Augmin deficiency in neural stem cells causes p53-dependent apoptosis and aborts brain development
Figures
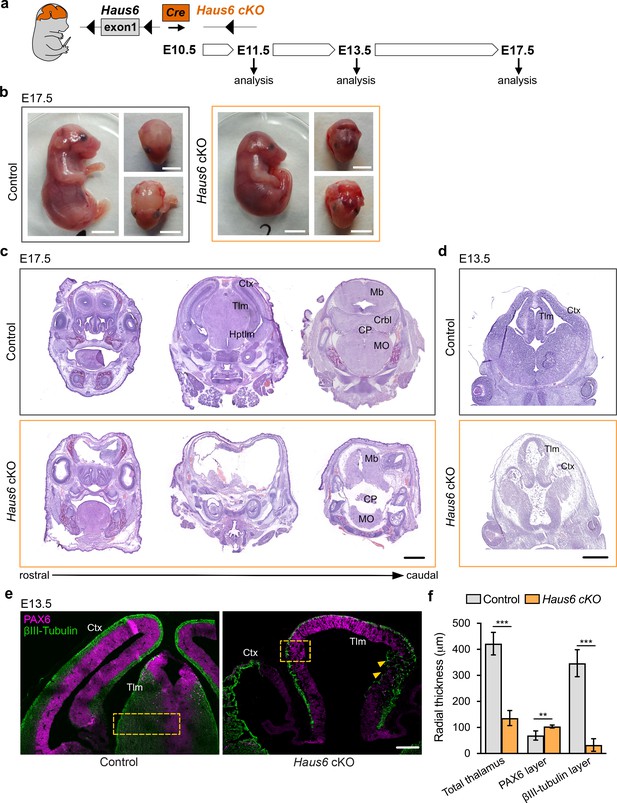
Loss of Haus6 aborts forebrain development.
(a) Schematic representation of the experimental strategy used to evaluate the role of augmin during mouse brain development through generation of brain-specific Nestin-Cre Haus6 cKO embryos. (b) Pictures of E17.5 control (Haus6fl/fl Nestin-Cre−) and Haus6 cKO (Haus6fl/fl Nestin-Cre+) embryos. (c, d) Coronal histological sections from (c) E17.5 and (d) E13.5 control and Haus6 cKO stained with hematoxylin-eosin. Different brain structures are labeled: Ctx (cortex), Tlm (thalamus), Hptlm (hypothalamus), Mb (midbrain), Crbl (cerebellum), MO (medulla oblongata), and CP (choroid plexus). (e) Representative images of the cortex (Ctx) and thalamus (Tlm) of E13.5 control (Haus6fl/wt Nestin-Cre+) and Haus6 cKO (Haus6fl/fl Nestin-Cre+) embryos. Coronal sections were stained against PAX6 (magenta – apical progenitors) and βIII-tubulin (green – neurons). Yellow arrowheads highlight regions of the thalamus where tissue disruption is observed in Haus6 cKO embryos. Yellow boxes indicate the regions used for quantifications in (f). (f) Quantification of the total radial thickness of the thalamus in E13.5 embryos and of layers formed by PAX6- and βIII-tubulin-positive cells. n=3 for control and n=5 for Haus6 cKO embryos. Plotted values are means, error bars show SD. **p<0.01, ***p<0.001 by two-tailed t-test. Scale bars: (b) 5 mm, (c) 1 mm, (d) 0.5 mm, and (e) 150 μm.
-
Figure 1—source data 1
Source data associated with Figure 1f.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig1-data1-v2.xlsx
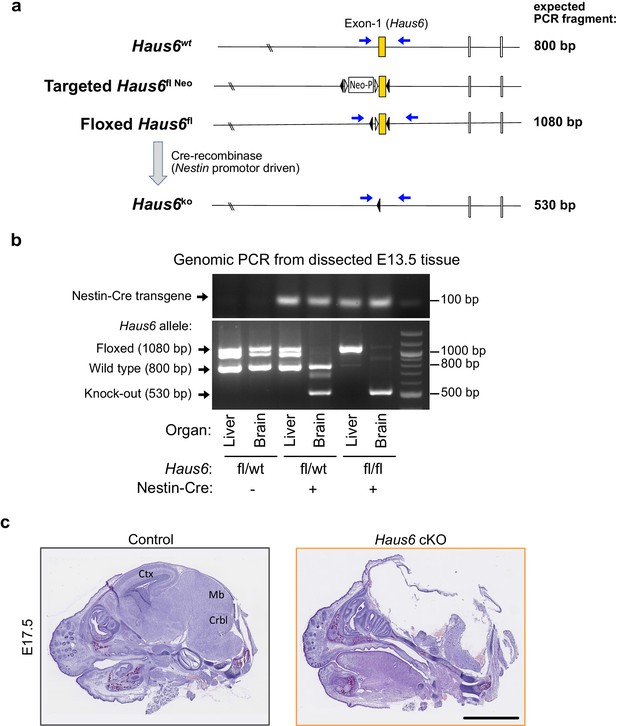
Forebrain structures are absent in Haus6 cKO embryos at E17.5.
(a) Schematic representation of the wild-type Haus6 allele (wt), the targeted allele (fl Neo), the floxed Haus6 allele (fl), and the Haus6 knockout allele (ko). Positions of exons 1–3, loxP sequences (black triangles), flippase recognition target (FRT) sequences (white triangles) and neomycin resistance cassette (Neo) are shown. Positions of polymerase chain reaction (PCR) primers are indicated by blue arrows and expected PCR fragments are indicated. (b) DNA gels showing genomic PCRs from dissected liver and forebrain of E13.5 embryos with different genotypes. In the upper gel, amplification of a 100 bp fragment using Nestin-Cre recombinase primers indicates the presence of the transgene. In the lower gel, amplification of a 1080, 800, and/or 570 bp fragment reveals the presence of Haus6 floxed, wild type, or knockout alleles, respectively, in the indicated genotypes and tissues. The band that is visible below the 1080 bp band in heterozygous samples is an artifact that results from heteroduplexes of floxed (1080 bp) and wild-type (530 bp) DNA strands due to extended regions of complementary. Genomic PCR products were run on a 3% agarose gel. (c) Sagittal histological sections from E17.5 control (Haus6fl/fl Nestin-Cre−) and Haus6 cKO (Haus6fl/fl Nestin-Cre+) embryos stained with hematoxylin-eosin. Scale bar: (c) 2.5 mm.
