Circular RNA repertoires are associated with evolutionarily young transposable elements
Figures
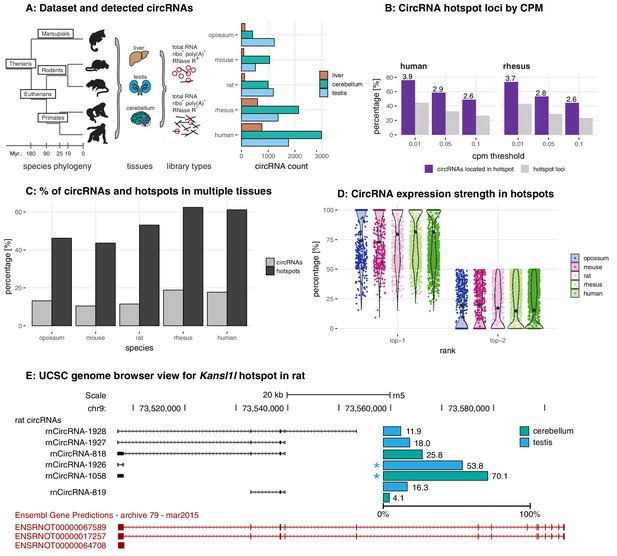
Study design, samples, datasets, and characterisation of circRNA properties and hotspots.
(A) Phylogenetic tree of species analysed in this study and detected circRNAs. CircRNAs were identified and analysed in five mammalian species (opossum, mouse, rat, rhesus macaque, human) and three organs (liver, cerebellum, testis). Each sample was split and one half treated with RNase R to enrich BSJs. A dataset of high confidence circRNAs was established, based on the enrichment of BSJs in RNase R-treated over untreated samples. To the right of the panel, the total number of circRNAs for each species in liver (brown), cerebellum (green), and testis (blue) is shown. (B) CircRNA hotspot loci by CPM (human and rhesus macaque). The graph shows, in grey, the proportion (%) of circRNA loci that qualify as hotspots and, in purple, the proportion (%) of circRNAs that originate from such hotspots, at three different CPM thresholds (0.01, 0.05, 0.1). The average number of circRNAs per hotspot is indicated above the purple bars. (C) Number of circRNA hotspot loci found in multiple tissues. The graph shows the proportion (%) of circRNAs (light grey) and of hotspots (dark grey) that are present in at least two tissues. (D) Contribution of top-1 and top-2 expressed circRNAs to overall circRNA expression from hotspots. The plot shows the contribution (%) that the two most highly expressed circRNAs (indicated as top-1 and top-2) make to the total circRNA expression from a given hotspot. For each plot, the median is indicated with a grey point. (E) Example of the Kansl1l hotspot in rat. The proportion (%) for each detected circRNA within the hotspot and tissue (cerebellum = green, testis = blue) are shown. The strongest circRNA is indicated by an asterisk. rnCircRNA-819 is expressed in testis and cerebellum.
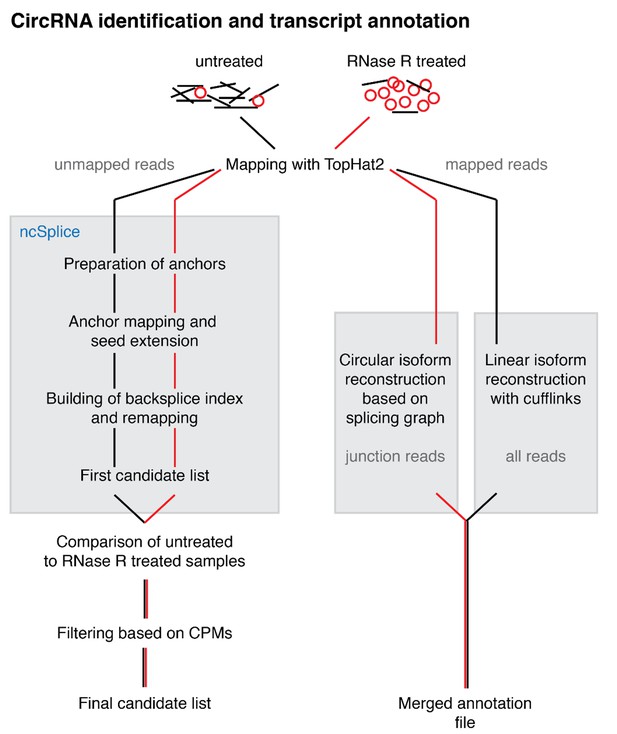
Overview of the reconstruction pipeline.
Overview of the reconstruction pipeline. CircRNA identification and transcript reconstruction. Unmapped reads from RNA-seq data were remapped and analysed with a custom pipeline. The reconstruction of circRNA transcripts was based on the junction enrichment after RNase R treatment. Further details on the pipeline are provided in the Materials and methods.
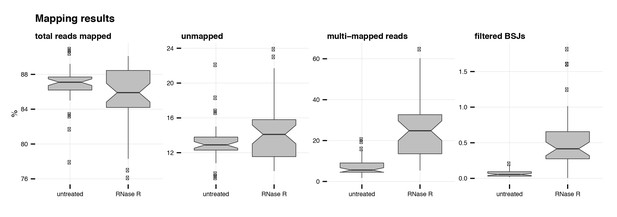
Mapping summary of RNA-seq reads.
Percentage of mapped, unmapped, multi-mapped and BSJ reads across all libraries in untreated and RNase R-treated conditions.
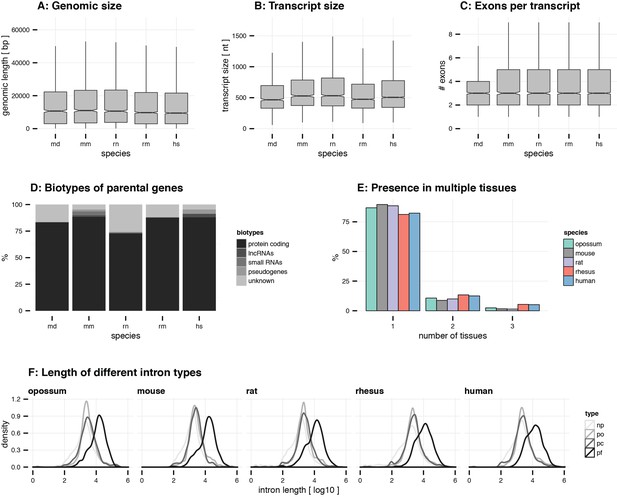
General circRNA properties.
(A) Genomic size. The genomic size (bp) of circRNAs is plotted for all species. (B) Transcript size. The transcript size (nt) of circRNAs is plotted for all species. (C) Exons per transcript. The number of exons in circRNAs is plotted for all species. For panel A-C, outliers are not plotted (abbreviations: md = opossum, mm = mouse, rn = rat, rm = rhesus macaque, hs = human). (D) Biotypes of parental genes. For each species, the frequency (%) of different biotypes in the circRNA parental genes was assessed using the ensembl annotation. CircRNA loci that were not found in the annotation were marked as ‘unknown’. (E) Presence in multiple tissues. For each species, the frequency (%) of circRNAs detected in one, two, or three tissues is plotted. (F) Length of different intron types. Distribution of median intron length (log10-transformed) is plotted for different intron types in each gene. Abbreviations: np = non-parental, po = parental-outside of circRNA, pf = parental-flanking of circRNA, pi = parental-inside of circRNA.
