Inter-tissue convergence of gene expression during ageing suggests age-related loss of tissue and cellular identity
Figures
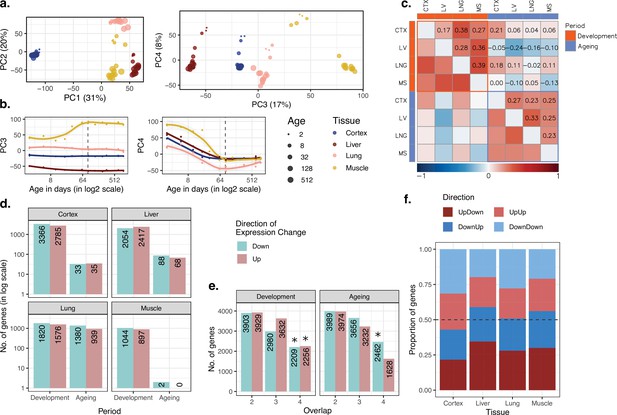
Data summary and age-related expression patterns.
(a) Principal components analysis (PCA) of expression levels of 15,063 protein-coding genes across four tissues of 16 mice. Values in parentheses show the variation explained by each component. (b) Age trajectories of PC3 (left) and PC4 (right). Spearman’s correlation coefficients between PC4 and age in each tissue in development range between 0.88 and 0.99 (see Figure 1—source data 1 for all tests). The dashed vertical line indicates 90 days of age, separating development and ageing periods. Age distribution of samples are given in Figure 1—figure supplement 1. (c) Similarity between the age-related gene expression changes (Spearman’s correlation coefficient between expression and age without a significance cutoff) across tissues in development and ageing. Similarities were calculated using Spearman’s correlation coefficient between expression-age correlations across tissues. CTX, cortex; LV, liver; LNG, lung; MS, muscle. (d) The number of significant age-related genes in each tissue (false discovery rate [FDR]-corrected p-value<0.1). (e) Shared age-related genes among tissues identified without using a significance cutoff. The x-axis shows the number of tissues among which age-related genes are shared. Significant overlaps are indicated with an asterisk (Figure 1—figure supplement 4). (f) The proportion of age-related expression change trends (no significance cutoff was used) in each tissue across the lifetime. UpDown: upregulation in development and downregulation in the ageing; DownUp: downregulation in development and upregulation in the ageing; UpUp: upregulation in development and upregulation in the ageing; DownDown: downregulation in development and downregulation in the ageing. We confirmed the robustness of the results using variance stabilising transformation (VST normalisation in Figure 1—figure supplement 10).
-
Figure 1—source data 1
Data summary, age-related expression patterns, and reversal patterns.
- https://cdn.elifesciences.org/articles/68048/elife-68048-fig1-data1-v4.xlsx
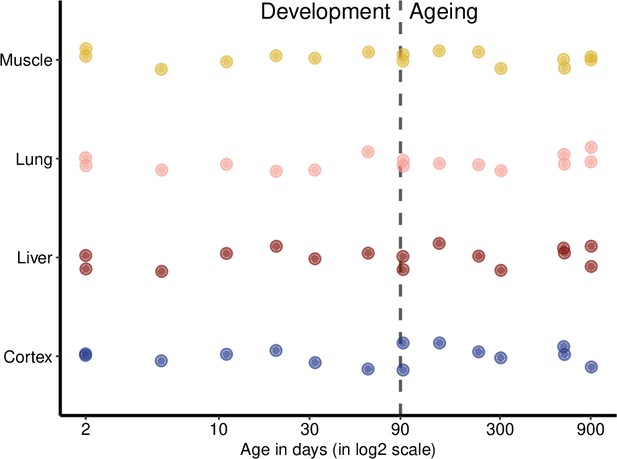
Age distribution of samples.
The x-axis shows the age in days in a log2 scale, and the y-axis lists different tissues. The period from 2- to 61-day-old mice is considered as postnatal development (referred to as development for brevity in the main text) and above 90-day-old as the ageing period. Random jitter was added on the y-axis to avoid overlap between points.
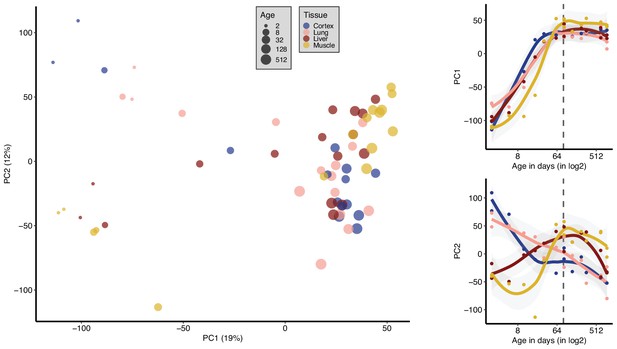
Principal components analysis (PCA) with all samples (tissue effect removed).
PCA using all samples (n = 16) after each tissue is standardised separately (i.e. gene expression values for individuals are scaled to mean = 0, sd = 1). PC1 (x-axis) and PC2 (y-axis) are plotted, and the variation explained by each PC is denoted within parentheses on each axis. The size of the points indicates the age, and the colour shows the tissue. The plots on the right show the correlations between the PCs (y-axis) and age (x-axis, on the log2 scale) in development and ageing. PC1-age Spearman’s correlation test during development (n = 7 mice); abs(ρdev) = [0.88, 0.99], nominal pdev < 0.01 for each tissue, same test for PC2 vs. age; abs(ρdev) = [0.30, 0.99], nominal pdev < 0.01 except muscle (Figure 1—source data 1).
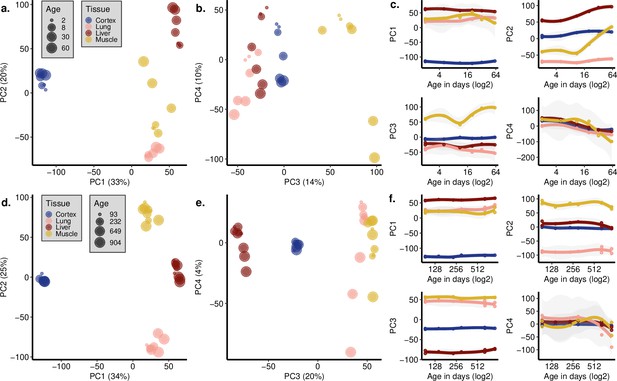
Principal components analysis (PCA) with development and ageing periods separately.
PCA using only the samples from the development period (2–61 days of age, n = 7) (a–c) and the ageing period (93–904 days of age, n = 9) (d–f). (a, d) PC1 (x-axis) vs. PC2 (y-axis) and (b, e) PC3 (x-axis) vs. PC4 (y-axis) are plotted, and the variation explained by each PC is denoted within parentheses on each axis. The size of the points indicates the age, and the colour shows the tissue. (c, f) Correlation between the PCs (y-axis) and age (x-axis, in the log2 scale) in development (c) and ageing (f). (c) Age effects can be observed in PC2 and PC4 in development: PC2-age Spearman’s correlation test, abs(ρ) = [0.72, 0.94], nominal p<0.05 in 3/4 tissues; PC4-age Spearman’s correlation test, abs(ρ) = [0.88, 0.99], nominal p<0.01 in all tissues. Inter-tissue transcriptome divergence can be observed as a trend in PC3–PC4 space (change in the mean Euclidean distance among tissues with age in PC1–4 space, ρ = 0.95, p=0.0008). (f) A small age effect can be observed in PC4 in ageing: PC4-age Spearman’s correlation test: abs(ρ) = [0.11, 0.77], nominal p<0.05 in 2/4 tissues. Inter-tissue transcriptome convergence can be observed as a subtle trend in PC1–4 space (change in mean Euclidean distance among tissues with age in PC1–4 spaces, ρ = −0.64, p=0.059). All PC-age correlation test results are given in Figure 1—source data 1.