-
Figure 1—figure supplement 1—source data 1
Original agarose gel images associated with Figure 1—figure supplement 1b.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig1-figsupp1-data1-v2.pdf
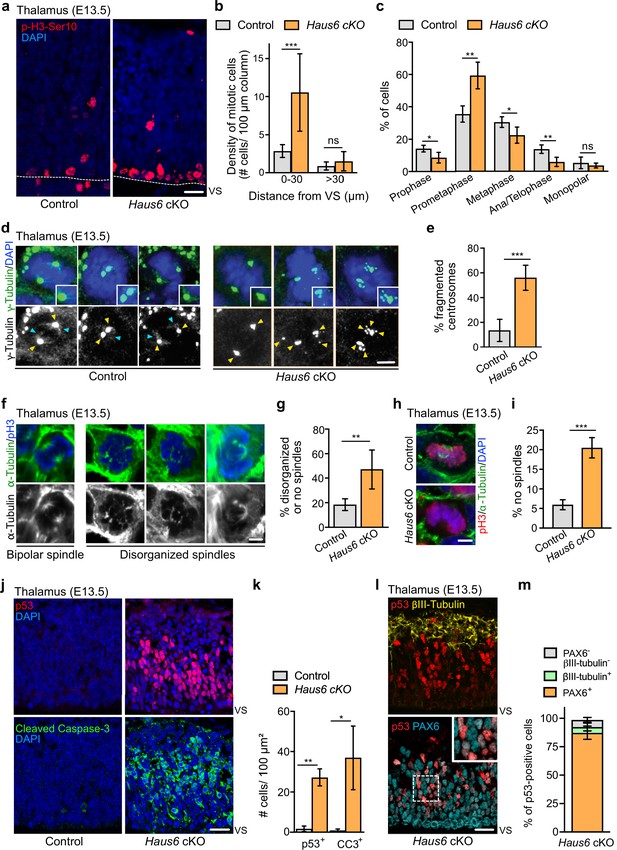
Augmin deficiency in neural progenitors impairs mitotic spindle assembly and induces p53 expression and apoptosis.
(a) Representative images of phospho-Histone H3 (pH3) positive mitotic cells in the thalamus of E13.5 control (Haus6fl/wt Nestin-Cre+) and Haus6 cKO (Haus6fl/fl Nestin-Cre+) embryos. Staining of the mitotic marker pH3-Ser10 in red and DAPI in blue. (b) Quantification of the density of mitotic cells close to the ventricular surface (VS) (<30 µm away) and in outer layers of the cortical plate (>30 µm away). n=4 for control and n=4 for Haus6 cKO embryos (total of 203 and 697 mitotic cells, respectively, in 2–4 sections per embryo). (c) Quantification of mitotic progenitors at different mitotic stages. n=4 for control and n=5 for Haus6 cKO embryos (total of 261 and 427 mitotic cells, respectively, in one section per embryo). (d) E13.5 control and Haus6 cKO coronal thalamus sections stained with antibodies against γ-tubulin (green) and DAPI to label DNA (blue). Yellow arrowheads point to γ-tubulin staining at spindle poles. Light blue arrowheads point to spindle-associated γ-tubulin staining. Insets are 1.4× magnifications of spindle poles in the example cells. (e) Quantification of the percentage of mitotic cells in (d) with fragmented centrosomes. n=5 for control and n=3 for Haus6 cKO embryos (total of 135 and 198 cells counted, respectively, 18–69 cells per embryo). (f) Coronal thalamus sections stained with antibodies against α-tubulin (green) and pH3 (blue). (g) Quantification of the percentage of mitotic progenitors from control and Haus6 cKO E13.5 embryos displaying disorganized or no spindles. n=5 for control and n=3 for Haus6 cKO embryos (total of 152 cells and 90 cells counted, respectively, 27–32 cells per embryo). (h) Examples of Haus6 cKO mitotic progenitors in the E13.5 thalamus in which spindle microtubules, stained with anti-α-tubulin antibodies, cannot be detected, whereas spindles are present in control cells. (i) Quantification of cells as in (h) without detectable spindle microtubules. n=3 for control and n=4 for Haus6 cKO embryos (total of 216 and 243 cells counted, respectively, 40–88 cells per embryo). (j) Representative images of control (Haus6fl/wt Nestin-Cre+) and Haus6 cKO (Haus6fl/fl Nestin-Cre+) coronal thalamus sections stained with an antibody against p53 (red – upper panel) and the apoptotic marker cleaved caspase-3 (green – lower panel). DNA is labeled with DAPI (blue). (k) Quantification of the density of p53- and cleaved caspase-3-positive cells in the E13.5 thalamus in brain sections as shown in (j). n=4 for control and n=3 for Haus6 cKO embryos for quantifications of p53 positive cells and n=3 for control and n=3 for Haus6 cKO embryos for cleaved caspase-3-positive cells (an area between 460 and 3950 µm2 quantified per embryo). (l) Representative images of Haus6 cKO coronal brain sections showing the thalamus stained for p53 (red), PAX6 (cyan), and βIII-tubulin (yellow). The inset is a 1.8× magnification of cells with nuclei staining positive for both p53 and PAX6. (m) Quantification of the percentage of Haus6 cKO cells showing induction of p53 and co-expressing PAX6 (orange), βIII-tubulin (light green), or none of these markers (gray). E13.5 thalamus regions, n=3 for different Haus6 cKO embryos (total of 1020 p53-positive cells). (b, c, e, g, i, k, m) Plotted values are means, error bars show SD. *p<0.05, **p<0.01, ***p<0.001 by two-tailed t-test. Scale bars: (a) 20 μm, (d, f, h) 3 μm, and (j, l) 25 μm.
-
Figure 2—source data 1
Source data associated with Figure 2b, c, e, g, i, k m.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig2-data1-v2.xlsx
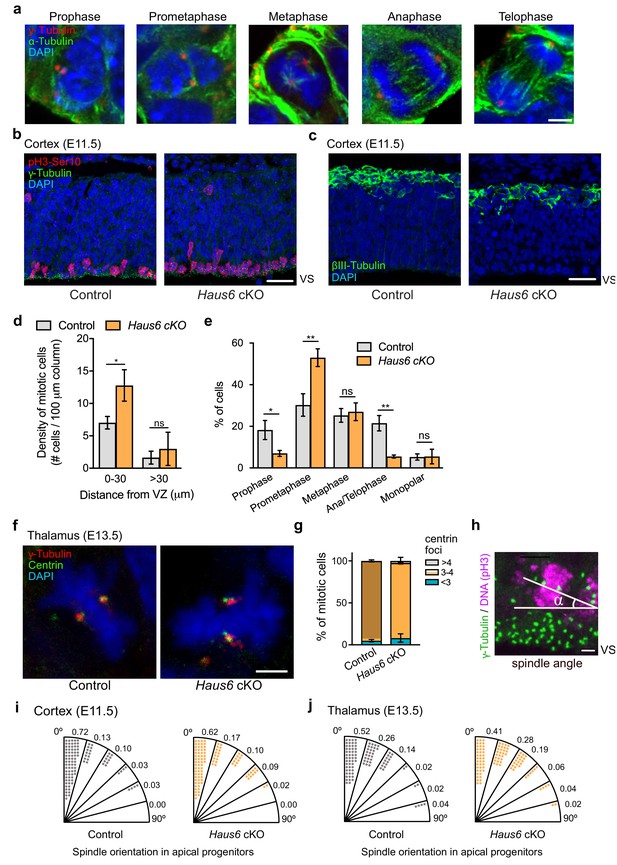
Haus6 cKO induces mitotic defects in the cortex at E11.5.