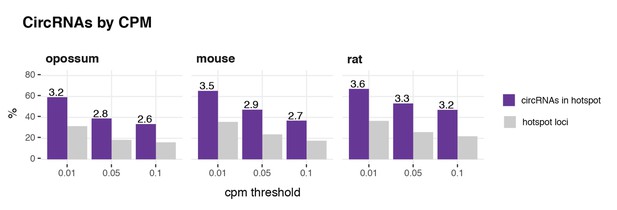
CircRNA hotspot loci by CPM (opossum, mouse, rat).
In grey, the proportion (%) of circRNA loci that qualify as hotspots and, in purple, the proportion (%) of circRNAs that originate from such hotspots, at three different CPM thresholds (0.01, 0.05, 0.1). The average number of circRNAs per hotspot is indicated above the purple bars.
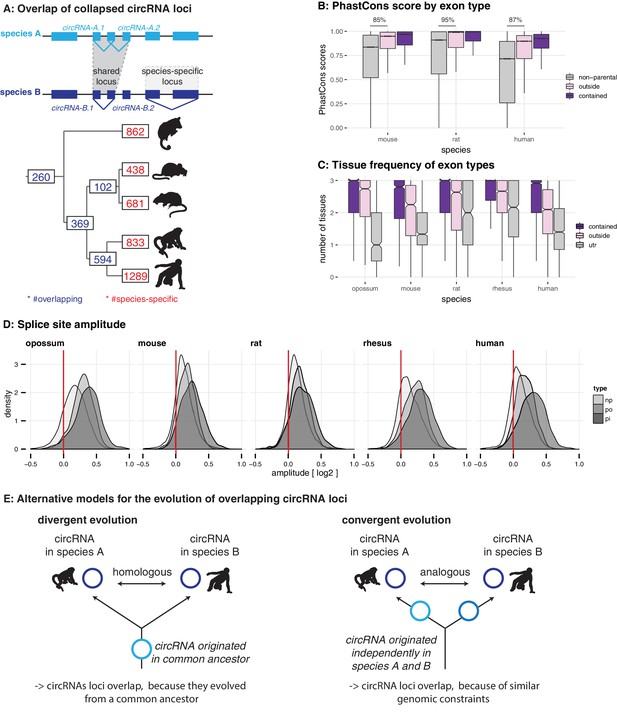
Evolutionary properties of circRNAs.
(A) CircRNA loci overlap between species. Upper panel: Schematic representation of the orthology definition used in our study. CircRNAs were collapsed for each gene, and coordinates were lifted across species. Lower panel: Number of circRNA loci that are species-specific (red) or circRNAs that arise from orthologous exonic loci of 1:1 orthologous genes (i.e. circRNAs sharing 1:1 orthologous exons) across lineages (purple) are counted. We note that in the literature, other circRNA ‘orthology’ definitions can be found, too. For example, assigning circRNA orthology simply based on parental gene orthology implies calling also those circRNAs ‘orthologous’ that do not share any orthologous exons, which directly argues against the notion of circRNA homology; that is, a common evolutionary origin (see Figure 2—figure supplement 1A). Overall, the orthology considerations we applied largely follow the ideas sketched out in Patop et al., 2019. (B) Distribution of phastCons scores for different exon types. PhastCons scores were calculated for each exon using the conservation files provided by ensembl. PhastCons scores for non-parental exons (grey), exons in parental genes, but outside of the circRNA (pink) and circRNA exons (purple) are plotted. The difference between circRNA exons and non-parental exons that can be explained by parental non-circRNA exons is indicated above the plot. (C) Mean tissue frequency of different exon types in parental genes. The frequency of UTR exons (grey), non-UTR exons outside of the circRNA (pink) and circRNA exons (purple) that occur in one, two, or three tissues was calculated for each parental gene. (D) Distribution of splice site amplitudes for different exon types. Distribution of median splice site GC amplitude (log2-transformed) is plotted for different exon types (np = non-parental, po = parental, but outside of circRNA, pi = parental and inside circRNA). Red vertical bars indicate values at which exon and intron GC content would be equal. (E) Different evolutionary models explaining the origins of overlapping circRNA loci.
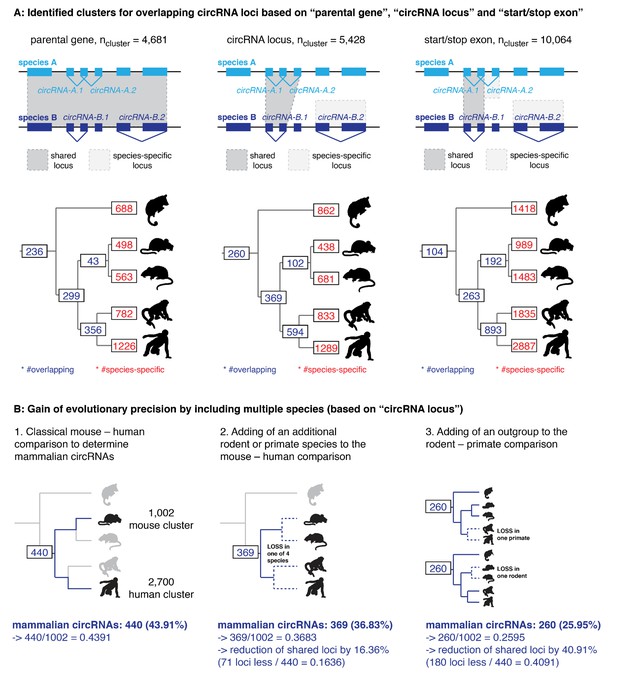
CircRNA loci overlap between species.
(A) Upper panel: The presence of circRNA in multiple species can be identified on the gene level (=‘parental gene’), based on the location of the circRNA within the gene (=‘circRNA locus’) or the overlap of the first and last exons of the circRNA (=‘start/stop exon’). Depending on the chosen stringency, the number of circRNA loci present in multiple species varies. For example: when considering the parental gene level (shown to the left), all four circRNAs depicted in the hypothetical example of this figure (circRNA-A.1, circRNA-A.2, circRNA-B.1, and circRNA-B.1) are located in the same orthologous locus. In contrast, when looking at the start and stop exons (right), only two circRNAs (circRNA-A.1 and circRNA-B.1) are generated from the same orthologous locus, whereas circRNA-A.2 and circRNA-B.2 - previously classified as ‘orthologous’ – are now found in different loci and labeled as species-specific. Depending on the classification, the number of shared circRNA loci thus differs and may influence the interpretation of results. Lower panel: For each classification, orthology clusters were counted and grouped by their overlap (in purple when present in primates, rodents, eutherians or therians; in red when species-specific). Please note that in our study, we apply the definition shown in the middle panels (which are identical to main Figure 2A) that considers exon overlap as relevant. (B) Figure shows the loss of shared circRNA loci (based on ‘circRNA locus’ definition) by adding additional species to the classical mouse-human comparison. All comparisons are made with mouse as reference to which the other loci are compared. The reduction of loci (%) by adding additional species is indicated below each figure.
