dash lines, A’, B’). The AF in the concave side of the Col2a1-Cre; Adgrg6f/f mice is elongated and composed with flatter lamellae, compared with that in Cre (-) control mice (white dash lines, A, B). No overt structural defects are observed in the EP and GP of the Col2a1-Cre; Adgrg6f/f mice (B’’’). Cre (-) control mice, n = 4 mice; scoliotic Col2a1-Cre; Adgrg6f/f mutant mice, n = 3 mice. (C–G) Immunohistochemistry (IHC) analyses of common anabolic markers of healthy IVD at P20. Col2a1-Cre; Adgrg6f/f mutant mice display reduced expression of SOX9 in AF, EP, and GP (green arrows, C’–D”), which is quantified in (G). Col2a1-Cre; Adgrg6f/f mice show a normal expression pattern of COLII compared with the controls (E, F). Cre (-) control mice, n = 6 mice; Col2a1-Cre; Adgrg6f/f mutant mice, n = 6 mice (three scoliotic mice, three non-scoliotic mice). Bars are plotted with mean and 95% CI. Each dot represents one mouse analyzed. Scoliotic Col2a1-Cre; Adgrg6f/f mutant mice are marked in red in (G). The statistical difference is evaluated by a two-tailed Student's t-test. The p-value for each comparison is shown. (H–M) IHC analyses of COLI at P20. COLI is mainly expressed in the I-AF (H, I) and some cells of the EP (J) in the control mice (indicated with red arrows). No obvious expression of COLI is observed in the O-AF or I-AF in the convex side of the mutant IVD (K), but robust expression is observed in the elongate I-AF in the concave side of the mutant IVD (red arrows, L). Notably, some cells in the GP of the Col2a1-Cre; Adgrg6f/f mice also express COLI (green arrows, M). Cre (-) control mice, n = 4 mice; scoliotic Col2a1-Cre; Adgrg6f/f mutant mice, n = 3 mice. Scale bars: 100 μm. AF: annulus fibrosis; I-AF: inner annulus fibrosis; O-AF: outer annulus fibrosis; EP: endplate; GP: growth plate; NP: nucleus pulposus.
-
Figure 3—source data 1
Quantification of SOX9 positive cells.
- https://cdn.elifesciences.org/articles/67781/elife-67781-fig3-data1-v3.xlsx
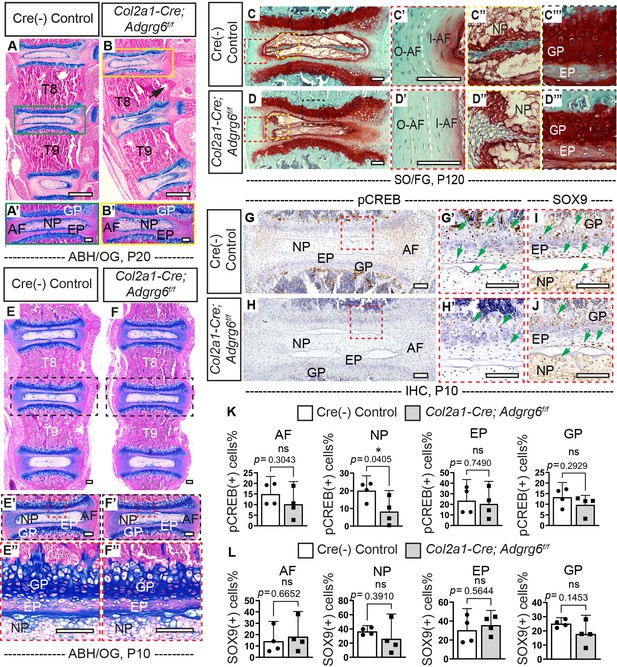
Loss of Adgrg6 in osteochondral progenitor cells leads to alternations in spinal elements.
(A–B’) Representative thoracic spine (T8–T9) and intervertebral disc (IVD) tissues of Cre (-) control and scoliotic Col2a1-Cre; Adgrg6f/f mutant mice stained with Alcian Blue Hematoxylin/Orange G (ABH/OG) at P20. The mutant IVD outside of the spine curve (B’) was comparable with the control IVD (A’). The black arrow indicates the cortical bone bulged to the concave side of the curvature in the apical region of the mutant spine (B). n = 4 mice for each group. (C–D’’’) Representative IVD tissues of Cre (-) control and scoliotic Col2a1-Cre; Adgrg6f/f mice stained with Safranin-O/Fast green (SO/FG) at P120. We can observe exacerbated phenotype of wedged IVD (D), more vertical lamellae in the I-AF and O-AF (white dash line, D’), sifted nucleus pulposus (D”), and disorganized EP and GP (D’’’) in the mutant mice. n = 3 mice for each group. (E–F’’) Representative thoracic spine (T8–T9) and IVD tissues of Cre (-) control and Col2a1-Cre; Adgrg6f/f mutant mice stained with ABH/OG at P10. The mutant mice show normal spine development and IVD structure compared with that of the control mice (E’–F’’). n = 4 mice for each group. (G–J) Immunohistochemistry (IHC) analyses of pCREB (G–H’) and SOX9 (I, J) on thoracic spine sections of Cre (-) control and Col2a1-Cre; Adgrg6f/f mutant mice at P10. pCREB expression was significantly reduced in the NP of the mutant mice (G–H’, K). The expression pattern of SOX9 is not changed in the mutant mice at this time point (I, J, L). The percentile of pCREB (+) cells and SOX9 (+) cells are quantified in (K) and (L), respectively. n = 4 mice for each group. The statistical difference is evaluated by a two-tailed Student's t-test. The p-value for each comparison is shown. ns: not significant. Scale bars: 100 μm in (A–J). AF: annulus fibrosis; I-AF: inner annulus fibrosis; O-AF: outer annulus fibrosis; EP: endplate; GP: growth plate; NP: nucleus pulposus.