(a) Representative images of apical progenitors at different mitotic stages. (b) E11.5 control (Haus6wt/wt Nestin-Cre+) and Haus6 cKO cortex (Haus6fl/fl Nestin-Cre+) stained with antibodies against the mitotic marker pH3-Ser10 (red), γ-tubulin (green), and DAPI to label DNA (blue). (c) Sections as in (b) were stained with an antibody against βIII-tubulin (green) and DAPI to label DNA (blue). (d) Quantification of the density of mitotic cells close to the ventricular surface (VS) (<30 µm away) and in the outer layers of the cortical plate (>30 µm away). (e) Quantification of mitotic stages as in (a) observed in cortical progenitors close to the VS. (d, e) n=3 for Nestin-Cre control and n=2 for Nestin-Cre Haus6 cKO individual embryos. (f) Control and Haus6 cKO mitotic progenitors in the E13.5 thalamus stained with antibodies against γ-tubulin and centrin and with DAPI to stain DNA. (g) Quantification of centrioles (centrin foci) in mitotic cells as in (f). The percentage of cells displaying the indicated number of centrin foci was plotted. n=3 for control and n=3 for Haus6 cKO embryos (total of 78 and 114 cells counted, respectively, 19–41 cells per embryo). (h) Image illustrating how spindle orientation was quantified based on the angle formed between the spindle axis and the ventricular lining. (i, j) Quantification of mitotic spindle angles in progenitors at meta/ana/telophase in E11.5 cortex (i) and E13.5 thalamus (j). Diagrams show the distribution of the spindle angle values between 0° and 90°, grouped in 15° intervals. Each dot represents 1% of the analyzed cells. E11.5 cortex: n=84 for control and n=44 for Haus6 cKO mitotic cells from four and two individual embryos, respectively. E13.5 thalamus: n=85 for control and n=113 for Haus6 cKO mitotic cells from four and five individual embryos, respectively. (d,e,g) Plots show mean values, error bars indicate SD. *p<0.05; **p<0.01 by two-tailed t-test. Scale bars: (a, f) 3 µm, (b, c) 30 µm, and (h) 2.5 µm.
-
Figure 2—figure supplement 1—source data 1
Source data associated with Figure 2—figure supplement 1d, e, g, i, j.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig2-figsupp1-data1-v2.xlsx

Haus6 cKO induces apoptosis and cell cycle arrest.
(a) Sections showing E11.5 control and Haus6 cKO E11.5 cortex stained with antibodies against p53 (red) and the apoptotic marker cleaved caspase-3 (green). DNA was stained with DAPI. (b) Immunohistochemistry sections showing E13.5 thalamus stained with antibodies against p21. n=3 for control and n=5 for Haus6 cKO embryos were analyzed (total of 40,460 and 41,063 cells per genotype, respectively, 3799–19,669 cells in 2–3 sections per embryo). (c) The percentage of p21-positive cells was quantified in sections of control and Haus6 cKO embryos. Plots show mean values, error bars indicate SD. *p<0.05 by two-tailed t-test. Scale bars: (a, b) 30 μm.
-
Figure 2—figure supplement 2—source data 1
Source data associated with Figure 2—figure supplement 2c.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig2-figsupp2-data1-v2.xlsx
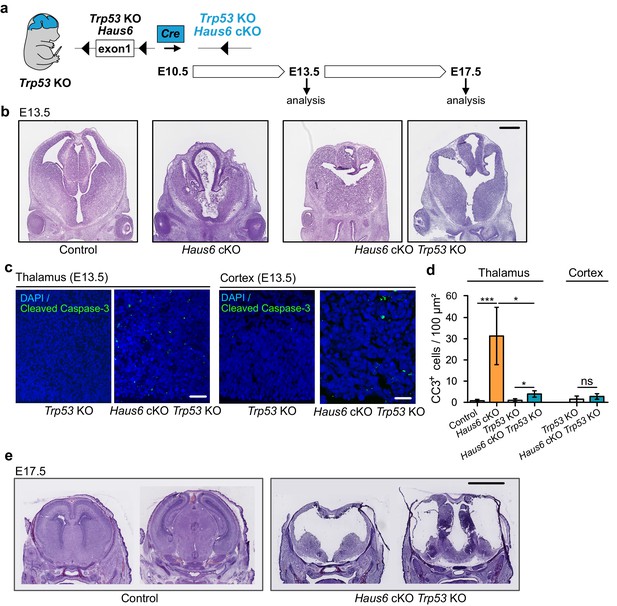
Co-deletion of Trp53 rescues apoptosis but not abortion of forebrain development.
(a) Schematic overview showing the experimental strategy used to generate Haus6 cKO Trp53 KO embryos, to test p53 dependency of the brain development phenotypes observed in Haus6 cKO embryos. (b) Coronal sections of E13.5 control (Haus6fl/wt Nestin-Cre+), Haus6 cKO (Haus6fl/fl Nestin-Cre+), and Haus6 cKO Trp53 KO (Haus6fl/fl Nestin-Cre+ Trp53−/−) embryos stained with hematoxylin-eosin. (c) Coronal sections of the thalamus and cortex of E13.5 Trp53 KO control and Haus6 cKO embryos stained against the apoptotic marker cleaved caspase-3 (green). DNA was labeled by DAPI (blue). (d) Quantification of the density of cleaved caspase-3 positive cells in the E13.5 thalamus and cortex in brain sections as shown in (c). n=5 for control, n=5 for Haus6 cKO, n=4 for Trp53 KO control, and n=3 for Haus6 cKO Trp53 KO embryos for quantifications in the thalamus, and n=4 for Trp53 KO control and n=4 for Haus6 cKO Trp53 KO embryos for quantifications in the cortex. Plotted values are means, error bars show SD. *p<0.05, **p<0.01, ***p<0.001 by two-tailed t-test. (e) Coronal sections of E17.5 control and Haus6 cKO Trp53 KO embryos stained with hematoxylin-eosin. Scale bars: (b) 0.5 mm; (c) 40 μm, 25 μm (cortex); and (e) 2 mm.