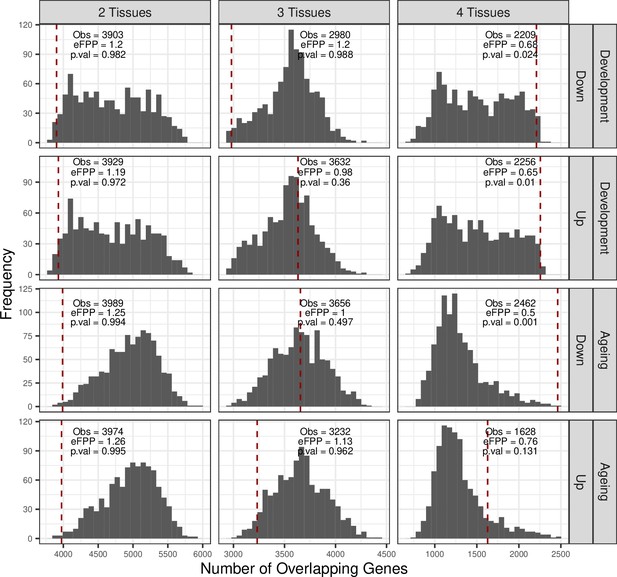
Permutation test results for shared expression trends among tissues.
Permutation test results of shared up/down genes across tissues for development and ageing periods. ‘Up’ and ‘down’ indicate positive and negative expression-age correlations (ρ), respectively. No significance cutoff was applied for choosing up/down genes in tissues (i.e. only considering ρ > 0 or ρ < 0). The null distributions are created by permuting individual ages and calculating expression-age correlations in each tissue, then summing the number of genes changing in the same direction in 2, 3, and 4 tissues. The red dashed lines show the observed values, also noted as ‘Obs.’ The estimated false-positive proportion (eFPP) was calculated as the ratio between the median expected value from the permutations and the observed value. p-Values were calculated as the proportion of permutations that are higher than or equal to the observed value.
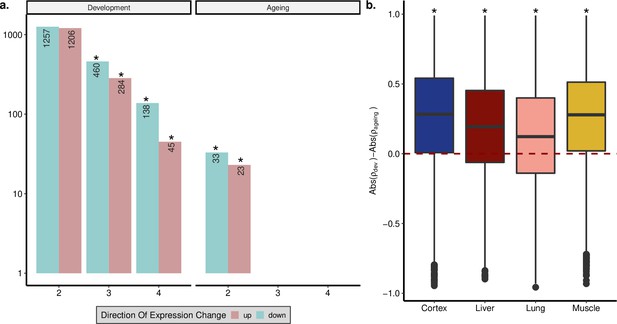
Shared age-related genes among tissues in development and ageing.
(a) Overlap between significant (false discovery rate [FDR]-corrected p-value<0.1) age-related gene sets among tissues. The x-axis shows the number of tissues compared; 2: overlap in two tissues, 3: overlap in three tissues, 4: overlap in four tissues. Cyan: downregulation with age; pink: upregulation with age. Significant overlaps (permutation test, p<0.05, see Figure 1—figure supplement 6 for test results) are indicated with asterisks. (b) The differences between the magnitude of age-related expression changes in development and ageing: (abs(ρdev)–abs(ρageing)), for each gene (n = 15,063 genes) in four tissues (Wilcoxon signed-rank test, p<10–16 for each tissue).
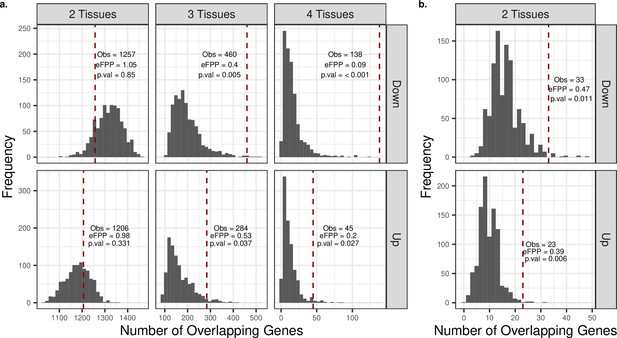
Permutation test results for significant trends shared among tissues.
Permutation test result for shared ‘up’ (or ‘down’) genes among tissues in development (a) and ageing (b). ‘Up’ and ‘down’ indicate positive and negative expression-age correlations (ρ), respectively. Significant up/down genes were chosen with false discovery rate [FDR]-corrected p-value<0.1, and their overlap across tissues was calculated. To create the null distributions, we chose as many up (or down) genes in permutations as the observed up (or down) genes in each tissue and then calculated the number of overlapping genes among tissues. The dashed red line shows the observed number of shared up (or down) genes between tissues, and estimated false-positive proportion (eFPP) was calculated as the ratio between the median expected value from the permutations and the observed value. ‘Obs’: number of genes displaying the same significant age-related change pattern among tissues. The p-value was calculated as the proportion of permutations that are higher than or equal to the observed value.
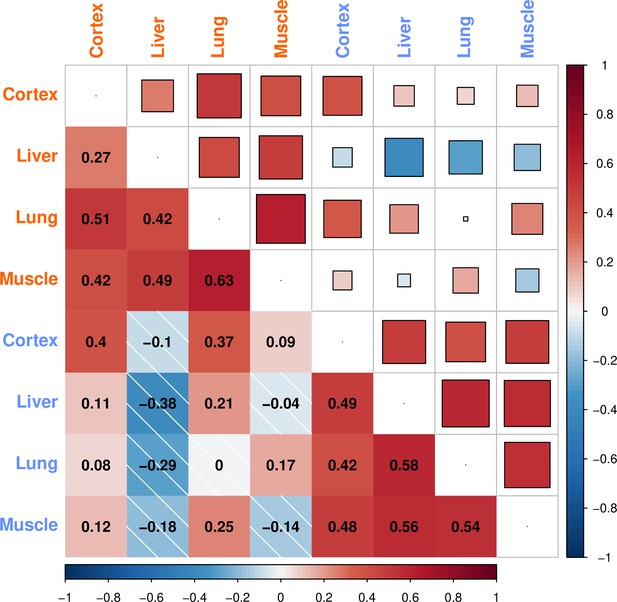
Similarities between age-related gene expression changes among tissues.
The similarity between the age-related gene expression changes (Spearman’s correlation coefficient between expression and age) across tissues in development and ageing. Similarities were calculated using Spearman’s correlations coefficient between expression-age correlations (with cutoff: |ρ| > 0.6) across tissues. No significance cutoff was used for expression change similarities. The intensity of the colours shows the magnitude of the correlation coefficient, where darker blue indicates a stronger negative correlation and darker red indicates a stronger positive correlation. Correlation values are written on the lower triangle. The colour of the tissue label indicates development (orange) and ageing (blue) datasets.