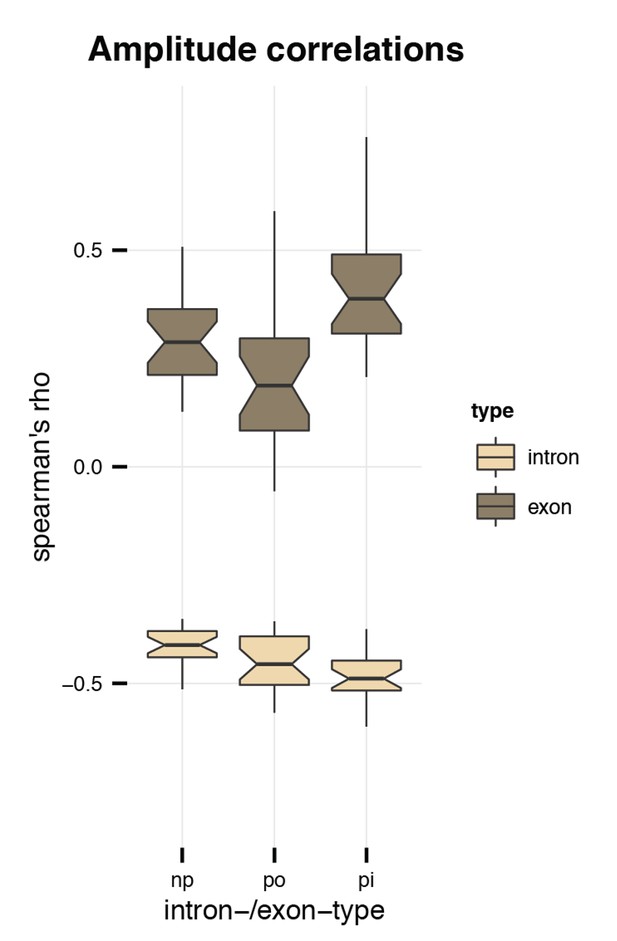
Amplitude correlations.
Plotted is the correlation (Spearman’s rho) between the amplitude and the GC content of introns (light brown) and exons (dark brown). Abbreviations: np = non-parental, po = parental, outside of circRNA, pi = parental, inside of circRNA.
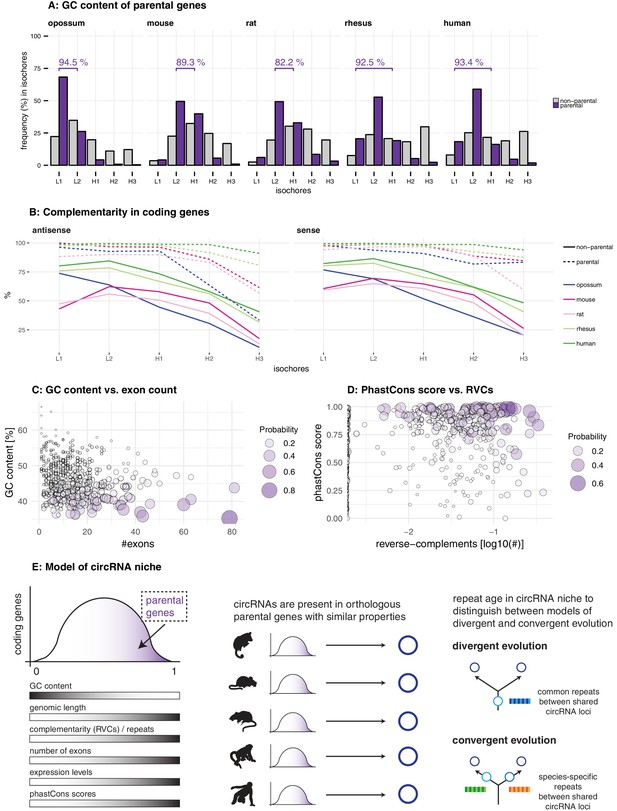
Characterisation of circRNA parental gene properties.
(A) GC content of parental genes. Coding genes were classified into L1-H3 based on their GC content, separately for non-parental (grey) and parental genes (purple). The percentage of parental genes in L1-L2 (opossum, mouse, rat) and L1-H1 (rhesus macaque, human) is indicated above the respective graphs. (B) Complementarity in coding genes. Each coding gene was aligned to itself in sense and antisense orientation using megaBLAST. The proportion of each gene involved in an alignment was calculated and plotted against its isochore. (C-D) Examples of parental gene predictors for linear regression models. A generalised linear model (GLM) was fitted to predict the probability of the murine coding gene to be parental, whereby x- and y-axis represent the strongest predictors. Colour and size of the discs correspond to the p-values obtained for 500 genes randomly chosen from all mouse coding genes used in the GLM. (E) Model of circRNA niche.
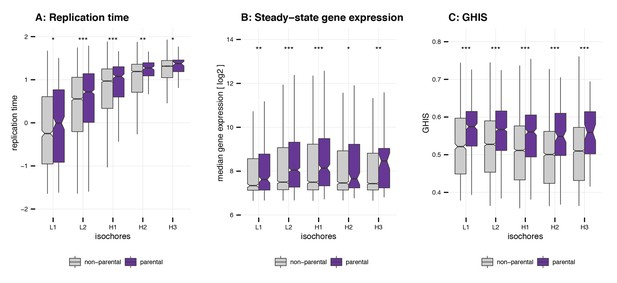
Replication time, gene expression steady-state levels and GHIS of human parental genes.
(A) Replication time of parental genes. Values for the replication time were used as provided in Koren et al., 2012. They were normalised to a mean of 0 and a standard deviation of 1. Differences between non-parental genes (ntotal = 18134) and parental genes (ntotal = 2058) were assessed by a one-tailed Mann-Whitney U test. (B) Gene expression steady-state levels of parental genes. Mean steady-state expression levels were used as provided in Pai et al., 2012. Differences between non-parental genes (ntotal = 14414) and parental genes (ntotal = 2058) were assessed by a one-tailed Mann-Whitney U test. (C) GHIS of parental genes. GHIS was used as provided in Steinberg et al., 2015. Differences between non-parental genes (ntotal = 17438) and parental genes (ntotal = 1995) were assessed by a one-tailed Mann-Whitney U test. (Note C-D: Outliers for all panels were removed prior to plotting. Significance levels: '***' < 0.001, '**' < 0.01, '*' < 0.05, 'ns' >= 0.05).
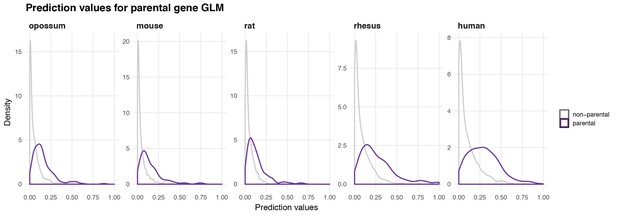
Distribution of prediction values for non-parental and parental circRNA genes.
The density of predicted values for non-parental (grey) and parental (purple) genes is plotted for each species based on the predictors identified by the GLM in each species.
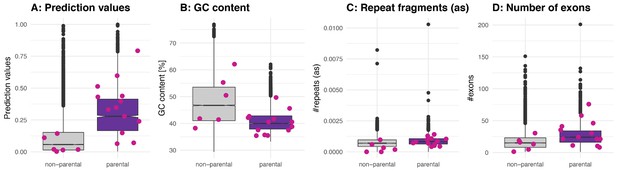
Properties of ‘functional circRNAs’ from literature.