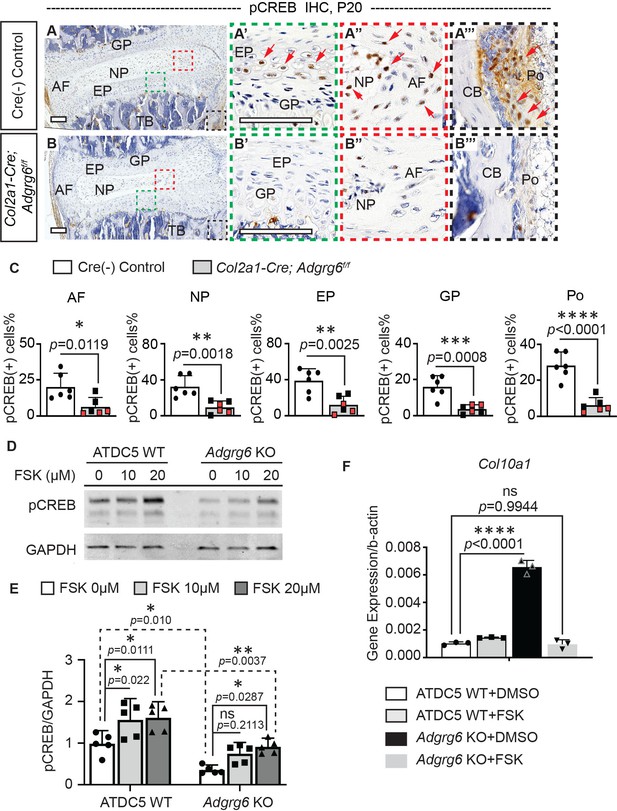
ADGRG6 regulates cAMP/CREB signaling-dependent gene expression in cartilaginous lineages.
(A–C) Immunohistochemistry (IHC) analyses of pCREB in mouse spine sections at P20. Cre (-) control mice exhibit pCREB-positive cells in AF, NP, EP, GP, and Po (red arrows, A–A’’’). Col2a1-Cre; Adgrg6f/f mutant mice show reduced expression of pCREB in all these tissues (B–B’’’). pCREB-positive cells are quantified in (C). Cre (-) control mice, n = 6 mice; Col2a1-Cre; Adgrg6f/f mutant mice, n = 6 mice (three scoliotic mice, three non-scoliotic mice). Bars are plotted with mean and 95% CI. Each dot represents one mouse analyzed. Scoliotic Col2a1-Cre; Adgrg6f/f mutant mice are marked in red in (C). The statistical difference is evaluated by a two-tailed Student's t-test. The p-values for each comparison are shown. (D, E) Representative western blot image (D) and densitometry of the western blot images (E) on pCREB in both wild-type (WT) and Adgrg6 KO ATDC5 cell lysates. The expression level of pCREB is significantly decreased in Adgrg6 KO cells compared with ATDC5 WT cells. Treatment with 20 μM of Forskolin (FSK) for 30 min can stimulate pCREB expression in both ATDC5 WT cells and Adgrg6 KO cells, though the induction level in Adgrg6 KO cells is significantly lower than that in ATDC5 WT cells (E). n = 5 biological replicates. Bars are plotted with mean and 95% CI. The statistical difference is evaluated by two-way ANOVA followed by Tukey’s multiple comparison test. The p-value for each comparison is shown. ns: not significant. (F) Real-time RT-PCR analyses of Col10a1 in ATDC5 WT cells and Adgrg6 KO cells, cultured with FSK (2 µM) or DMSO control for 7 days with maturation medium. The increased expression of Col10a1 in Adgrg6 KO cells was rescued by FSK treatment. n = 3 biological replicates, and the representative result is shown. Bars were plotted with mean and 95% CI. The statistical difference is evaluated by one-way ANOVA followed by Tukey's multiple comparison test. The p-value for each comparison is shown. ns: not significant. Scale bars: 100 μm. AF: annulus fibrosis; EP: endplate; GP: growth plate; NP: nucleus pulposus; TB: trabecular bone; CB: cortical bone; Po: periosteum.