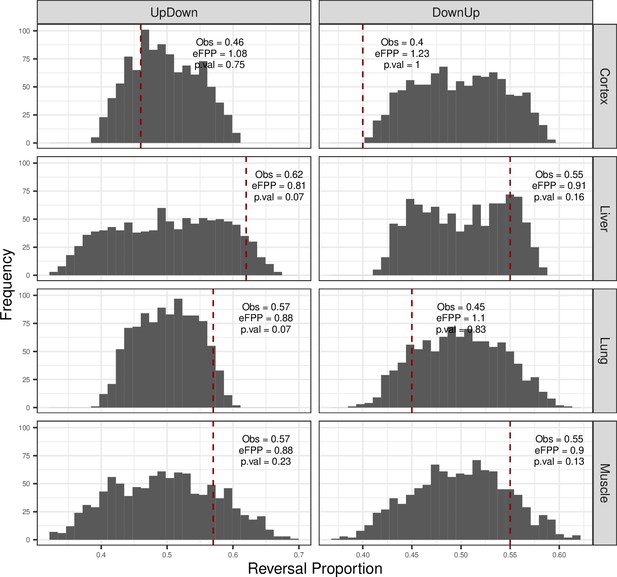
Permutation test results for reversal patterns in each tissue.
Permutation test result for up-down and down-up reversal genes in each tissue. Developmental up- (or down-) genes, that is, genes with expression-age ρ > 0 (or ρ < 0), were kept constant, and the age labels of the individuals in the ageing period were permuted (Materials and methods). No significance cutoff was used in choosing genes. The dashed red line shows the observed (‘Obs’) up-down (or down-up) proportions in tissues, and estimated false-positive proportion (eFPP) was calculated as the median expected value of the permutations divided by the observed value. p-Values were calculated as the proportion of permutations that are higher than or equal to the observed value. Left panel: up-down reversal proportions were calculated as UD/(UD + UU). Right panel: down-up reversal proportions were calculated as DU/(DU + DD).
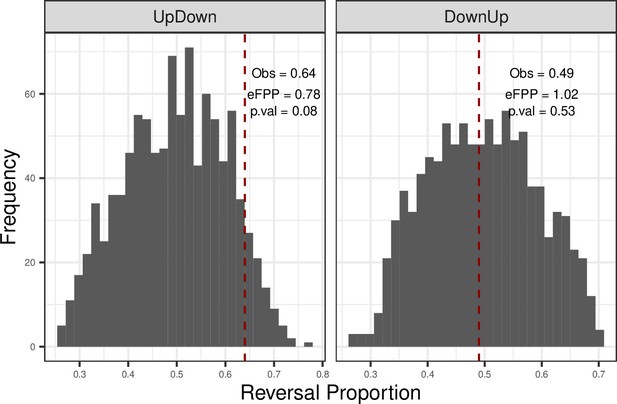
Permutation test results for shared reversals among tissues.
Permutation test result for shared up-down (or down-up) reversal genes across tissues. Developmental up- (or down-) genes were kept constant (among 2255 shared up-genes and 2209 shared down-genes in development), and the age labels of the individuals in the ageing period were permuted (Materials and methods). The dashed red line shows the observed (‘Obs’) up-down (or down-up) proportions shared among tissues, and estimated false-positive proportion (eFPP) was calculated as the median of the permutations divided by the observed value. p-Values were calculated as the proportion of permutations that are higher than or equal to the observed value. Left panel: up-down reversal proportions were calculated as UD/(UD + UU). Right panel: down-up reversal proportions were calculated as DU/(DU + DD).
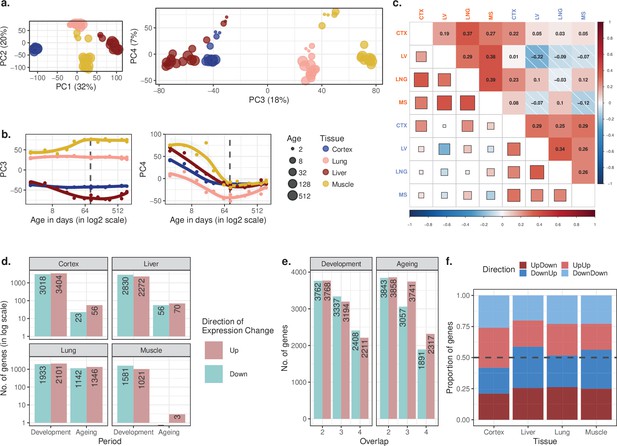
Replication of Figure 1 results using variance stabilising transformation (VST) normalisation.
To confirm the robustness of the results to the choice of normalisation method, the analysis was repeated using an alternative normalisation approach (VST) implemented in the DESeq2 package (see Materials and methods). (a) Principal components analysis (PCA) of expression levels of 14,973 protein-coding genes across four tissues of 16 mice. Values in parentheses show the variation explained by each component. (b) Age trajectories of PC3 (left) and PC4 (right). Spearman’s correlation coefficients between PC4 and age in each tissue in development range between 0.58 and 0.99 (see Figure 1—source data 1 for all tests). The dashed vertical line indicates 90 days of age, separating development and ageing periods. (c) Similarity between the age-related gene expression changes (Spearman’s correlation coefficient between expression and age without a significance cutoff) across tissues in development and ageing. Similarities were calculated using Spearman’s correlation coefficient between expression-age correlations across tissues. CTX, cortex; LV, liver; LNG, lung; MS, muscle. (d) The number of significant age-related genes in each tissue (false discovery rate [FDR]-corrected p-value<0.1). (e) Shared age-related genes among tissues identified without using a significance cutoff. The x-axis shows the number of tissues among which age-related genes are shared. (f) The proportion of age-related expression change trends in each tissue across the lifetime. No significance cutoff was used. UpDown: upregulation in development and down-regulation in the ageing; DownUp: downregulation in development and upregulation in the ageing; UpUp: upregulation in development and upregulation in the ageing; DownDown: downregulation in development and downregulation in ageing.
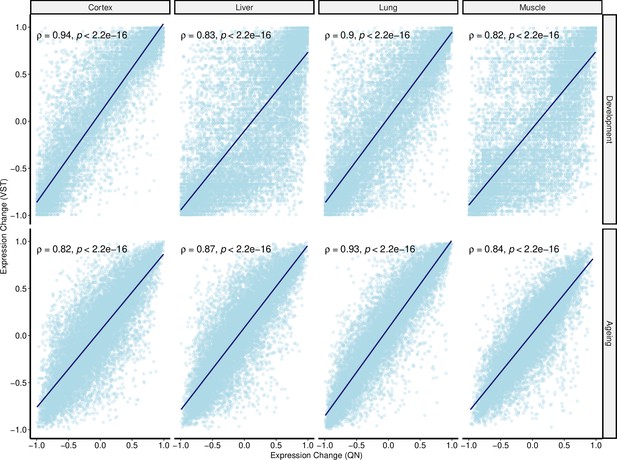
Correlation between quantile normalised (QN) and variance stabilising transformation (VST) normalisation methods using age-related expression changes.