(A) Prediction values of linear regression model for human circRNA parental and non-parental genes as previously defined (Materials and methods). Functional circRNAs as described in Chen, 2020 are plotted in pink on top of the boxplot and are separated by whether they are in a non-parental or parental gene. (B-D) GC content, repeat fragments (in antisense, normalised by genomic length of parental gene) and number of exons for human non-parental and parental circRNA genes; values for functional circRNAs are plotted in pink. Parental genes of functional circRNAs listed in Chen, 2020, which were identified in our study: SHPRH, ZNF609, GCN1L1, HIPK2, HIKP3, ZNF91, BIRC6, FOXO3, MBNL1, ASAP1, PAN3, SMARCA5, ITCH.
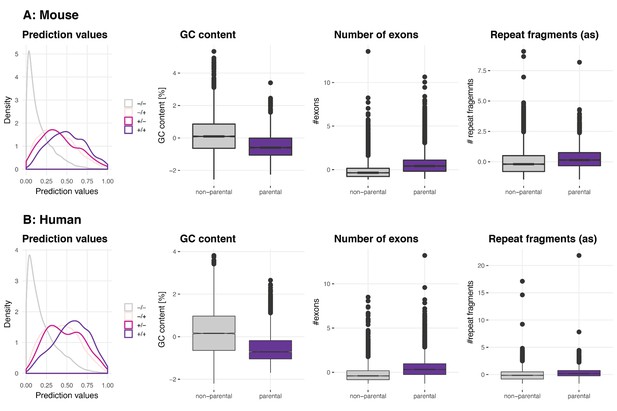
Validation of parental gene GLM on Werfel et al. dataset.
(A) Mouse. To assess the parental gene properties identified by this study, the generalised model was used to predict circRNA parental genes on data from an independent study. The density plot ‘Prediction values’ shows the predicted values for non-parental genes in both datasets (Werfel et al., 2016 and data from this publication, n = 11963, in grey and labeled as -/-), parental genes only present in the Werfel dataset (n = 2843, light pink, labeled as -/+), parental genes only present in this study’s underlying dataset (n = 210, dark pink, labeled as +/-) and parental genes that were present in both datasets (n = 638, purple, labeled as +/+). The plots ‘GC content’, ‘Number of exons’ and ‘Repeat fragments (as)’ (the latter normalised by the genomic length of the parental gene) show the properties of circRNA parental genes (highlighted in purple) as identified by Werfel et al. (B) Human. Same plot outline as for mouse. The number of non-parental genes in both datasets is n = 10591; 2724 parental genes are only present in the Werfel dataset and 356 parental genes only in our dataset. The overlap between both datasets is n = 1666.
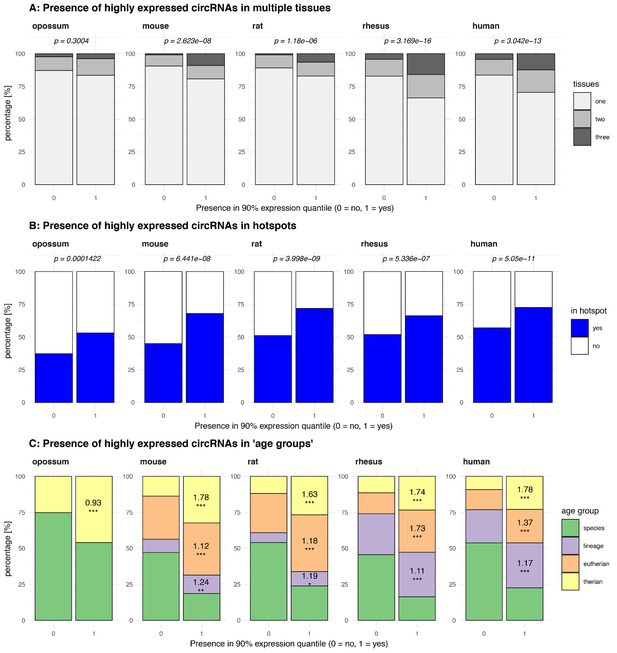
Properties of highly expressed circRNAs.
(A) Presence of highly expressed circRNAs in multiple tissues. Plot shows the percentage (%) of circRNAs from the 90% expression quantile (nopossum = 158, nmouse = 156, nrat = 217, nrhesus = 340, nhuman = 471), which is present in one, two or three of the tissues analysed compared to circRNAs outside the 90% expression quantile. For each species, distributions were compared using Fisher’s exact test, p-values are shown above each barplot. (B) Presence of highly expressed circRNAs in hotspots. Plot shows the percentage (%) of circRNAs from the 90% expression quantile, which is found in a hotspot compared to circRNAs outside the 90% expression quantile. For each species, distributions were compared using Fisher’s exact test, p-values are shown above each barplot. (C) Presence of highly expressed circRNAs in ‘age groups’. Plot shows the percentage (%) of circRNAs from the 90% expression quantile, which is present in different ‘age groups’ compared to circRNAs outside the 90% expression quantile. Age groups were defined as whether circRNA is species-specific (age = 1), lineage-specific (age = 2), eutherian (age = 3) or shared across all therian species (age = 4). Log-odds ratio and significance levels (significance levels based on p-value: ‘***’ < 0.001, ‘**’ < 0.01, ‘*’ < 0.05, ‘ns’ >= 0.05) were calculated using a generalised linear model (see Supplementary file 10) and are shown for the respective age groups and species.
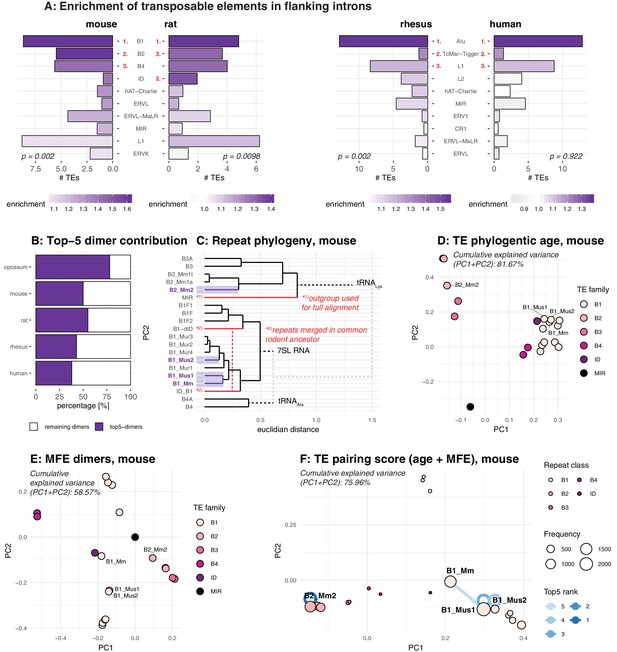
Analysis of the repeat landscape of circRNA parental genes.