-
Figure 3—source data 1
Source data associated with Figure 3d.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig3-data1-v2.xlsx
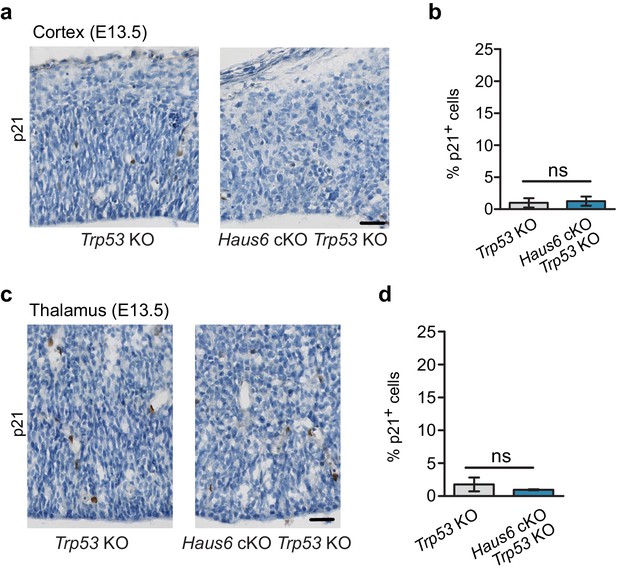
Expression of p21 in augmin-deficient progenitors is rescued by co-deletion of Trp53.
(a) Immunohistochemistry sections showing Trp53 KO (control) and Haus6 cKO Trp53 KO cortex at E13.5 stained with antibodies against p21. (b) Quantification of p21-expressing cells as in (a). The percentage of p21-positive cells was plotted. n=3 for Trp53 KO and n=4 for Haus6 cKO Trp53 KO embryos were analyzed (total of 40,502 and 22,555 cells per genotype, respectively, 3577–16,763 cells in three sections per embryo). (c) Immunohistochemistry sections showing Trp53 KO (control) and Haus6 cKO Trp53 KO thalamus at E13.5 stained with antibodies against p21. (d) Quantification of p21-expressing cells as in (c). The percentage of p21-positive cells was plotted. n=3 for Trp53 KO and n=3 for Haus6 cKO Trp53 KO embryos were analyzed (total of 43,072 and 41,063 cells per genotype, respectively, 2746–23,924 cells in 2–3 sections per embryo). (b, d) Plots show mean values, error bars indicate SD. Scale bars: (a, c) 30 μm.
-
Figure 3—figure supplement 1—source data 1
Source data associated with Figure 3—figure supplement 1b, d.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig3-figsupp1-data1-v2.xlsx
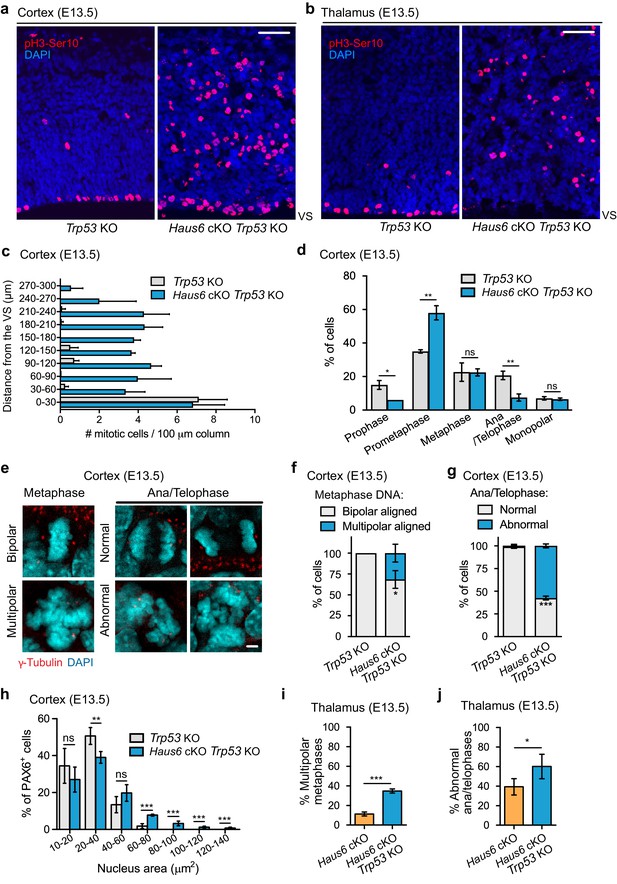
Co-deletion of Trp53 exacerbates mitotic defects caused by augmin deficiency.
(a, b) Representative coronal sections of the (a) cortex and the (b) thalamus of E13.5 Trp53 KO control (Haus6fl/wt Nestin-Cre-Trp53−/−) and Haus6 cKO Trp53 KO embryos (Haus6fl/fl Nestin-Cre+Trp53−/−). Sections were stained with Ser10-phospho-Histone H3 antibody (pH3-Ser10) (red – mitotic cells) and DAPI to stain DNA (blue). (c) Quantification of the density of progenitors undergoing mitosis in the cortex at the indicated distances in µm from the ventricular surface (VS). n=3 for Trp53 KO control and n=2 for Haus6 cKO Trp53 KO embryos (total of 171 and 485 cells, respectively, from four sections per embryo). (d) Quantification of mitotic progenitors at the indicated mitotic stages. n=3 for Trp53 KO control and n=2 for Haus6 cKO Trp53 KO embryos (total of 442 and 443 mitotic cells counted, respectively, 140–248 cells per embryo). (e) Examples of normal, bipolar mitotic stages, and of stages with multipolar and other abnormal configurations in the cortex of E13.5 control and Haus6 cKO Trp53 KO embryos, respectively. Coronal sections were stained with an antibody against γ-tubulin to label spindle poles (red) and DAPI (cyan) to label DNA. (f) Quantification of the percentage of metaphase cells displaying aligned chromosomes with bipolar (white) and multipolar (blue) configuration in the cortex of embryos with the indicated genotypes. n=3 for Trp53 KO control and n=2 for Haus6 cKO Trp53 KO embryos (total of 100 and 97 metaphases counted, respectively, 27–51 metaphases per embryo). (g) Quantification of the percentage of normal and abnormal ana/telophases in the cortex of embryos with the indicated genotypes. n=3 for Trp53 KO control and n=2 for Haus6 cKO Trp53 KO embryos (total of 91 and 33 ana/telophases counted, respectively, 16–34 per embryo). (h) Quantification of the nucleus area in interphase PAX6-positive progenitors in the cortex of E13.5 Trp53 KO control and Haus6 cKO Trp53 KO embryos. n=5 for Trp53 KO control and n=4 for Haus6 cKO Trp53 KO embryos (330–2012 nuclei per embryo). (i) Quantification of the percentage of metaphase cells displaying aligned chromosomes with multipolar configuration in the thalamus of embryos with the indicated genotypes. n=4 for Trp53 KO control and n=3 for Haus6 cKO Trp53 KO embryos (total of 156 and 161 metaphases counted, respectively, 32–81 per embryo). (j) Quantification of the percentage of abnormal ana/telophases in the thalamus of embryos with the indicated genotypes. n=4 for Trp53 KO control and n=3 for Haus6 cKO Trp53 KO embryos (total of 103 and 90 ana/telophases counted, respectively, 17–39 per embryo). (c, d, f, g, h, i, j) Plots show mean values, error bars indicate SD. *p<0.05, **p<0.01, ***p<0.001 by two-tailed t-test. Scale bars: (a, b) 50 μm and (e) 5 μm.