Spearman’s correlation coefficient between expression trajectories of QN (x-axis) and VST (VST method from DESeq2 package, y-axis) normalised data. Expression trajectories were calculated using Spearman’s correlation coefficient between age and expression level for each gene in both periods (ndev = [14,705, 14,710], nageing = [14,689, 14,710]). Blue lines represent the regression lines.
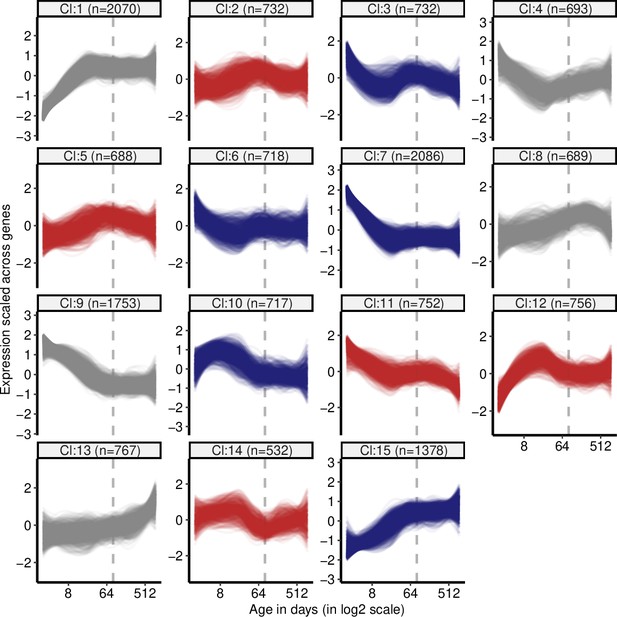
Clustering of genes by expression levels in cortex tissue.
k-means clustering (k = 15) of genes (15,063) using expression levels in cortex tissue. Numbers in the parentheses show the number of genes in each cluster. Expression levels of genes were scaled across samples (mean = 1, sd = 0) before clustering. The optimal number of clusters was determined with gap statistics (see Materials and methods). Clusters enriched among divergence-convergence (DiCo) genes compared to all other clusters are indicated with red colour, and the ones depleted among DiCo genes are indicated with blue colour. The list of genes belonging to each cluster and their enrichment among DiCo genes are given in Figure 1—source data 1.
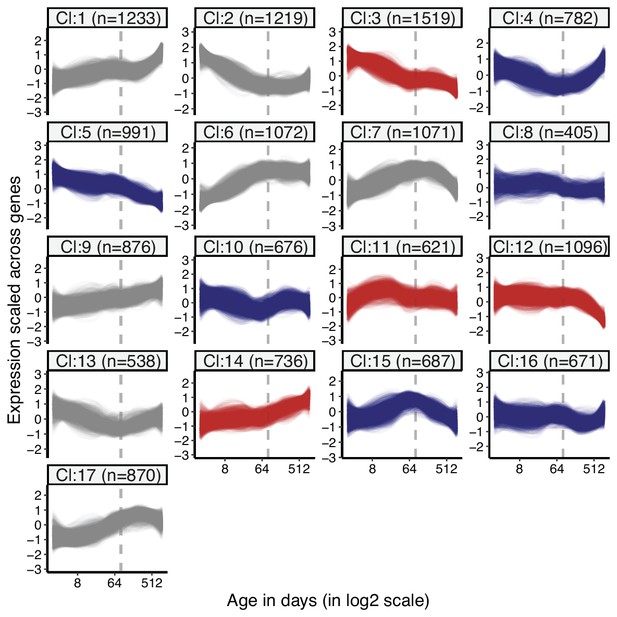
Clustering of genes by expression levels in lung tissue.
k-means clustering (k = 17) of genes (15,063) using expression levels in lung tissue. Numbers in the parentheses show the number of genes in each cluster. Expression levels of genes were scaled across samples (mean = 1, sd = 0) before clustering. The optimal number of clusters was determined with gap statistics (see Materials and methods). Clusters enriched among divergence-convergence (DiCo) genes are indicated with red colour, and the ones depleted among DiCo genes are indicated with blue colour. The list of genes belonging to each cluster and their enrichment among DiCo genes are given in Figure 1—source data 1.
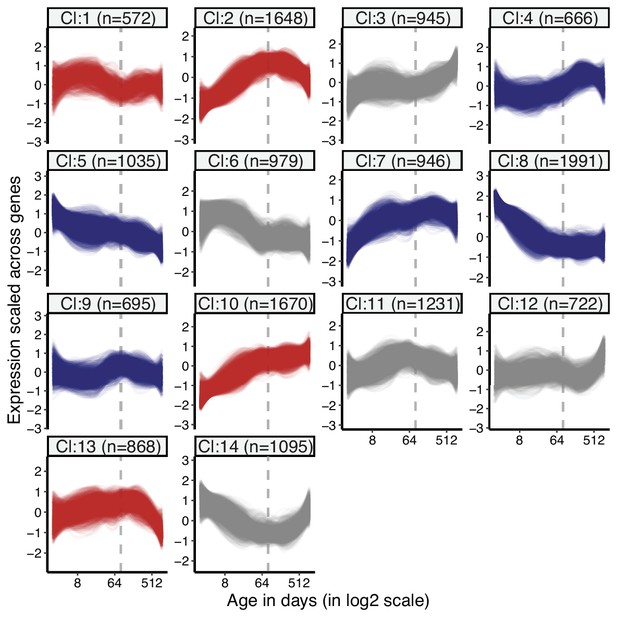
Clustering of genes by expression levels in liver tissue.
k-means clustering (k = 14) of genes (15,063) using expression levels in liver tissue. Numbers in the parentheses show the number of genes in each cluster. Expression levels of genes were scaled across samples (mean = 1, sd = 0) before clustering. The optimal number of clusters was determined with gap statistics (see Materials and methods). Clusters enriched among divergence-convergence (DiCo) genes are indicated with red colour, and the ones depleted among DiCo genes are indicated with blue colour. The list of genes belonging to each cluster and their enrichment among DiCo genes are given in Figure 1—source data 1.
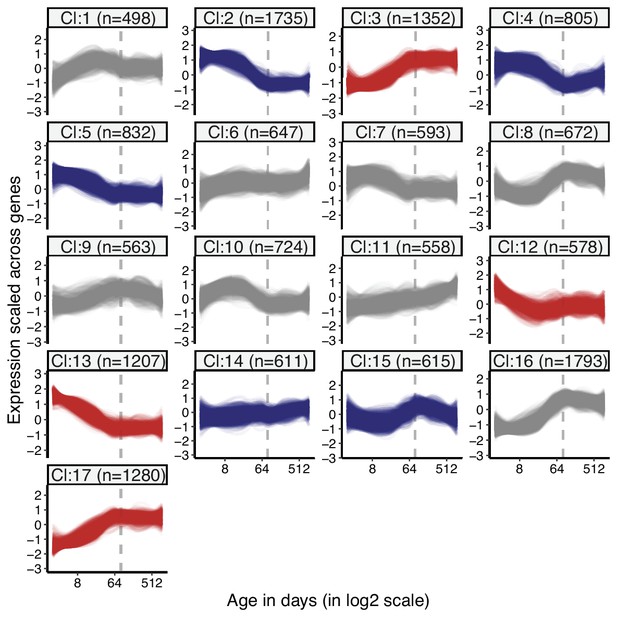
Clustering of genes by expression levels in muscle tissue.
k-means clustering (k = 17) of genes (15,063) using expression levels in muscle tissue. Numbers in the parentheses show the number of genes in each cluster. Expression levels of genes were scaled across samples (mean = 1, sd = 0) before clustering. The optimal number of clusters was determined with gap statistics (see Materials and methods). Clusters enriched among divergence-convergence (DiCo) genes are indicated with red colour, and the ones depleted among DiCo genes are indicated with blue colour. The list of genes belonging to each cluster and their enrichment among DiCo genes are given in Figure 1—source data 1.