-
Figure 4—source data 1
Quantification of pCREB expression.
- https://cdn.elifesciences.org/articles/67781/elife-67781-fig4-data1-v3.xlsx
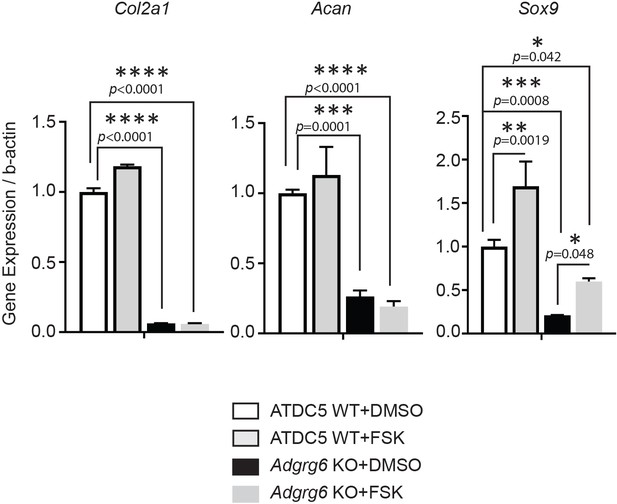
Forskolin treatment can partially rescue the dysregulation of Sox9 expression but not Col2a1 and Acan expression in Adgrg6 KO cells.
Real-time RT-PCR analyses of Col2a1, Acan, and Sox9 in wild-type ATDC5 cells (ATDC5 WT) and Adgrg6 knockout ATDC5 cells (Adgrg6 KO), cultured with 2 µM of Forskolin (FSK) or DMSO control for 7 days with maturation medium. The decreased expression of Col2a1 and Acan in Adgrg6 KO cells was not rescued by FSK treatment. The decreased expression of Sox9 was partially rescued by FSK treatment. n = 3 biological replicates, and the representative result is shown. Bars are plotted with mean and SD. The statistical difference is evaluated by one-way ANOVA followed by Tukey's multiple comparison test. The p-value for each comparison is shown.
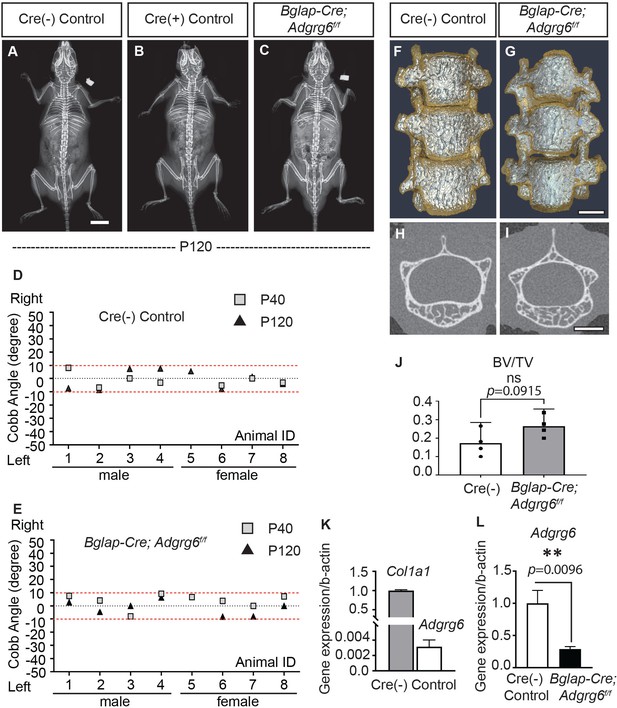
Loss of Adgrg6 in mature osteoblast lineages is dispensable of adolescent idiopathic scoliosis (AIS) development.
(A–C) Representative X-ray images of Cre (-) control (A), Cre (+) control (B), and Bglap-Cre; Adgrg6f/f mutant (C) mice at P120. (D, E) Longitudinal analyses of Cobb angle values of Cre (-) control mice (D) and Bglap-Cre; Adgrg6f/f mice (E) at P40 and P120. Cre (-) control mice, n = 8 mice at P40 and P120; Bglap-Cre; Adgrg6f/f mutant mice, n = 8 and 7 at P40 and P120, respectively. Thresholds of scoliosis (Cobb angle >10°) are indicated with two red dot lines. No Bglap-Cre; Adgrg6f/f mice showed scoliosis at P40 (0/8) or P120 (0/7) (E). (F–J) MicroCT scanning of the thoracic region of the spine shows normal morphology of the vertebral bodies in both Cre (-) controls (F) and the Bglap-Cre; Adgrg6f/f mice (G). Transverse sections of the microCT three-dimensional reconstruction of the thoracic vertebral body show a comparable bone mass in the control (H) and mutant mice (I). The bone volume per total volume (BV/TV) of the control and mutant mice is shown in (J). n = 4 mice for each group. Bars are plotted with mean and 95% CI. The statistical difference is evaluated by a two-tailed Student's t-test. The p-value is shown. ns: not significant. (K, L) Real-time RT-PCR analysis of RNA isolated from long bone shows that the expression of Adgrg6 is very low in bony tissues compared with the expression of Col1a1 (K). However, the expression of Adgrg6 was efficiently knockdown in Bglap-Cre; Adgrg6f/f mice (L). RNA was isolated and pooled from three mice of each experimental group. Bars are plotted with mean and SD. The statistical difference is evaluated by a two-tailed Student's t-test. The p-value for each comparison is shown. Scale bars: 10 mm in (A); 1 mm in (G) and (I).