(A) Enrichment of TEs in flanking introns for mouse, rat, rhesus macaque and human. The number of TEs was quantified in both intron groups (circRNA flanking introns and length- and GC-matched control introns). Enrichment of TEs is represented by colour from high (dark purple) to low (grey). The red numbers next to the TE name indicate the top-3 enriched TEs in each species. Enrichment was assessed using a Wilcoxon Signed Rank Test; p-values are indicated at the bottom of each plot. (B) Top-5 dimer contribution. The graph shows the proportion of top-5 dimers (purple) vs. other, remaining dimers (white) to all predicted dimers in flanking introns. Top-5 dimers thus account for 78, 50, 55, 43, and 38% of all dimers in opossum, mouse, rat, rhesus macaque and human, respectively. (C) Phylogeny of mouse TEs. Clustal-alignment based on consensus sequences of TEs. Most recent TEs are highlighted. (D) PCA for phylogenetic age of mouse TE families. PCA is based on the clustal-alignment distance matrix for the reference sequences of all major SINE families in mouse with the MIR family used as an outgroup. TEs present in the top-5 dimers are labelled. (E) PCA based on binding affinity of mouse TE families. PCA is based on the minimal free energy (MFE) for all major SINE families in mouse with the MIR family used as an outgroup. TEs present in the top-5 dimers are labelled. (F) PCA for TE pairing score of mouse dimers. PCA is based on a merged and normalised score, taking into account binding strength of the dimer structure (=MFE) and phylogenetic distance. Absolute frequency of TEs is visualised by circle size. TEs present in the five most frequent dimers (top-5) are highlighted by blue lines connecting the two TEs engaged in a dimer (most frequent dimer in dark blue = rank 1). If the dimer is composed of the same TE family members, the blue line loops back to the TE (=blue circle).
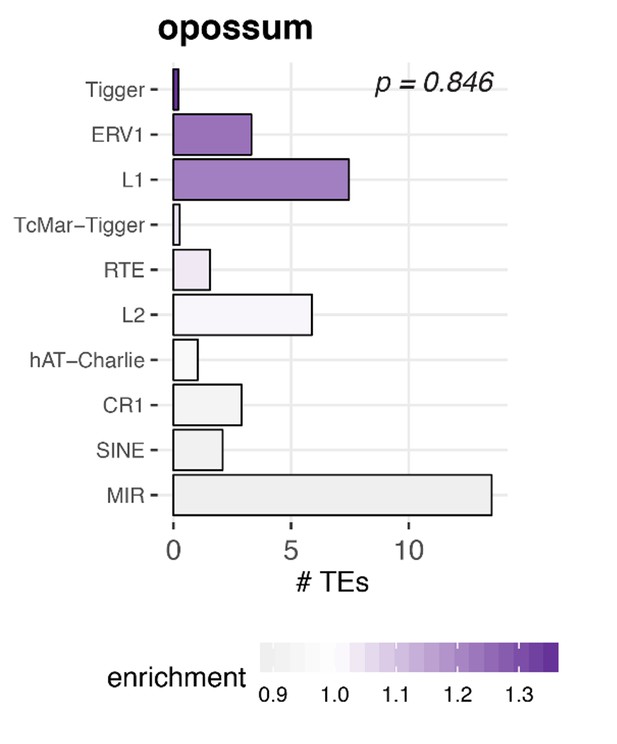
Enrichment of transposable elements in flanking introns for opossum.
The number of transposable elements was quantified in both intron groups (circRNA flanking introns and length- and GC-matched control introns). Enrichment of transposable elements is represented by colour from high (dark purple) to low (grey). The frequency distributions of TEs in background and flanking introns were compared using a Wilcoxon Signed Rank Test; p-value is shown in the upper right corner.
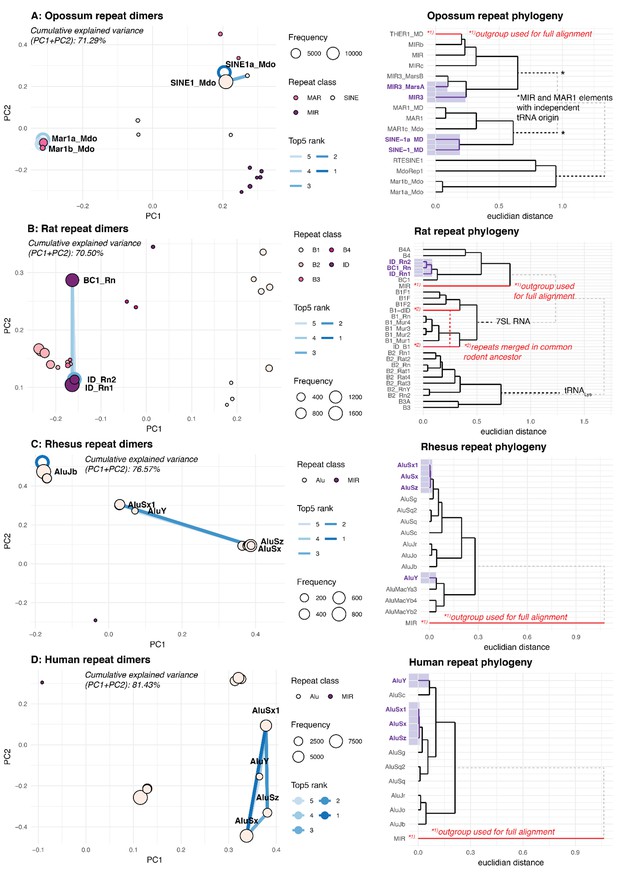
PCA and phylogeny of opossum, rat, rhesus macaque, and human repeat dimers.
(A) Opossum. Panel A shows the PCA for dimer clustering based on a merged and normalised score, taking into account binding phylogenetic distance, binding capacity of TEs to each other and absolute frequency. Absolute frequency is also represented by circle size. The top-ranked dimers are indicated. Circles around the discs represent cases where the TE binds to itself. Furthermore, a phylogeny of opossum transposable elements is shown, the top-5 dimers are highlighted with purple shading. Phylogenetic trees are based on multiple alignments with Clustal-Omega. Several TE families have independent origins, which cannot be taken into account with Clustal-Omega. These cases are indicated by a grey, dotted line and TE origins – if known – have been manually added. We deemed this procedure sufficiently precise, given that the aim was to only visualise the general relationship of TEs. TEs used as outgroups, as well TEs that merged are indicated with a red line. (B-D) Same analysis as in Panel A, but for rat, rhesus macaque, and human, respectively.
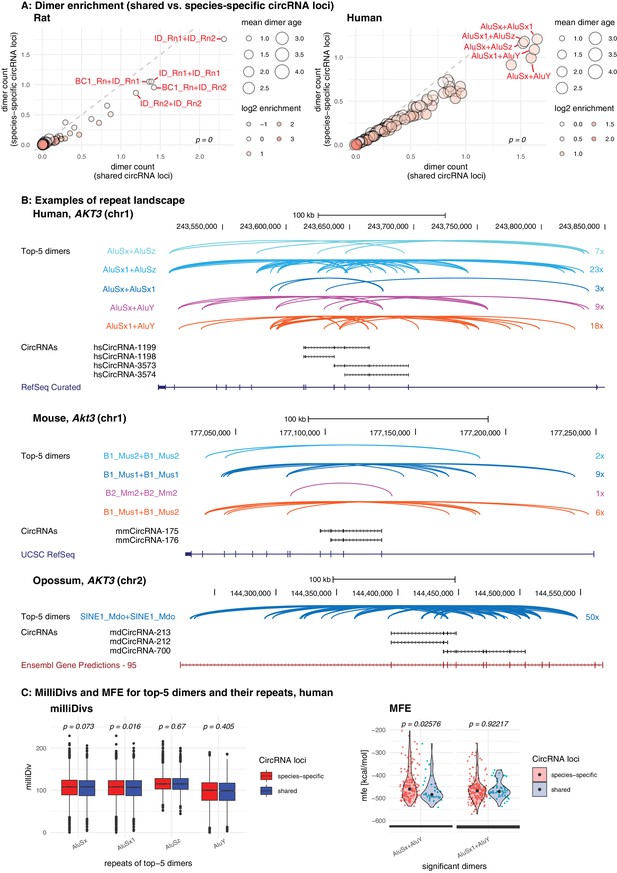
Repeat analysis and dimer potential of shared and species-specific parental genes.