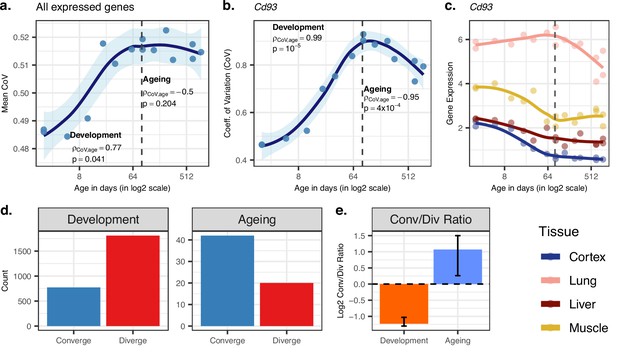
Age-related change in gene expression variation among tissues estimated with coefficient of variation (CoV).
(a) Transcriptome-wide mean CoV trajectory with age. Each point represents the mean CoV value of all protein-coding genes (15,063) for each mouse (n = 15) except the one that lacks expression data in the cortex. (b) Age effect on CoV value of the Cd93 gene which has the highest rank for the divergence-convergence (DiCo) pattern in four tissues (Materials and methods). CoV increases during development and decreases during ageing, indicating expression levels show DiCo patterns among tissues. (c) Expression trajectories of the gene Cd93 in four tissues. (d) The number of significant CoV changes with age (false discovery rate [FDR]-corrected p-value<0.1) during development (left, nconverge = 772, ndiverge = 1809) and ageing (right, nconverge = 42, ndiverge = 20). Converge: genes showing a negative correlation (ρ) between CoV and age; diverge: genes showing a positive correlation between CoV and age. (e) Log2 ratio of convergent/divergent genes in development and in ageing. The graph represents only genes showing significant CoV changes (FDR-corrected p-value<0.1, given in panel d). Error bars represent the range of log2 ratios calculated from leave-one-out samples using the jackknife procedure (Materials and methods, values are given in Figure 2—source data 1).
-
Figure 2—source data 1
All the data related to divergence-convergence (DiCo) pattern: age-related coefficient of variation (CoV) change of genes, pairwise tissue expression correlations, analysis of independent datasets; GSE34378 (Jonker et al.), GSE132040 (Schaum et al.), and GTEx.
- https://cdn.elifesciences.org/articles/68048/elife-68048-fig2-data1-v4.xlsx
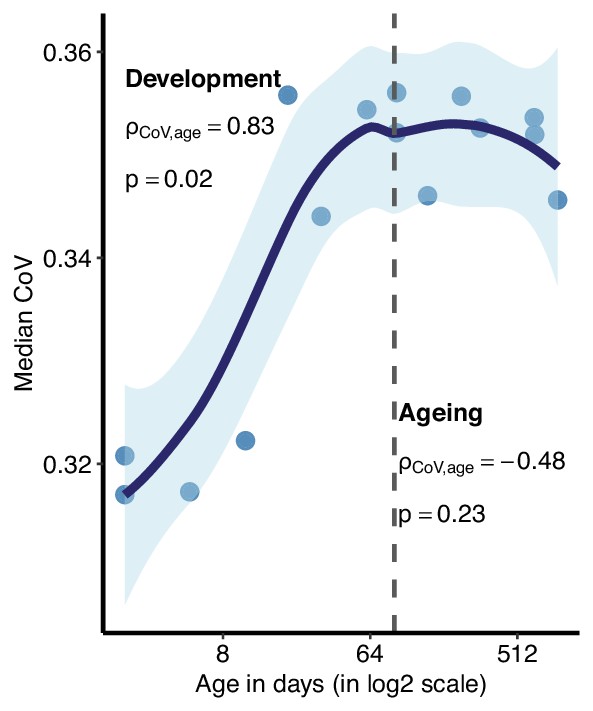
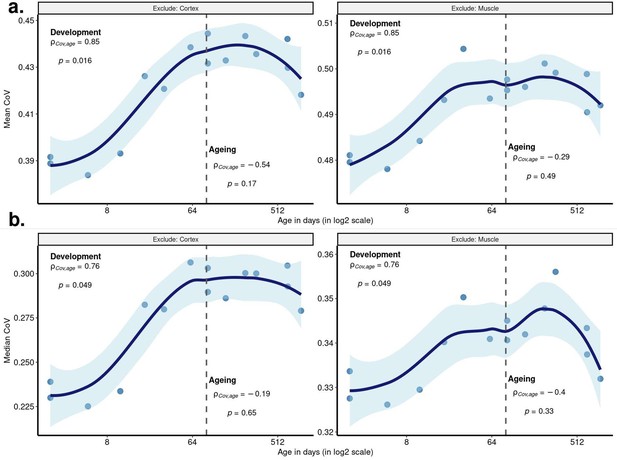
Age-related change in coefficient of variation (CoV) summarised across genes using median CoV values.
Each point represents the median CoV value (instead of the mean given in Figure 2a) of all protein-coding genes (15,063) for each mouse except the one that lacks expression data in the cortex (n = 15). x-axis is in log2 scale. The dashed grey line shows the start of the ageing period. The Spearman’s correlation coefficient and p-value for each period are indicated separately on the plot.
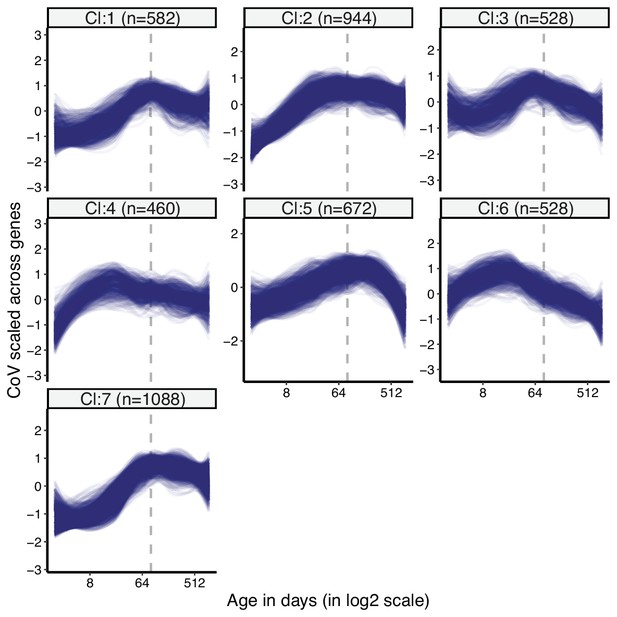
Clustering of divergence-convergence (DiCo) genes by expression variations (coefficient of variation [CoV]) among tissues.
k-means clustering (k = 7) of DiCo genes (4802) using CoV values. Numbers in the parentheses show the number of genes in each cluster. CoV values were scaled across genes (mean = 1, sd = 0) before clustering. The optimal number of clusters was determined with gap statistics (Materials and methods). The list of genes belonging to each cluster and their age-related CoV change correlations are given in Figure 2—source data 1.