-
Figure 5—source data 1
Characterization of mice with Adgrg6 ablation in mature osteoblast lineages.
- https://cdn.elifesciences.org/articles/67781/elife-67781-fig5-data1-v3.xlsx
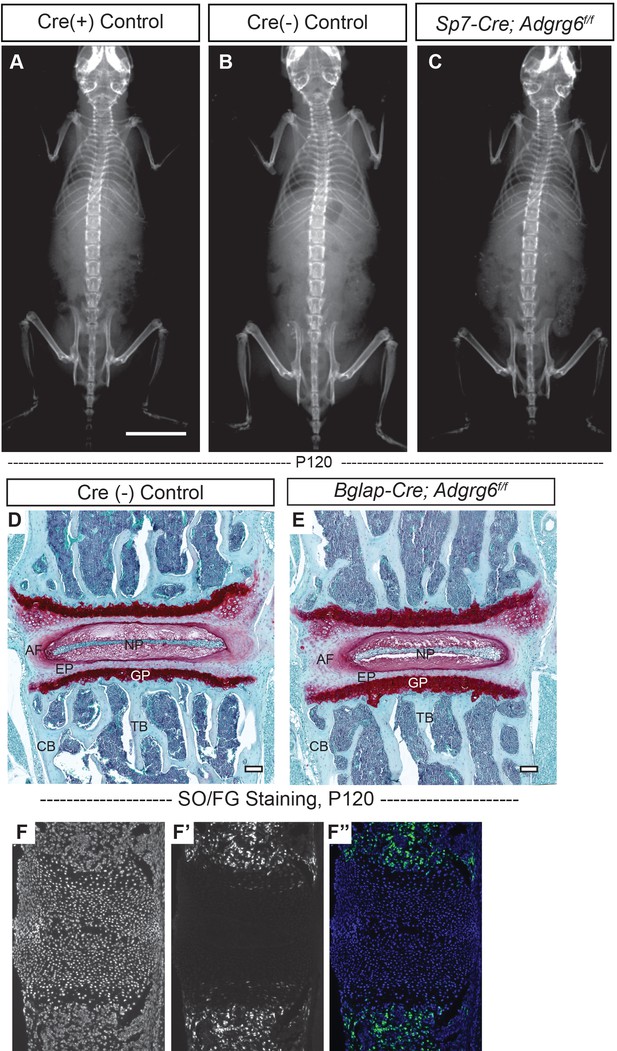
Loss of Adgrg6 in osteoblast lineages leads to no scoliosis or spinal deformity.
(A–C) Representative X-ray images of Cre (+) control (A), Cre (-) control (B), and Sp7-Cre; Adgrg6f/f mutant (C) mice at P120. n = 6, 5, 9 for Cre (+) control mice, Cre (-) control mice, and Sp7-Cre; Adgrg6f/f mice, respectively. (D, E) Representative intervertebral disc (IVD) and vertebral body sections of Cre (-) control and Bglap-Cre; Adgrg6f/f mutant mice stained with Safranin-O/Fast green (SO/FG) at P120. (F–F’’) Midline sectioned P1 mouse spine showing Sp7-Cre-GFP expression specific to the vertebral body (F’, F’’), counterstained with DAPI (F). n = 3 mice for each group. Scale bars: 10 mm in (A–C); 100 μm in (D, E). AF: annulus fibrosis; EP: endplate; GP: growth plate; NP: nucleus pulposus; TB: trabecular bone; CB: cortical bone.