-
Figure 4—source data 1
Source data associated with Figure 4c, d, f, g, h, i, j.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig4-data1-v2.xlsx
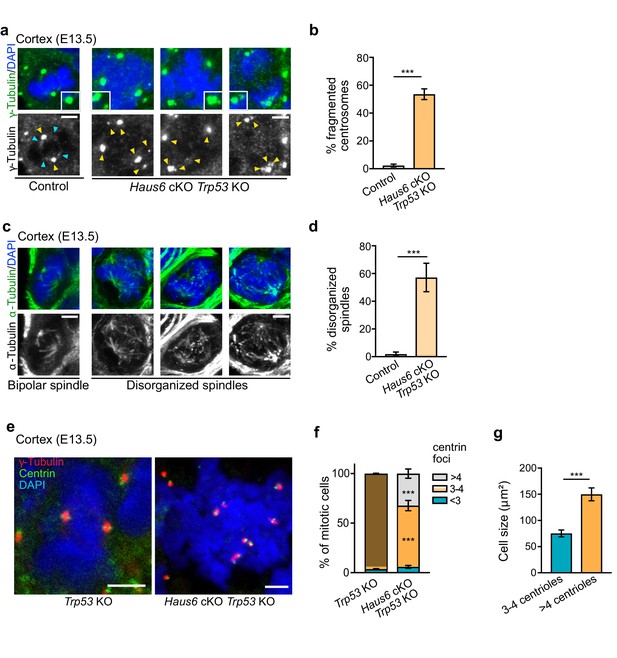
Spindle defects persist in Haus6 cKO Trp53 KO cells.
(a) E13.5 control and Haus6 cKO Trp53 KO coronal cortex sections stained with antibodies against γ-tubulin (green) and DAPI to label DNA (blue). Yellow arrowheads point to γ-tubulin staining at spindle poles. Light blue arrowheads point to spindle-associated γ-tubulin staining. Insets are 1.5× magnifications of spindle poles in the example cells. (b) Quantification of the percentage of mitotic cells in (a) with fragmented centrosomes. n=4 for Trp53 KO and n=3 for Haus6 cKO Trp53 KO embryos (total of 215 and 254 cells counted, respectively, 30–129 cells per embryo). (c) Coronal cortex sections from control and Haus6 cKO Trp53 KO E13.5 embryos stained with antibodies against α-tubulin (green) to reveal spindle microtubules and DAPI to stain DNA (blue). (d) Quantification of the percentage of mitotic progenitors in (c) displaying disorganized spindles. n=4 for Trp53 KO and n=3 for Haus6 cKO Trp53 KO embryos (total of 215 and 254 cells counted, respectively, 30–129 cells per embryo). (e) Sections as in (c) were stained with antibodies against γ-tubulin and centrin, and with DAPI to stain DNA. Note that control and Haus6 cKO Trp53 KO cells are shown at different scales due to the increased size of Haus6 cKO Trp53 KO cells with extra centrosomes. (f) Quantification of the number of centrioles (centrin foci) at spindle poles in mitotic cells as in (e). The percentage of cells with the indicated number of centrioles was plotted. n=3 for Trp53 KO and n=3 for Haus6 cKO Trp53 KO embryos (total of 82 and 125 cells counted, respectively, 25–62 cells per embryo). (g) For the two categories of cells in (f) that have 3–4 or more than 4 centrin foci, respectively, the area occupied by the cells was measured and means were plotted. A total of 62 and 60 mitotic cells, respectively, in n=3 for Trp53 KO Haus6cKO embryos. (b, d, f, g) Plotted values are means, error bars show SD. ***p<0.001 by two-tailed t-test. Scale bars: (a, c, e) 3 μm.
-
Figure 4—figure supplement 1—source data 1
Source data associated with Figure 4—figure supplement 1b, d, f, g.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig4-figsupp1-data1-v2.xlsx
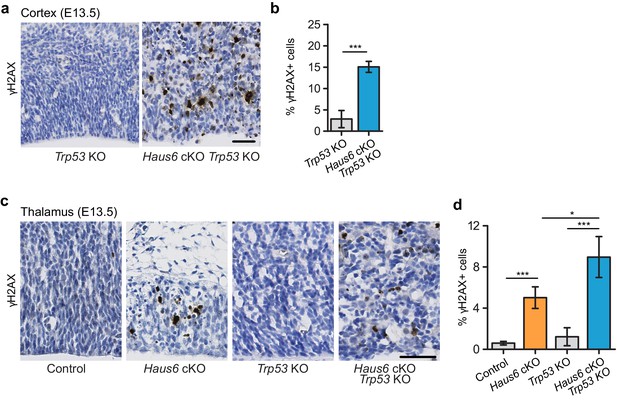
Impaired mitosis in augmin-deficient progenitors causes DNA damage.