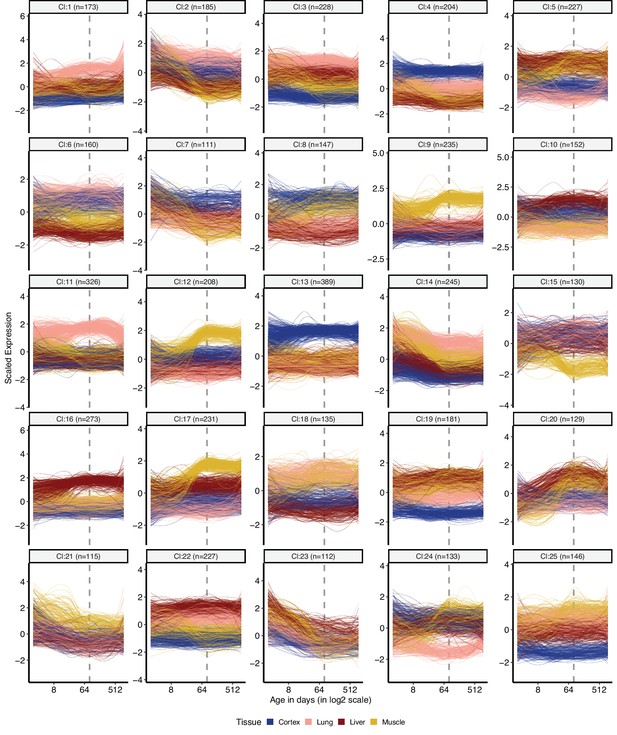
Clustering of divergence-convergence (DiCo) genes by expression levels in tissues.
k-means clustering (k = 25) of DiCo genes (n = 4802) using gene expression levels. Numbers in the parentheses show the number of genes in each cluster. Expression levels of genes were scaled across tissues (mean = 1, sd = 0) before clustering. The optimal number of clusters was determined with gap statistics (Materials and methods). The list of genes belonging to each cluster and their age-related CoV change correlations are given in Figure 2—source data 1.
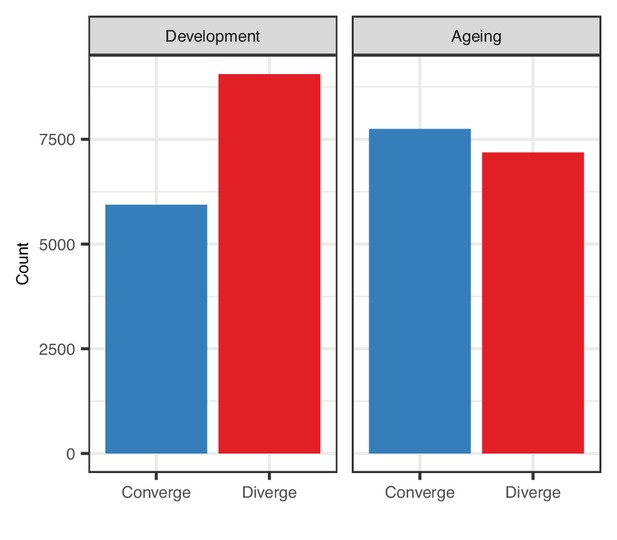
Number of genes with inter-tissue divergence and convergence tendencies in development and ageing.
The number of coefficient of variation (CoV) changes with age (without a significance cutoff) during development and ageing. Converge: genes showing negative correlation (ρ < 0) between CoV and age; diverge: genes showing positive correlation (ρ > 0) between CoV and age (development: nconverge = 5939, ndiverge = 9058; ageing: nconverge = 7748, ndiverge = 7187).
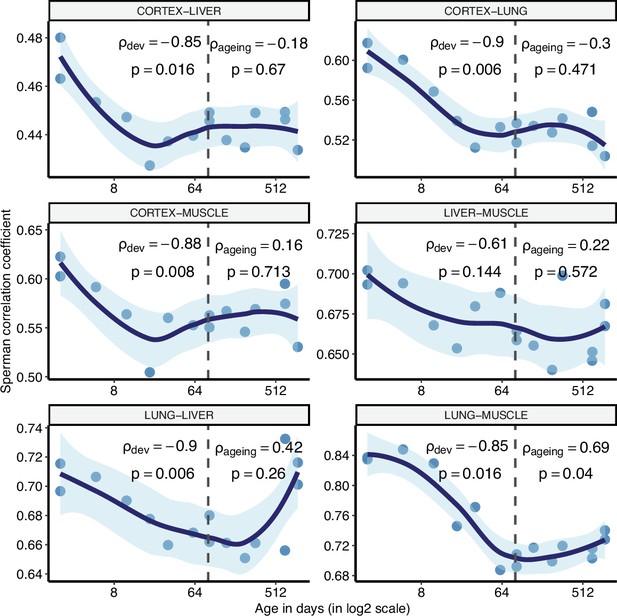
Pairwise tissue expression correlations.
Age-related changes in pairwise Spearman’s correlation coefficients for the expression levels (y-axis) between tissues of the same individual mouse in our dataset. The dashed grey line indicates the start of the ageing period. The Spearman’s correlation coefficients and p-values for each period are indicated separately on the plot.
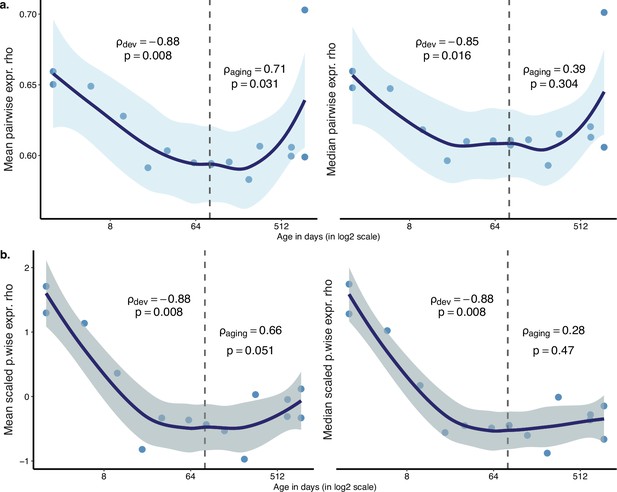
Summary of pairwise expression correlations among tissues.
Age-related change in the mean (left) or the median (right) pairwise expression correlations among tissues. Each point represents the mean (left) or the median (right) of pairwise expression correlations among tissues of the same mouse (mean/median values are calculated from Figure 2—figure supplement 5). (a) Absolute expression correlations were used to calculate the mean or the median. (b) Expression correlations were scaled within each tissue pair (mean = 1, sd = 0) before calculating the mean and median. The Spearman’s correlation coefficients and p-values for each period are indicated separately on the plot.