(A) Dimer enrichment in shared vs. species-specific repeats in rat and human (see Figure 5—figure supplement 1 for other species). The frequency (number of detected dimers in a given parental gene), log2-enrichment (shared vs. species-specific) and mean age (defined as whether repeats are species-specific: age = 1, lineage-specific: age = 2, eutherian: age = 3, therian: age = 4) of the top-100 most frequent and least frequent dimers in parental genes with shared and species-specific circRNA loci in rat and human were analysed. The frequency is plotted on the x- and y-axis, point size reflects the age and point colour the enrichment (blue = decrease, red = increase). Based on the comparison between shared and species-specific dimers (using a Wilcoxon Signed Rank Test), the top-5 dimers defined by frequency and enrichment are highlighted and labelled in red. (B) Species-specific dimer landscape for the Akt3 gene in human, mouse and opossum. UCSC genome browser view for the parental gene, circRNAs and top-5 dimers (as defined in panel B). Start and stop positions of each dimer are connected via an arc. Dimers are grouped by composition represented by different colours, the number of collapsed dimers is indicated to the right-side of the dimer group. Only dimers that start before and stop after a circRNAs are shown as these are potentially those that can contribute to the hairpin structure. The human Akt3 gene possesses two circRNA clusters. For better visualisation, only the upstream cluster is shown. (C) Degradation rates (MilliDivs) and minimal free energy (MFE) for top-5 dimers in human. MilliDiv values for all repeats composing the top-5 dimers (defined by their presence in all parental genes) were compared between parental genes of species-specific (red) and shared (blue) circRNA loci in human (see Figure 5—figure supplement 3 for other species). A Wilcoxon Signed Rank Test was used to compare dimers between parental genes with shared and species-specific circRNA loci, with p-values plotted above the boxplots. MFE values were compared between the least degraded dimers in parental genes of species-specific (red) and shared (blue) circRNA loci. MFE values were calculated using the genomic sequences of all top-5 dimers. For each parental gene, the least degraded dimer (based on its mean milliDiv value) was then chosen which let to a strong enrichment of only a subset of the top-5 dimers (in this case AluSx+AluY and AluSx1+AluY). If enough observations for a statistical test were present, the two distributions (shared/species-specific) were compared using a Student’s t-Test and plotted as violin plots with p-values above the plot.
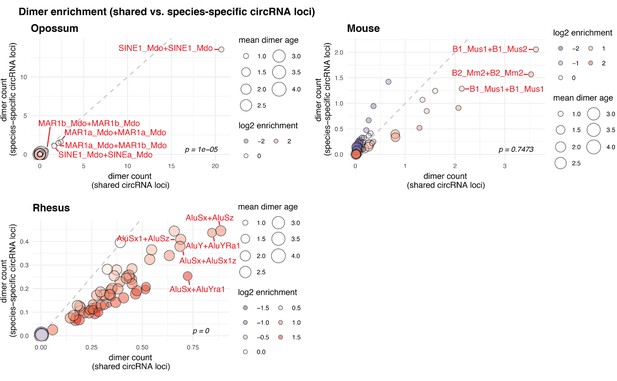
Contribution of species-specific repeats to the formation of shared circRNA loci.
Dimer enrichment in shared and species-specific repeats in opossum, mouse and rhesus macaque. The frequency (number of detected dimers in a given parental gene), log2-enrichment (shared vs. species-specific) and mean age (defined as whether repeats are species-specific: age = 1, lineage-specific: age = 2, eutherian: age = 3, therian: age = 4) of the top-100 most frequent and least frequent dimers in parental genes with shared and species-specific circRNA loci in opossum, mouse and rhesus macaque were analysed and compared with a Wilcoxon Signed Rank Test. Frequencies are plotted on the x- and y-axis, point size reflects the age and point colour the enrichment (blue = decrease, red = increase). Based on the comparison between shared and species-specific dimers, the top-5 dimers defined by frequency and enrichment are highlighted and labelled in red.
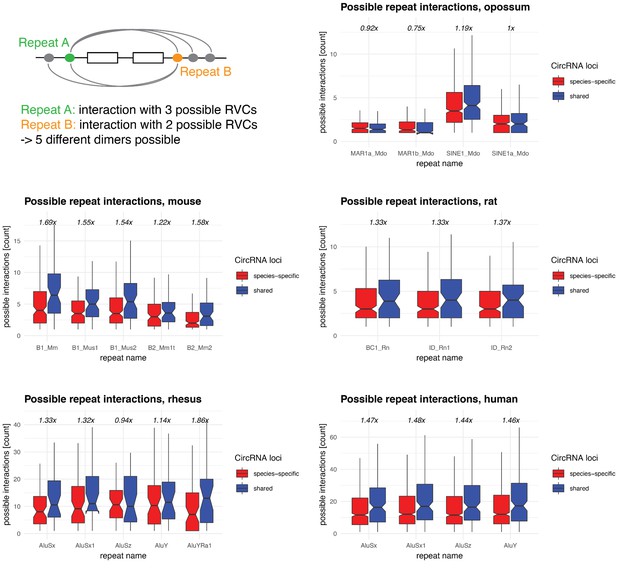
Repeat interaction landscape in shared vs. species-specific circRNA loci.
Upper left: graphical representation of possible repeat interactions (=dimers that can be formed) across RVCs. Afterwards: Frequency distribution of possible interactions of a given repeat (from the top-5 dimers, based on Figure 5A and Figure 5—figure supplement 1) in parental genes of species-specific (red) and shared (blue) circRNA loci in opossum, mouse, rat, rhesus macaque, and human. The enrichment of possible interactions (shared vs. species-specific, based on each distribution’s median) is indicated above each plot.
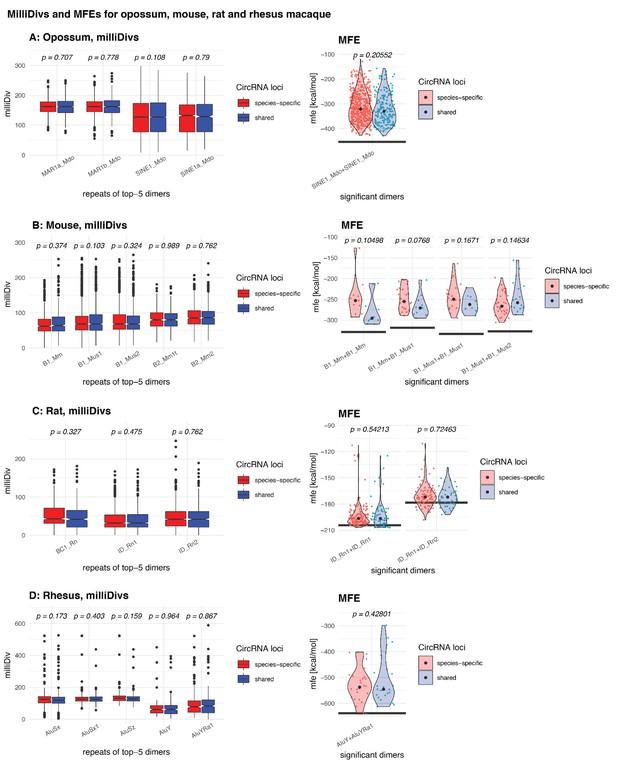
MilliDivs and MFE for dimers in shared and species-specific circRNA loci.