(a) Representative images of a region of the cortex of E13.5 Trp53 KO control (Haus6fl/wt Nestin-Cre+Trp53−/−) and Haus6 cKO Trp53 KO (Haus6fl/fl Nestin-Cre+ Trp53−/−) embryos. Coronal sections were stained by immunohistochemistry with an antibody against γH2AX (brown). (b) Quantification of the percentage of cells overexpressing γH2AX in the E13.5 cortex. n=4 for Trp53 KO control and n=4 for Haus6 cKO Trp53 KO embryos (total of 9874 and 14,506 cells counted, respectively, two sections per embryo). (c) Representative images of the region of the thalamus of E13.5 control (Haus6fl/wt Nestin-Cre+ Trp53+/+), Haus6 cKO (Haus6fl/fl Nestin-Cre+ Trp53+/+), Trp53 KO control, and Haus6 cKO Trp53 KO embryos. (d) Quantification of γH2AX-positive cells in the E13.5 thalamus in embryos of the indicated genotypes. n=3 for control, n=4 for Haus6 cKO, n=4 for Trp53 KO, and n=3 for Haus6 cKO Trp53 KO embryos (total of 10,773, 5433, 23,384, 20,602 cells counted, respectively, two sections per embryo). (b, d) Plots show mean values, error bars indicate SD. *p<0.05, **p<0.01, ***p<0.001 by two-tailed t-test. Scale bars: (a, c) 45 µm.
-
Figure 5—source data 1
Source data associated with Figure 5b, d.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig5-data1-v2.xlsx
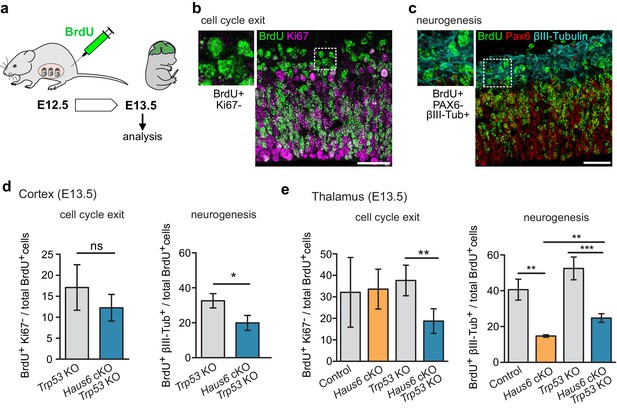
The production of neurons is reduced in Haus6 cKO and Haus6 cKO Trp53 KO brains.
(a) Schematic depicting the experimental procedure of BrdU injection and analysis. (b) Examples of brain sections stained with antibodies against BrdU and the proliferation marker Ki67. Identification of BrdU-labeled cells that did not display Ki67 staining is shown. (c) Examples of brain sections stained with antibodies against BrdU, the progenitor marker PAX6, and the neuronal marker βIII-tubulin. Identification of BrdU-labeled cells that did not express PAX6 but were positive for βIII-tubulin is shown. (d) Quantifications of cells in E13.5 cortex of Trp53 KO (control) and Haus6 cKO Trp53 KO embryos stained as in (b and c). The percentage of BrdU-positive cells showing the indicated staining was plotted. For cell cycle exit analysis, n=4 for Trp53 KO and n=4 for Haus6 cKO Trp53 KO embryos were analyzed (total of 8730 and 7361 BrdU positive cells per genotype, respectively, 1381–2942 cells in 2–3 sections per embryo). For analysis of neurogenesis, n=4 for Trp53 KO and n=3 for Haus6 cKO Trp53 KO embryos were analyzed (total of 13,194 and 8139 BrdU positive cells per genotype, respectively, 1750–4614 in 3–6 sections per embryo). (e) Quantifications of cells in E13.5 thalamus of control and Haus6 cKO, and Trp53 KO (control) and Haus6 cKO Trp53 KO embryos stained as in (b and c). The percentage of BrdU-positive cells showing the indicated staining was plotted. For cell cycle exit analysis, n=3 for control, n=3 for Haus6 cKO, n=4 for Trp53 KO, and n=4 for Haus6 cKO Trp53 KO embryos were analyzed (total 9050, 5923, 12,663, 8610 of BrdU positive cells per genotype, respectively, 1159–4106 cells in 2–3 sections per embryo). For analysis of neurogenesis, n=3 for control, n=3 for Haus6 cKO, n=4 for Trp53 KO, and n=3 for Haus6 cKO Trp53 KO embryos were analyzed (total of 11,749, 5301, 8927 and 4377 BrdU positive cells per genotype, respectively, 1363–5087 cells in 2–5 sections per embryo). (d,e) Plots show mean values, error bars indicate SD. *p<0.05, **p<0.01, ***p<0.001 by two-tailed t-test. Scale bars: (b, c) 40 μm.
-
Figure 6—source data 1
Source data associated with Figure 6d, e.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig6-data1-v2.xlsx
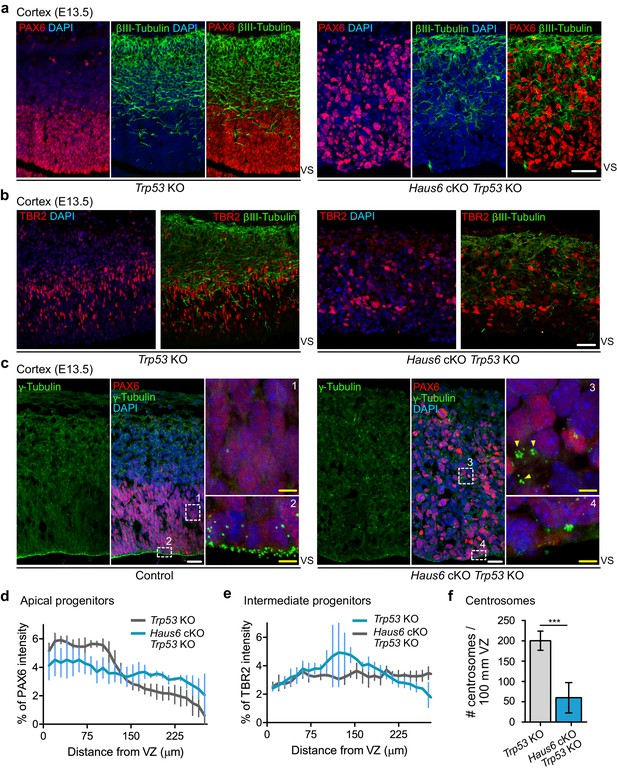
Co-deletion of Haus6 and Trp53 leads to loss of cortical layering.