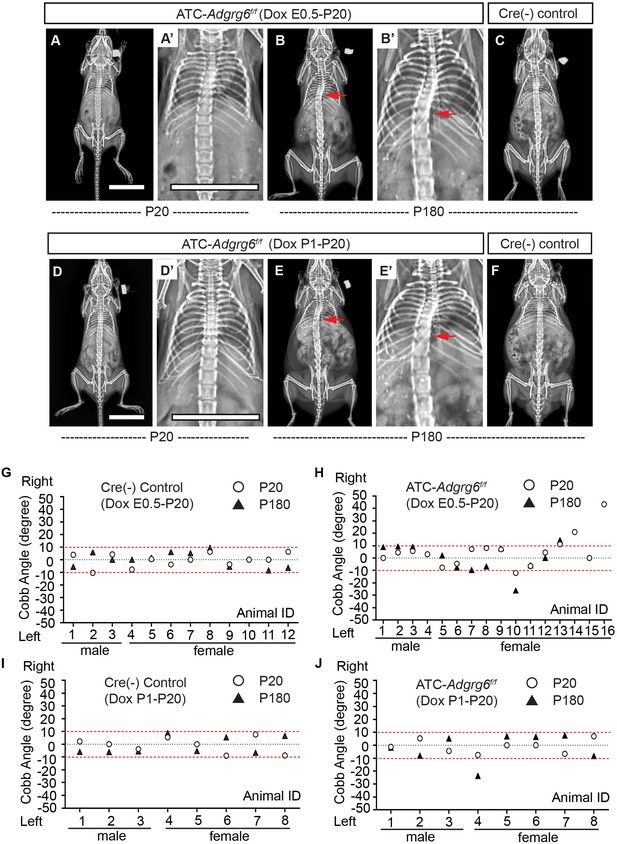
Ablation of Adgrg6 in cartilaginous tissues leads to scoliosis in the mouse.
(A–F) Representative X-ray images of Cre (-) control and ATC; Adgrg6f/f mutant mice at P20 and P180. ATC; Adgrg6f/f mice (Dox induction from E0.5–P20) analyzed at P20 and P180 are shown in (A, A’) and (B, B’), respectively. ATC; Adgrg6f/f mice (Dox induction from P1–P20) were analyzed at P20 (D, D’) and P180 (E, E’). Corresponding Cre (-) control mice analyzed at P180 is shown in (C) and (F). Scoliosis is indicated with red arrows in (B, B’) and (E, E’). (G–J) Longitudinal analyses of Cobb angle values of Cre (-) control mice and ATC; Adgrg6f/f mice at P20 and P180. For embryonic induction (E0.5–P20), n = 12 mice for Cre (-) controls at P20 and P120; n = 16 and 12 for ATC; Adgrg6f/f mice at P20 and P180, respectively. For perinatal induction (P1–P20), n = 8 mice for Cre (-) control at P20 and P180; n = 8 for ATC; Adgrg6f/f mice at P20 and P180. Thresholds of scoliosis (Cobb angle >10°) are indicated with two red dot lines. Scale bars: 10 mm.
-
Figure 6—source data 1
Cobb angle measurements of mice with Adgrg6 ablation in cartilaginous tissues.
- https://cdn.elifesciences.org/articles/67781/elife-67781-fig6-data1-v3.xlsx
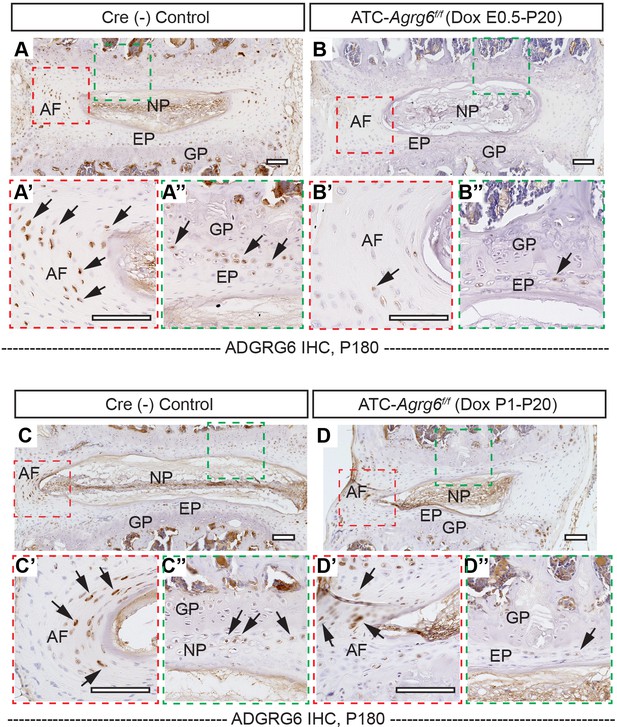
ADGRG6 expression is dramatically reduced in ATC; Adgrg6f/f mice.
Representative immunohistochemistry (IHC) analyses of ADGRG6 on spine sections of Cre (-) control mouse (induced from E0.5–P20, A–A’’’; induced from P1–P20, C–C’’’) and ATC; Adgrg6f/f mouse (induced from E0.5–P20, B–B’’’); induced from P1–P20 (D–D’’’). ADGRG6 expression is dramatically reduced in ATC; Adgrg6f/f mice with both induction strategies. Note that there is more retention of ADGRG6 expression in the AF of the mutant mice when induced from P1–P20 (black arrows, D’) than induced from E0.5–P20 (B’). n = 3 mice for each group. Scale bars: 100 μm. AF: annulus fibrosis; EP: endplate; GP: growth plate; NP: nucleus pulposus.