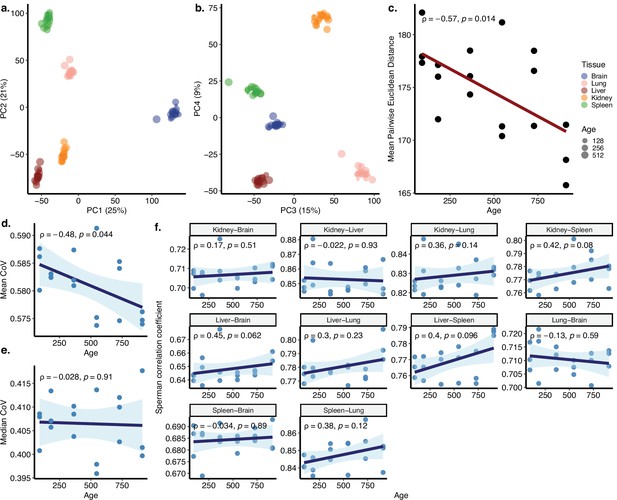
Coefficient of variation (CoV) and pairwise correlation analysis of Jonker dataset.
(a, b) Principal components analysis (PCA) of expression values of 17,661 protein-coding genes across five tissues (brain [cortex], liver, lung, kidney, spleen) of 18 individuals in the Jonker dataset (contains samples only from the ageing period). Values in parentheses show the variance explained by each PC. (c) The change in mean pairwise Euclidean distance between the PC values for the tissues of the same individuals (y-axis) with age (x-axis). Transcriptome-wide (d) mean and (e) median CoV changes with age across five tissues. The x-axis shows age in days. Each point represents the mean or median CoV value of all protein-coding genes for each individual. (f) Associationbetween age (x-axis) and gene expression correlations of each individual in pairwise tissues (y-axis). Spearman’s correlation coefficient and p-values are indicated in each plot.
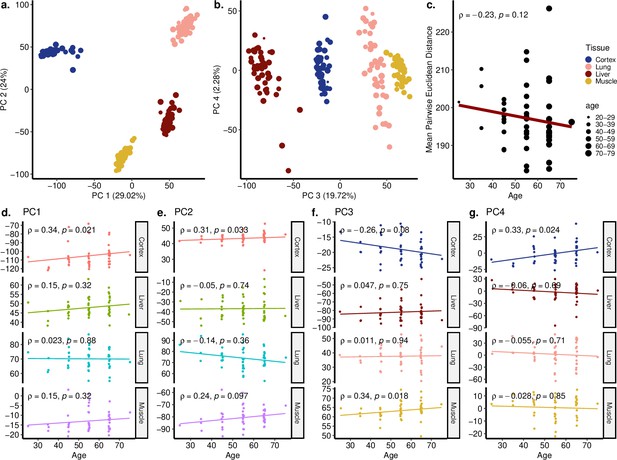
Principal components analysis (PCA) of GTEx dataset covering cortex, liver, lung, and muscle tissues.
(a, b) PCA of expression values of 16,197 genes across four tissues (cortex, liver, lung, muscle) of 47 individuals in GTEx. Values in parentheses show the variance explained by each PC. (c) The change in mean pairwise Euclidean distance between the PC values for the tissues of the same individuals (y-axis) with age (x-axis). (d–g) Association between the first four PCs (y-axis) and age (x-axis). The tissue and age of the samples are indicated by the colour and size of the points, respectively. Spearman’s correlation test results are indicated in each plot.
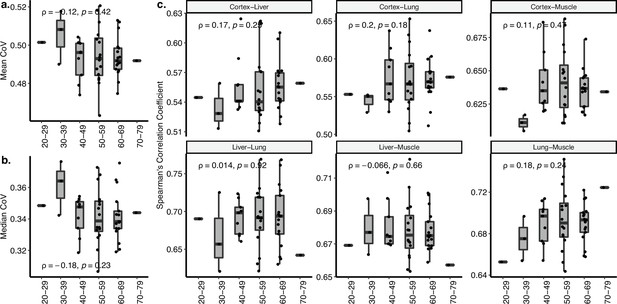
Coefficient of variation (CoV) and pairwise correlation analysis of GTEx dataset covering cortex, liver, lung, and muscle tissues.
(a, b) Transcriptome-wide mean (a) and median (b) CoV change with age across four tissues (cortex, liver, lung, muscle) in GTEx. Each point represents the mean or median CoV value of all protein-coding genes (16,197) for each individual (n = 47) in GTEx. Spearman’s correlation coefficients and p-values are also presented in the plot. (c) The change in pairwise Spearman’s correlation coefficient between gene expression values of the same individual across ages (y-axis) with age (x-axis). Spearman’s correlation coefficient and p-values between the pairwise tissue correlations and age are also presented in each plot.
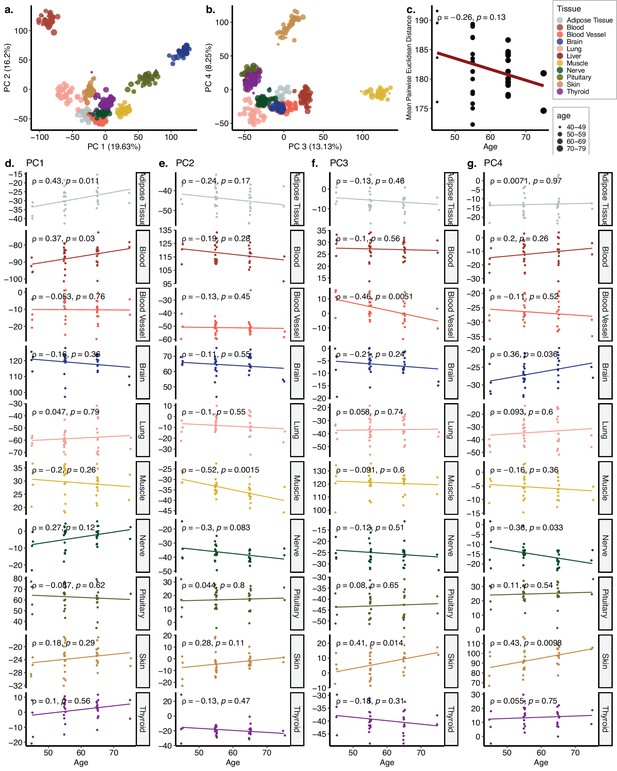
Principal components analysis (PCA) of GTEx dataset with 10 tissues.
(a, b) PCA of expression values of 16,290 genes across 10 tissues of 35 individuals in GTEx. Values in parentheses show the variance explained by each PC. (c) The change in mean pairwise Euclidean distance between the PC values for the tissues of the same individuals (y-axis) with age (x-axis). (d–g) Association between the first four PCs (y-axis) and age (x-axis). The tissue and age of the samples are indicated by the colour and size of the points, respectively.