(a, b) Representative images of the E13.5 cortex from Trp53 KO control (Haus6fl/wt Nestin-Cre+Trp53−/−) and Haus6 cKO Trp53 KO (Haus6fl/fl Nestin-Cre+Trp53−/−) embryos stained with antibodies against PAX6 (a) or TBR2 (b) (red) and the neuronal marker βIII-tubulin (green). DNA was stained with DAPI. (c) Representative images of E13.5 cortex from control (Haus6fl/fl Nestin-Cre-Trp53−/−) and Haus6 cKO Trp53 KO (Haus6fl/fl Nestin-Cre+Trp53−/−) embryos. Coronal sections were co-stained with antibodies against γ-tubulin (green) and PAX6 (red). DNA was stained with DAPI. Magnifications of the boxed regions labeled with 1, 2, 3, and 4 are shown. In the magnified region labeled with 4, yellow arrowheads point to ectopic clusters of interphase centrosomes. (d, e) Distribution of PAX6 and TBR2 staining in sections as in (a and b), respectively. Intensity values were averaged into 9.8-µm-thick bins and plotted as the percentage of total intensity. Lines connect mean values and error bars display SD. (d) n=5 for Trp53 KO and n=4 for Haus6 cKO Trp53 KO embryos. (e) n=2 for Trp53 KO and n=2 for Haus6 cKO Trp53 KO embryos. (f) Quantification of the density of centrosome number at the ventricular surface of the cortex of E13.5 Trp53 KO and Haus6 cKO Trp53 KO embryos. n=4 for Trp53 KO and n=4 for Haus6 cKO Trp53 KO embryos. Plots show mean values and error bars display SD. ***p<0.001 by two-tailed t-test. Scale bars: (a, b) 35 μm and (c) white – 25 µm, yellow – 5 µm.
-
Figure 7—source data 1
Source data associated with Figure 7d, e, f.
- https://cdn.elifesciences.org/articles/67989/elife-67989-fig7-data1-v2.xlsx
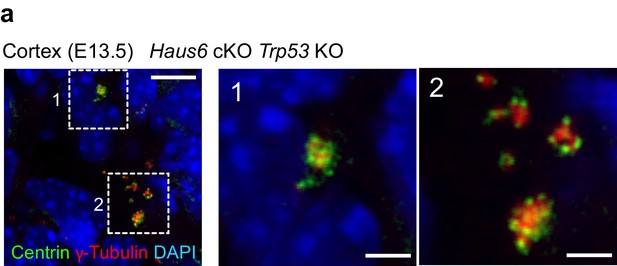
Identification of interphase centrosome clusters in Haus6 cKO Trp53 KO cortex.
E135 cortical sections were stained with antibodies against γ-tubulin and the centriolar marker centrin. Two regions containing multiple, clustered interphase centrosomes are shown as magnifications. Scale bars: 5 μm and 2 μm (magnifications).
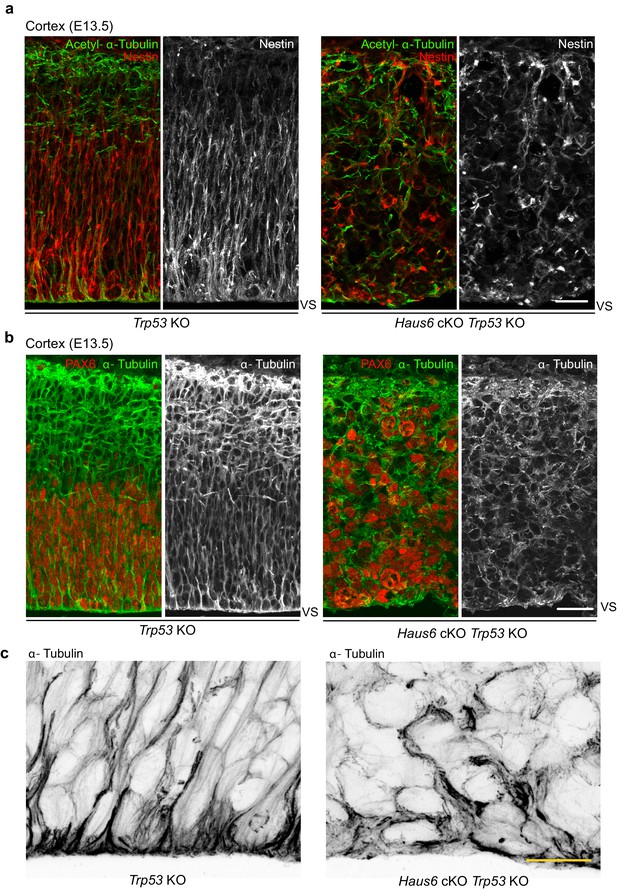
Co-deletion of Haus6 and Trp53 disrupts polarity in surviving progenitors.
(a) Representative images of E13.5 cortical sections from Trp53 KO control (Haus6fl/fl Nestin-Cre− Trp53−/−) and Haus6 cKO (Haus6fl/fl Nestin-Cre+ Trp53−/−) embryos stained with antibodies against the apical progenitor marker nestin (red/white) and acetylated α-tubulin, a marker for stable microtubules (green). (b) Representative images of E13.5 cortical sections from Trp53 KO control (Haus6fl/fl Nestin-Cre− Trp53−/−) and Haus6 cKO Trp53 KO (Haus6fl/fl Nestin-Cre+ Trp53−/−) embryos stained with antibodies against α-tubulin (green/white) and the apical progenitor marker PAX6 (red). (c) Magnification of the apical region of the cortex of E13.5 Trp53 KO control and Haus6 KO Trp53 KO embryos showing microtubules stained with α-tubulin antibody. Scale bars: (a) 25 μm, (b) 35 μm, and (c) 15 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Haus6 | NCBI gene | Gene ID: 230376 | |
| Strain, strain background (M. musculus) | Nestin-Cre Haus6 cKO | This paper | See Materials and methods | |
| Strain, strain background (M. musculus) | Nestin-Cre Haus6 cKO Trp53 KO | This paper | See Materials and methods | |
| Strain, strain background (M. musculus) | Haus6 floxed Neo (Haus6fl-Neo) | RIKEN http://www2.clst.riken.jp/arg/mutant%20mice%20list.html | CDB1218K | |
| Strain, strain background (M. musculus) | Haus6 floxed (Haus6fl) | RIKEN http://www2.clst.riken.jp/arg/mutant%20mice%20list.html | CDB1354K RRID:IMSR_RBRC09630 | |
| Strain, strain background (M. musculus) | C57BL/6-Tg(CAG-flpe)36Ito/ItoRbrc | RIKEN (Kanki et al., 2006) | RRID:IMSR_RBRC01834 | |
| Strain, strain background (M. musculus) | B6.Cg-Tg(Nes-cre)1Kln/J | Gift from Maria Pia Cosma (originally from Jackson Laboratories) | RRID:IMSR_JAX:003771 | |
| Strain, strain background (M. musculus) | Trp53-deficient mice (B6.129S2-Trp53tm1Tyj/J) | Jackson Laboratories | RRID:IMSR_JAX:002101 | |
| Antibody | Anti-α-tubulin (mouse monoclonal) | Sigma-Aldrich | #3873T | IF (1:500) |
| Antibody | Anti-acetylated α-tubulin (mouse monoclonal) | Sigma-Aldrich | #T6793 RRID:AB_477585 | IF (1:500) |
| Antibody | Anti-βIII-tubulin (rabbit polyclonal) | Abcam | #ab18207 RRID:AB_444319 | IF (1:1000) |