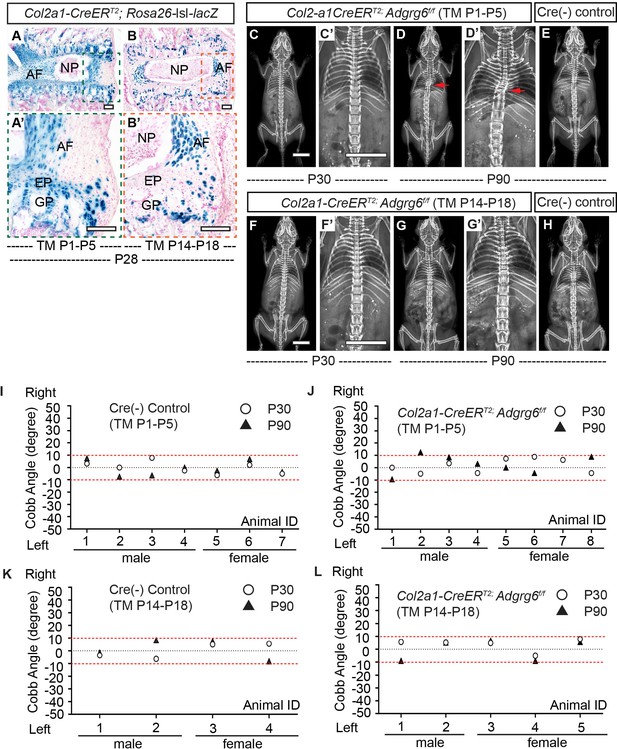
Loss of Adgrg6 in cartilaginous tissues leads to low penetrance of scoliosis.
(A–B’) β-Galactosidase staining of Col2a1-CreERT2; Rosa26-lsl-lacZ spine sections that were inducted from P1–P5 (A, A’) or P14–P18 (B, B’) at P28. Note that Col2a1-CreERT2 targets EP, GP, and inner AF, but does not target NP or outer AF with neither induction strategy (A’, B’). (C–H) Representative X-ray images of Col2a1-CreERT2; Adgrg6f/f mutant mice (TM induced from P1–P5 or P14–P18) at P30 (C, C’ or F, F’, respectively) and P90 (D, D’ or G, G’, respectively). Corresponding Cre (-) controls at P90 are shown in (E) and (H). Scoliosis is indicated with red arrows in (D, D’). (I–L) Longitudinal analyses of Cobb angle values of Cre (-) control mice and Col2a1-CreERT2; Adgrg6f/f mice at P30 and P90 with two induction strategies. For TM induced from P1–P5, n = 7 mice for Cre (-) controls at both P30 and P90; n = 8 and 7 mice for Col2a1-CreERT2; Adgrg6f/f mice at P30 and P90, respectively. For TM induction from P14–P18, n = 4 mice for Cre (-) controls at both P30 and P90; n = 5 for Col2a1-CreERT2; Adgrg6f/f mice at both P30 and P90. Thresholds of scoliosis (Cobb angle >10°) are indicated with two red dot lines. Scale bars: 100 μm in (A–B’); 10 mm in (C, C’) and (F, F’). AF: annulus fibrosis; EP: endplate; GP: growth plate; NP: nucleus pulposus.
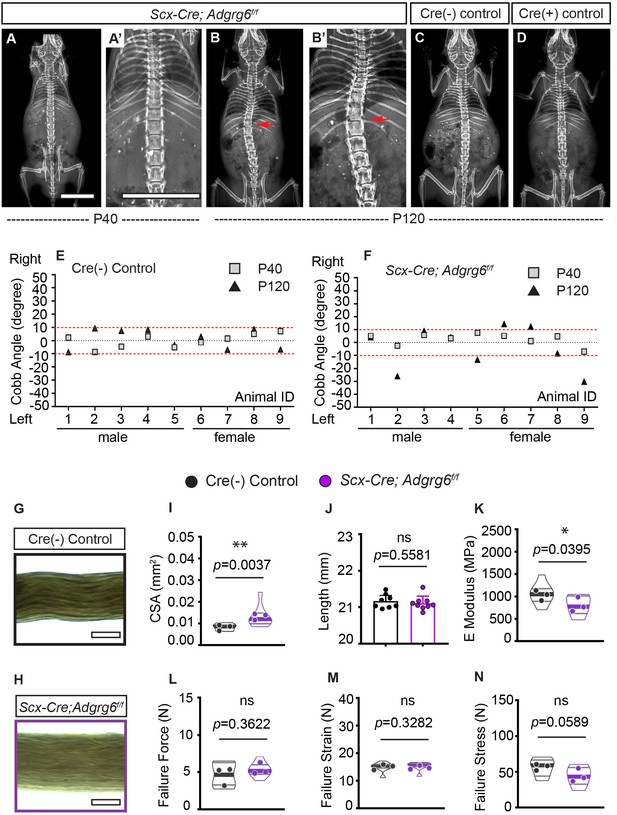
Ablation of Adgrg6 in dense connective tissues leads to late-onset scoliosis and compromised biomechanical properties of the tendons.