Left panel of each species: MilliDiv values were compared between parental genes of species-specific (red) and shared (blue) circRNA loci using a Student’s t-Test (alternative = ‘less’) with corresponding p-values plotted above each boxplots. Since dimers are composed of two repeats, the mean milliDiv value between both repeats was taken. Right panel of each species: Violin Plots depicting the minimal free energy (MFE) of genomic sequences for dimers in species-specific (red) and shared (blue) circRNA loci. For each gene, the ‘least degraded dimer’ was chosen to calculate its MFE value leading to a strong enrichment of only a few of the top-5 dimers (see Materials and methods). The ‘maximum’ MFE possible, which is based on the dimer formed by each TE’s reference sequence (downloaded from RepBase [Bao et al., 2015]), is depicted with a grey line below each pair of violin plots. Each distribution’s median is indicated with a grey point. MFE values between species-specific and shared circRNA loci were compared with a Student’s t-Test; corresponding p-values are indicated above each pair of violin plots.
Tables
Generalised linear model predicting the probability of coding genes to be a parental gene.
A generalised linear model was fitted to predict the probability of coding genes to be a parental gene (nopossum = 18807, nmouse = 22015, nrat = 11654, nrhesus = 21891, nhuman = 21744). The model was trained on 80% of the data (scaled values, cross-validation, 1000 repetitions). Only the best predictors were kept and then used to predict probabilities for the remaining 20% of data points (validation set, shown in table). Genomic length, number of exons and GC content are based on the respective ensembl annotations; number of repeats in antisense and sense orientation to the gene was estimated using the RepeatMasker annotation, phastCons scores taken from UCSC (not available for opossum and rhesus macaque) and expression levels and the tissue specificity index based on Brawand et al., 2011. An overview of all log-odds ratios and p-values calculated in the validation set of each species is provided in the table, further details can be found in Supplementary file 6. Abbreviations: md = opossum, mm = mouse, rn = rat, rm = rhesus macaque, hs = human. Significance levels: ‘***’ < 0.001, ‘**’ < 0.01, ‘*’ < 0.05, ‘ns’ >= 0.05.
| Predictor | Log-odds range (significance) | Species with significant predictor |
|---|---|---|
| Genomic gene length (bp) | rn: 0.26 (***) rm: 0.17 (***) hs: 0.26 (***) md, mm: ns | rn, rm, hs |
| Number of exons | md: 0.45 (***) mm: 0.38 (***) rn: 0.30 (***) rm: 0.42 (***) hs: 0.32 (***) | md, mm, rn, rm, hs |
| GC content | md: -1.84(***) mm: -1.09(***) rn: -0.72(***) rm: -1.44(***) hs: -1.42(***) | md, mm, rn, rm, hs |
| Repeat fragments (antisense) | md: 0.28 (**) mm: 0.20 (**) rm: 0.59 (***) rn, hs: ns | md, mm, rm |
| Repeat fragments (sense) | hs: 0.58 (***) md, mm, rn, rm: ns | hs |
| PhastCons scores | mm: 0.58 (***) rn: 0.51 (***) hs: 0.45 (***) | mm, rn, hs |
| Mean expression levels | md: 0.34 (**) rm: 0.38 (***) hs: 0.25 (**) mm, rn: ns | md, rm, hs |
| Tissue specificity index | md, mm, rn, rm, hs: ns | - |
Overview of external programmes.
| Programme | Version |
|---|---|
| Blast | 2.2.29+ |
| BEDTools | 2.17.0 |
| Bowtie2 | 2.1.0 |
| Clustal Omega | 1.2.4 |
| Cufflinks | 2.1.1 |
| FastQC | 0.10.1 |
| Mcl | 14.137 |
| R | 3.0 and 3.1 |
| Ruby | 2.0 and 2.1 |
| SAMTools | 0.1.19 |
| TopHat2 | 2.0.11 |
| ViennaRNA | 2.1.8 |
Ensembl genome versions and annotation files for each species.
| Species | Genome | Annotation |
|---|---|---|
| Opossum | monDom5 | ensembl release 75, feb 2014 |
| Mouse | mm10 | ensembl release 75, feb 2014 |
| Rat | rn5 | ensembl release 75, feb 2014 |
| Rhesus macaque | rheMac2 | ensembl release 77, oct 2014 |
| Human | hg38 | ensembl release 77, oct 2014 |
Removed regions during mapping.
| Species | Tissue | Chromosome | Start | Stop | Strand |
|---|---|---|---|---|---|
| Rhesus macaque | Testis | 7 | 164261343 | 164283671 | + |
| Rhesus macaque | Testis | 7 | 22010814 | 22092409 | - |
| Rhesus macaque | Testis | 19 | 52240850 | 52288425 | - |
| Rhesus macaque | Testis | 19 | 59790996 | 59834798 | + |
| Rhesus macaque | Testis | 19 | 59790996 | 59847609 | + |
| Human | Testis | 2 | 178535731 | 178600667 | + |
| Human | Testis | 7 | 66429678 | 66490107 | - |
| Human | Testis | 9 | 97185441 | 97211487 | - |
| Human | Testis | 12 | 97492460 | 97561047 | + |
| Human | Testis | 14 | 100913431 | 100949596 | + |
| Human | Testis | 18 | 21765771 | 21849388 | + |
Analysis of circRNA databases.
Evolutionary analyses are sensitive and prone to the amplification of noise. We thus used a comprehensive dataset including samples from wild-type tissues and the same sex. In addition, as pointed out by Reviewer 2 “without a mock control, [it] is impossible […] to determine the actual resistance to the degradation by the exonuclease and hence determine which potential backsplicing junctions are really coming from circular molecules.” As most of the samples reported in the public database are not RNase R-treated, none of them could provide sufficiently trustworthy circRNA annotations for our study.
| Species | Sample types, circRNA calling | Expression | |
|---|---|---|---|
| circAtlas Wu et al. Genome Biol 2020 PMID: 32345360 | Human, mouse, rhesus, rat, pig, chicken | 1,070 RNA-seq samples collected from 19 normal tissues Identification with CIRI2, DCC, find_circ, CIRCexplorer2 No RNase R-treated samples, but circRNAs need to be detected (1) by at least two of the tools and (2) junctions be covered with a minimum of two independent reads (-> identification on a per sample basis) CircRNA numbers shown in the database for each tissue are pooled, meaning there are for example 225’000 circRNAs in 39 human brain samples -> no separation by region, or minimum overlap (e.g in half of the samples analysed) | No comprehensive expression data provided (i.e. only numerical values for the top 30 circRNAs). For cases where biological samples are pooled (same tissue), the maximal expression value across all samples is used to define high expressors. However, due to strong heterogeneity in expression levels, mean values are frequently magnitudes lower. |
| Comment: The circRNAs reported are not consistently expressed in the tissue. For the brain, it is for example sufficient for a circRNA to be detected in one out of the 39 brain samples, leading to the high numbers reported. This is reflected in the expression variation observed within the same tissue samples. The authors report the maximal observed value, but the tissue mean seems to be at least a magnitude lower. In addition, no RNase R-treated samples were used for validation. We considered this database inappropriate for our study due to the lack of RNase R-treated expression data, which we consider a more important indicator for a circRNA than the detection by two independent identification methods. Note also statement from publication: “Using this method, we found that the vast majority of circRNAs (an average of 61.7%) could be detected only in one species, with only 797 circRNAs shared by all species, in agreement with previous reports on the highly species-specific expression of circRNAs” | |||
| circBase Glažar et al. RNA 2014 PMID: 25234927 | Human, mouse, worm, fly | Mostly cell lines. Mouse cerebellum is the only tissue comparable to our study. The authors report 2,407 circRNAs for this tissue (but: only one cerebellum sample was used in study). | Ranking of circRNAs according to “Scores” that are difficult to transform to expression levels. |
| Comment: Due to the high number of cell lines samples, we did not consider this database. In addition, only human and mouse samples are present, yet we were looking for an evolutionary dataset that included multiple mammalian species. | |||
| CIRCpedia Zhang et al. Genome Res 2016 PMID: 27365365 | Human, mouse, rat, zebra fish, fly, worm | 180 RNA-seq datasets (at first sight, mostly cell lines). One detection tool used. Only 20 out of the 180 samples are RNase Rtreated samples. 4 out of the 20 RNase Rtreated samples are from a mammalian species. | |
| Comment: This database was not used because it mainly contains cell line samples and only a very low number of RNase R-treated samples. | |||
Additional files
-
Supplementary file 1
Sample overview.