| Antibody | Anti-βIII-tubulin (mouse monoclonal) | BioLegend | #801201 RRID:AB_2313773 | IF (1:1000) |
| Antibody | Anti-cleaved caspase-3 (rabbit monoclonal) | Novus Biologicals | #MAB835 RRID:AB_2243951 | IF (1:500) |
| Antibody | Anti-BrdU (mouse monoclonal) | Abcam | #ab8955 RRID:AB_306886 | IF (1:750) |
| Antibody | Anti-γ-tubulin (mouse monoclonal, clone TU-30) | ExBio | #ab27074 RRID:AB_2211240 | IF (1:500) |
| Antibody | Anti-γ-tubulin (rabbit monoclonal) | Sigma-Aldrich | #T5192 RRID:AB_261690 | IF (1:500) |
| Antibody | Anti-Ki67 (rabbit polyclonal) | Abcam | #ab15580 RRID:AB_443209 | IF (1:750) |
| Antibody | Anti-nestin (mouse monoclonal) | Cell signaling | #4760 RRID:AB_2235913 | IF (1:300) |
| Antibody | Anti-p53 (mouse monoclonal) | Cell signaling | #CST2524S RRID:AB_331743 | IF (1:500) |
| Antibody | Anti-PAX6 (mouse monoclonal) | BioLegend | #901301 RRID:AB_2565003 | IF (1:300) |
| Antibody | Anti-phosphorylated-Histone H3 (rabbit polyclonal) | Millipore | #06-570 RRID:AB_310177 | IF (1:1000) |
| Antibody | Anti-TBR2 (rabbit polyclonal) | Abcam | #ab23345 RRID:AB_778267 | IF (1:200) |
| Antibody | Anti-phospho-histone H2AX (Ser139) (mouse monoclonal, clone JBW301) | Millipore | #05-636 RRID:AB_309864 | IHC (1:500) |
| Antibody | Anti-p21 (rat monoclonal, HUGO291) | Abcam | #ab107099 RRID:AB_10891759 | IHC (1:500) |
| Antibody | Anti-centrin-Alexa 488 (rabbit polyclonal) | Homemade (Andrew Holland) (Phan et al., 2021) | IF (1:500) | |
| Antibody | Anti-mouse IgG Alexa 488 (goat polyclonal) | Life Technologies | #A11029 RRID:AB_138404 | IF (1:500) |
| Antibody | Anti-mouse IgG1 Alexa 488 (goat polyclonal) | Life Technologies | #A21121 RRID:AB_2535764 | IF (1:500) |
| Antibody | Anti-mouse IgG1 Alexa 568 (goat polyclonal) | Life Technologies | #A21124 RRID:AB_2535766 | IF (1:500) |
| Antibody | Anti-mouse IgG1 Alexa 633 (goat polyclonal) | Life Technologies | #A21052 RRID:AB_2535719 | IF (1:500) |
| Antibody | Anti-mouse IgG2a Alexa 488 (goat polyclonal) | Life Technologies | #A21131 RRID:AB_2535771 | IF (1:500) |
| Antibody | Anti-rabbit IgG Alexa 488 (goat polyclonal) | Life Technologies | #A11034 RRID:AB_2576217 | IF (1:500) |
| Antibody | Anti-rabbit IgG Alexa 568 (goat polyclonal) | Life Technologies | #A11036 RRID:AB_10563566 | IF (1:500) |
| Antibody | Anti-rabbit IgG Alexa 633 (goat polyclonal) | Life Technologies | #A21071 RRID:AB_141419 | IF (1:500) |
| Antibody | Anti-mouse IgG HRP conjugated (goat polyclonal) | Dako-Agilent | #P0447 RRID:AB_2617137 | IHC (1:500) |
| Antibody | Rabbit IgG polyclonal isotype control (rabbit polyclonal) | Abcam | #ab27478 RRID:AB_2616600 | IHC (1:500) |
| Antibody | Mouse IgG1 (NCG01) isotype control (mouse monoclonal) | Abcam | #ab81032 RRID:AB_2750592 | IHC (1:500) |
| Antibody | Mouse IgG2a isotype control (eBM2a) (mouse monoclonal) | Invitrogen | #14-4724-82 RRID:AB_470114 | IHC (1:500) |
| Sequence-based reagent | mAug6KO_FW | This paper | Genomic PCR primer Haus6 | 5′-CAACCCGAGCAACAGAAACC-3′ |
| Sequence-based reagent | mAug6KO_Rev | This paper | Genomic PCR primer Haus6 | 5′-CCTCCCACCAACTACAGACC-3′ |
| Sequence-based reagent | olMR1084 | This paper | Genomic PCR primer Cre | 5′-GCGGTCTGGCAGTAAAAACTATC-3′ |
| Sequence-based reagent | olMR1085 | This paper | Genomic PCR primer Cre | 5′-GTGAAACAGCATTGCTGTCACTT-3′ |
| Sequence-based reagent | olMR7338 | This paper | Genomic PCR primer control | 5′-CTAGGCCACAGAATTGAAAGATCT-3′ |
| Sequence-based reagent | olMR7339 | This paper | Genomic PCR primer control | 5′-GTAGGTGGAAATTCTAGCATCATCC-3′ |
| Software and algorithm | GraphPad Prism | GraphPad Software Inc | RRID:SCR_002798 | |
| Software and algorithm | QuPath | Queens University (Belfast,UK) | RRID:SCR_018257 | |
| Software and algorithm | FIJI (ImageJ) | NIH | RRID:SCR_002285 | |
| Other | Hematoxylin | Dako-Agilent | S202084 | |
| Other | 5-Bromo-2′-deoxyuridine (BrdU) | Sigma-Aldrich | B5002 | Injected peritoneally to pregnant females at a final concentration of 120 mg/kg of animal weight |
| Other | EnVision Flex Antibody Diluent | Dako-Agilent | K800621 | |
| Other | Envision Flex Wash buffer | Dako-Agilent | K800721 | |
| Other | 3-3′-diamino-benzidine | Dako-Agilent | K3468 | |
| Commercial assay or kit | Mouse on mouse (M.O.M) Immuno-detection Kit | Vector Laboratories | BMK-2202 RRID:AB_2336833 |