(A–D) Representative X-ray images of Cre (-) control and Scx-Cre; Adgrg6f/f mutant mice. Scx-Cre; Adgrg6f/f mice were analyzed at P40 (A, A’) and P120 (B, B’). Cre (-) control and Cre (+) control mice analyzed at P120 are shown in (C) and (D), respectively. Scoliosis is indicated with red arrows in (B, B’). (E, F) Longitudinal analyses of Cobb angle values of Cre (-) control mice and Scx-Cre; Adgrg6f/f mice at P40 and P120. Cre (-) control mice, n = 9 mice at both P40 and P120; Scx-Cre; Adgrg6f/f mutant mice, n = 9 at both P40 and P120. Thresholds of scoliosis (Cobb angle >10°) are indicated with two red dot lines. (G–N) Biomechanical characterization of Cre (-) control and Scx-Cre; Adgrg6f/f mutant tendons at 12 weeks. Representative phase-contrast images of tail fascicles isolated from Cre (-) control and Scx-Cre; Adgrg6f/f mutant mice are shown in (G) and (H). Quantification of fascicles’ cross-sectional area (CSA) (I), initial length (J), elastic modulus (E Modulus) (K), failure force (L), failure strain (M), and failure stress (N) are also shown. n = 8–9 fascicles isolated from three mice. For (I) and (K–N), n = 8–9 fascicles (violin plots) isolated from three different mice (close circles). For (J), n = 8 and 9 fascicles isolated from Cre (-) control mice or Scx-Cre; Adgrg6f/f mutant mice, respectively. Bars are plotted with mean and 95% CI. Cross-sectional area (I) (mean: Cre [-] control: 0.008 [95% CI 0.0069, 0.0095]; Scx-Cre; Adgrg6f/f: 0.013 [95% CI 0.0094, 0.0169]) and elastic modulus (K) (mean: Cre [-] control: 1060 [95% CI 863.8, 1256]; Scx-Cre; Adgrg6f/f: 810.9 [95% CI 644.6, 977.3]) were significantly different between control and mutant groups. The statistical difference is evaluated by unpaired t-test with Welch's correction for (J, K, L, N), and with Mann–Whitney test for non-normally distributed data (I, M). The p-value for each comparison is shown. ns: not significant. Scale bars: 10 mm in (A, A’); 100 μm in (G, H).
-
Figure 7—source data 1
Cobb angle measurements and biomechanical testing of mice with Adgrg6 ablation in dense connective tissues.
- https://cdn.elifesciences.org/articles/67781/elife-67781-fig7-data1-v3.xlsx
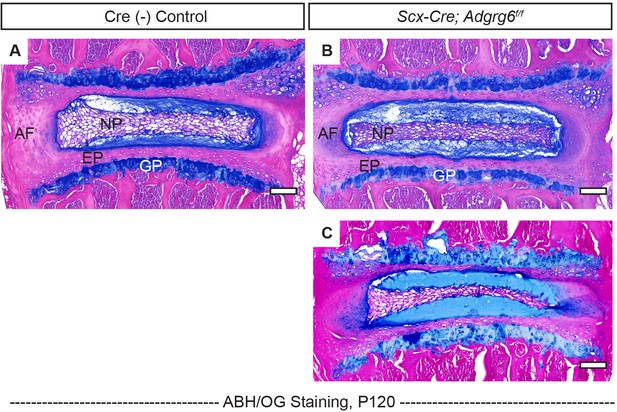
Scx-Cre; Adgrg6f/f mice display mildly wedged intervertebral discs (IVDs) within the curve.