Summary of organism, tissue, age and sex for each sample; last column shows the RNA Quality Number (RQN) for the extracted RNA.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp1-v4.docx
-
Supplementary file 2
Filtering steps and reduction of circRNAs candidates during the identification pipeline.
Description of the different filtering steps applied to generate a high confidence circRNA dataset based on the comparison of untreated and RNase R-treated samples. The number of unique BSJs left after each filtering step is shown for each tissue (see Materials and methods, section Generation of high confidence circRNA candidates from the comparison of RNase R-treated vs. -untreated samples); mouse was chosen as representative example.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp2-v4.docx
-
Supplementary file 3
Detected back splice junctions (BSJs) across samples.
Table summarises the total number of detected BSJs after the filtering step in each species. The percentage of BSJs that are unique to one, two, three or more than three samples of the same species is shown.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp3-v4.docx
-
Supplementary file 4
Total number of circRNAs in different species and tissues.
Indicated is the total number of different circRNAs that were annotated in each of the tissues across species.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp4-v4.docx
-
Supplementary file 5
Mean amplitude correlations.
Spearman’s rank correlation for the GC amplitude and GC content of introns and exons are calculated for each isochore and species. The mean correlation between the GC amplitude and GC content of introns and exons is shown for different splice sites relative to the circRNA.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp5-v4.docx
-
Supplementary file 6
GLM summary for presence of parental genes.
A generalised linear model was fitted to predict the probability of coding genes to be a parental gene (nopossum = 18807, nmouse = 22015, nrat = 11654, nrhesus = 21891, nhuman = 21744). The model was trained on 80% of the data (scaled values, cross-validation, 1000 repetitions, shown in rows labeled as ‘prediction’). Only the best predictors were kept and then used to predict probabilities for the remaining 20% of data points (validation set, shown in rows labeled as ‘validation’). Log-odds ratios, standard error and 95% confidence intervals (CI) for the validation set have been (beta) standardised.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp6-v4.docx
-
Supplementary file 7
GLM summary for ‘sharedness’ of hotspots.
A generalised linear model was fitted to predict the probability of a hotspot to be present across multiple species (nopossum = 872, nmouse = 848, nrat = 665, nrhesus = 1682, nhuman = 2022). Reported log-odds ratios, standard error and 95% confidence intervals (CI) are (beta) standardised.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp7-v4.docx
-
Supplementary file 8
GLM summary for circRNA hotspots among parental genes.
A generalised linear model was fitted to predict the probability of circRNA hotspots among parental genes; parental genes were filtered for circRNAs that were either species-specific or occurred in orthologous loci across therian species (nopossum = 869, nmouse = 503, nrat = 425, nrhesus = 912, nhuman = 1213). The model was trained on 80% of the data (scaled values, cross-validation, 1000 repetitions, shown in rows labeled as ‘prediction’). Only the best predictors were kept and then used to predict probabilities for the remaining 20% of data points (validation set, shown in rows labeled as ‘validation’). Log-odds ratios, standard error and 95% confidence intervals (CI) for the validation set have been (beta) standardised.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp8-v4.docx
-
Supplementary file 9
Analysis of highly expressed circRNAs.
Highly expressed circRNAs were defined as the circRNAs present in the 90% expression quantile of a tissue in a species. Per species, the circRNAs in the 90% expression quantiles from each of the three tissues were then pooled for further analysis (nopossum = 158, nmouse = 156, nrat = 217, nrhesus = 340, nhuman = 471) and their properties compared to circRNAs outside the 90% expression quantile. Highly expressed circRNAs are designated ‘1’, others ‘0’. Differences in genomic length, circRNA length, exon number and GLM model performance were assessed with a Student’s t-Test; p-values are indicated in the table (ns = non-significant).
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp9-v4.docx
-
Supplementary file 10
GLM for highly expressed circRNAs based on ‘age groups’.
A generalised linear model was fitted on the complete dataset to predict the probability of parental genes of highly expressed circRNAs to be produce circRNAs in multiple species (nopossum = 869, nmouse = 844, nrat = 661, nrhesus = 1673, nhuman = 2016). The ‘sharedness’ definition is based on the phylogeny of species as: present in only one species, in rodents (mouse, rat) or primates (rhesus, human), eutherian species (rodents + at least one primate, or primates + at least one rodent) and therian species (opossum + rodents + at least one primate, or opossum + primates + at least one rodents). Log-odds ratios, standard error, 95% confidence intervals (CI) and p-values are shown.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp10-v4.docx
-
Supplementary file 11
Frequency and enrichment of top-5 dimers in shared and species-specific circRNA loci.
The total number of detected top-5 dimers in shared and species-specific circRNA loci as well as their enrichment after correction for co-occurrence in multiple RVCs (see Materials and methods) are shown. Loci were normalised by the number of detected genes in each category before calculating the enrichment of dimers in shared over species-specific loci. The number of parental genes in both categories is shown below the species name. For mouse, only the top-3 dimers, which are outside the 95% frequency quantile, are shown (see Materials and methods). For rhesus, the analysis could only be done on a subset of genes due to lifting uncertainties between the rheMac2 and the rheMac3 genome (see Materials and methods).
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp11-v4.docx
-
Supplementary file 12
CircRNA annotation file for opossum.
A gtf-file with all circRNA transcripts including the transcript and exon coordinates.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp12-v4.gtf
-
Supplementary file 13
CircRNA annotation file for mouse.
A gtf-file with all circRNA transcripts including the transcript and exon coordinates.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp13-v4.gtf
-
Supplementary file 14
CircRNA annotation file for rat.
A gtf-file with all circRNA transcripts including the transcript and exon coordinates.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp14-v4.gtf
-
Supplementary file 15
CircRNA annotation file for rhesus macaque.
A gtf-file with all circRNA transcripts including the transcript and exon coordinates.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp15-v4.gtf
-
Supplementary file 16
CircRNA annotation file for human.
A gtf-file with all circRNA transcripts including the transcript and exon coordinates.
- https://cdn.elifesciences.org/articles/67991/elife-67991-supp16-v4.gtf
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/67991/elife-67991-transrepform-v4.pdf