(A–C) Representative IVD tissues of Cre (-) control (A) and Scx-Cre; Adgrg6f/f mutant mice (B, C) stained with Alcian Blue Hematoxylin/Orange G (ABH/OG) at P120. IVD outside of the curve and within the curve is shown in (B) and (C), respectively. Cre (-) control mice, n = 3 mice; Scx-Cre; Adgrg6f/f mutant mice, n = 3 mice (two scoliotic mice, one non-scoliotic mice). Scale bars: 100 μm. AF: annulus fibrosis; EP: endplate; GP: growth plate; NP: nucleus pulposus.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Adgrg6 | GenBank | Gene ID: 215798 | |
| Genetic reagent (Mus musculus) | Adgrg6 conditional knockout | Taconic | Model #: TF0269 | Adgrg6f/f |
| Genetic reagent (Mus musculus) | B6.129S4-Gt(ROSA)26Sortm1Sor/J | Jackson Laboratory | Stock no.: 003474 | Rosa26-lsl-lacZ |
| Genetic reagent (Mus musculus) | ATC | Dy et al., 2012 | A gift from Dr. Véronique Lefebvre | |
| Genetic reagent (Mus musculus) | Col2a1-Cre | Long et al., 2001 | A gift from Dr. Fanxin Long | |
| Genetic reagent (Mus musculus) | Col2a1-CreERT2 | Chen et al., 2007 | A gift from Dr. Matthew Hilton | |
| Genetic reagent (Mus musculus) | Scx-Cre | Blitz et al., 2009 | A gift from Dr. Ronen Schweitzer | |
| Genetic reagent (Mus musculus) | B6.FVB-Tg(BGLAP-cre)1Clem/J | Jackson Laboratory | Stock no.: 019509 | Bglap-Cre |
| Genetic reagent (Mus musculus) | B6.Cg-Tg(Sp7-tTA,tetO-EGFP/cre)1Amc/J | Jackson Laboratory | Stock no.: 006361 | Sp7-Cre |
| Cell line (Mus musculus) | ATDC5 | Sigma | Sigma: 99072806 | |
| Cell line (Mus musculus) | Adgrg6 KO ATDC5 | Liu et al., 2019 | CRISPR constructed to knock down the expression of Adgrg6 in ATDC5 cells | |
| Antibody | Anti-GPCR GPR126 (ADGRG6) (rabbit polyclonal) | Abcam | ab117092 | IHC 1:500 |
| Antibody | Anti-SOX9 (rabbit Polyclonal) | EMD Millipore | AB5535 | IHC 1:200 |
| Antibody | Anti-collagen II Ab-2 (clone 2B1.5) (mouse monoclonal) | Thermo Scientific | MS235B | IHC 1:100 |
| Antibody | Recombinant anti-collagen I antibody (rabbit monoclonal) | Abcam | ab138492 | IHC 1:1000 |
| Antibody | Phospho-CREB (Ser133) (87G3) (rabbit monoclonal) | Cell Signaling | #9198 | IHC: 1:200 WB: 1:1000 |
| Antibody | GAPDH (14C10) | Cell Signaling | #2118 | WB: 1:2500 |
| Antibody | IRDye 800CW Goat anti-Rabbit IgG Secondary Antibody | LI-COR | P/N: 926-32211 | WB: 1:10,000 |
| Sequence-based reagent | b-actin-F | Sigma | qPCR primers | AGATGTGGATCAGCAAGCAG |
| Sequence-based reagent | b-actin-R | Sigma | qPCR primers | GCGCAAGTTAGGTTTTGTCA |
| Sequence-based reagent | Col1a1-F | Sigma | qPCR primers | GCATGGCCAAGAAGACATCC |
| Sequence-based reagent | Col1a1-R | Sigma | qPCR primers | CCTCGGGTTTCCACGTCTC |
| Sequence-based reagent | Adgrg6-F | Sigma | qPCR primers | CCAAAGTTGGCAATGAAGGT |
| Sequence-based reagent | Adgrg6-R | Sigma | qPCR primers | GCTGGATCAGGTAGGAACCA |
| Sequence-based reagent | Col10a1-F | Sigma | qPCR primers | CTTTGTGTGCCTTTCAATCG |
| Sequence-based reagent | Col10a1-R | Sigma | qPCR primers | GTGAGGTACAGCCTACCAGTTTT |
| Sequence-based reagent | Col2a1-F | Sigma | qPCR primers | ACTGGTAAGTGGGGCAAGAC |
| Sequence-based reagent | Col2a1-R | Sigma | qPCR primers | CCACACCAAATTCCTGTTCA |
| Sequence-based reagent | Acan-F | Sigma | qPCR primers | CGTGTTTCCAAGGAAAAGGA |
| Sequence-based reagent | Acan-R | Sigma | qPCR primers | TGTGCTGATCAAAGTCCAG |
| Sequence-based reagent | Sox9-F | Sigma | qPCR primers | AGGAAGCTGGCAGACCAGTA |
| Sequence-based reagent | Sox9-R | Sigma | qPCR primers | CGTTCTTCACCGACTTCCTC |
| Chemical compound, drug | Forskolin | Sigma | Sigma: F6886 | |
| Software, algorithm | GraphPad Prism software | GraphPad Prism (https://graphpad.com) |
Additional files
-
Supplementary file 1
Chondroitin sulfate digestion profile and total hyaluronan content in Cre (-) control and Col2a1-Cre; Adgrg6f/f mutant mice at P20.
Data shows µg estimated for total reaction volume; w/w percentage. CS: chondroitin sulfate; HA: hyaluronan; GAG: glycosaminoglycan; ND: not detected.
- https://cdn.elifesciences.org/articles/67781/elife-67781-supp1-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67781/elife-67781-transrepform-v3.docx