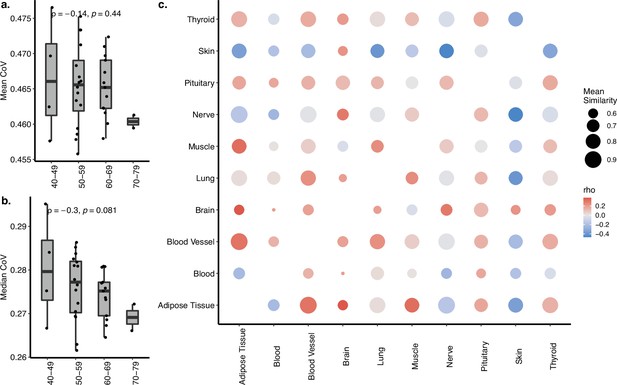
Coefficient of variation (CoV) and pairwise correlation analysis of GTEx dataset with 10 tissues.
(a, b) Transcriptome-wide mean (a) and median (b) CoV change with age across 10 tissues in GTEx. Each point represents the mean or median CoV value of all protein-coding genes (16,290) for each individual (n = 35) in GTEx. Spearman’s correlation coefficients and p-values are also presented in the plot. (c) Age-related changes in pairwise Spearman’s correlation coefficient between gene expression values of the same individual. The colour of points shows the correlations between age and pairwise correlations, where darker red colour indicates an increased correlation with age and darker blue indicates a decreased correlation. The size of points shows the mean similarity (correlation) between tissues using all ages. None of the correlations is significant after multiple testing correction (using Benjamini–Hochberg [BH]).
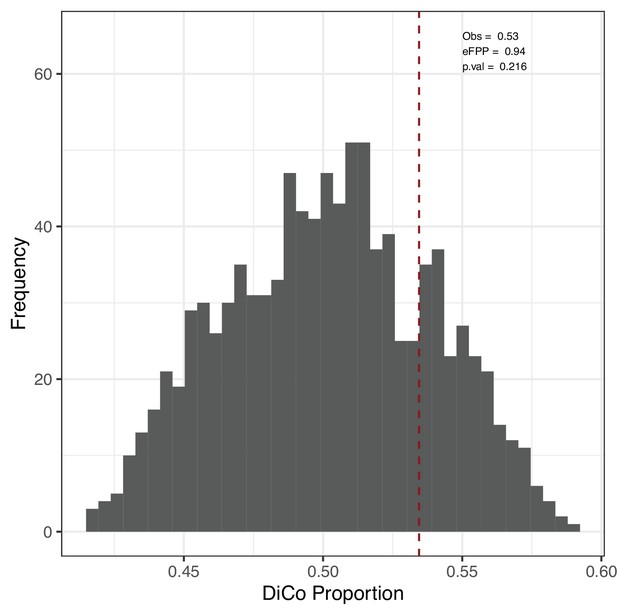
Permutation test result for the proportion of divergence-convergence (DiCo) genes.
DiCo genes (n = 4802) were tested with a permutation-based test explained in Materials and methods. We kept the divergent genes (n = 9058) in development constant and permuted age labels of individuals in the ageing period. Then, we calculated the DiCo proportion among those genes in permutations. ‘Obs‘: observed DiCo proportion (Obs = 4802/9058, i.e. DiCo/(DiCo + Di–); Di~: divergence across lifetime). Estimated false-positive proportion (eFPP) was calculated as the median expected proportion divided by the observed value. The p-value was calculated as the proportion of permutations that are higher than or equal to the observed value.
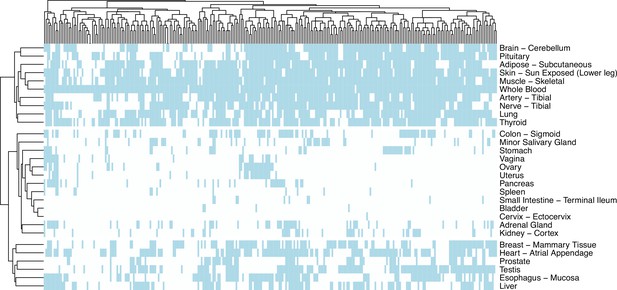
Clustering of tissues by the presence of samples from the same individuals.
Heatmap showing whether individuals (columns) have samples (light blue colour) in tissues (y-axis).
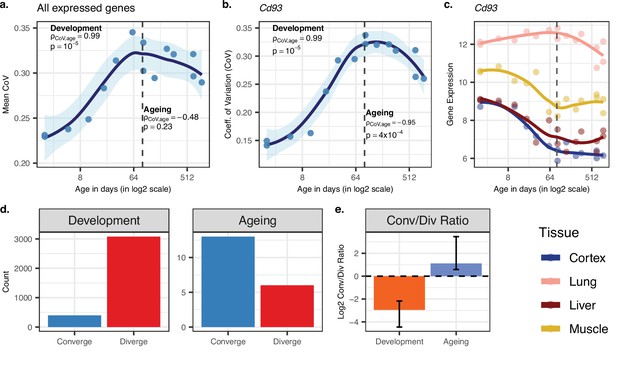
Reproducing Figure 2 results with variance stabilising transformation (VST) normalisation.
(a) Transcriptome-wide mean coefficient of variation (CoV) trajectory with age. Each point represents the mean CoV value of all protein-coding genes (14,973) for each mouse (n = 15) except the one that lacks expression data in the cortex. (b) Age effect on CoV value of the Cd93 gene which has the highest rank for the divergence-convergence (DiCo) pattern in four tissues (Materials and methods). CoV increases during development and decreases during ageing, indicating expression levels show DiCo patterns among tissues. (c) Expression trajectories of the gene Cd93 in four tissues. (d) The number of significant CoV changes with age (false discovery rate [FDR]-corrected p-value<0.1) during development (left, nconverge = 398, ndiverge = 3,078) and ageing (right, nconverge = 13, ndiverge = 6). Converge: genes showing a negative correlation (ρ) between CoV and age; diverge: genes showing a positive correlation between CoV and age. (e) Log2 ratio of convergent/divergent genes in development and in ageing. The graph represents only genes showing significant CoV changes (at FDR-corrected p-value<0.1, given in panel d). Error bars represent the range of log2 ratios calculated from leave-one-out samples in jackknife procedure.
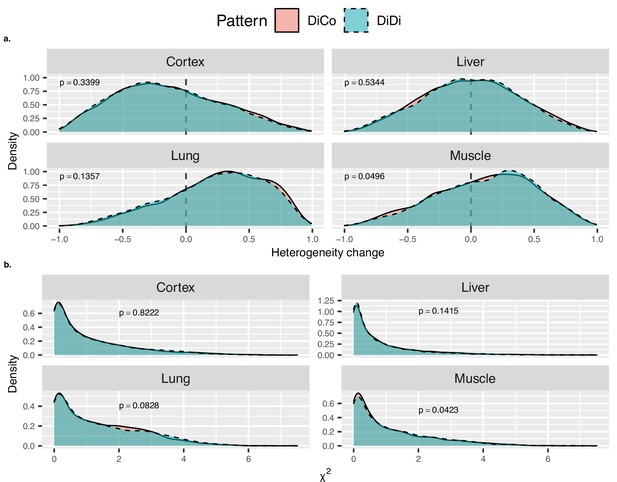
Effect of heteroscedasticity to divergence-convergence (DiCo) pattern.
Two different heteroscedasticity tests were performed to compare DiCo (n = 4802) vs. divergent-divergent (DiDi) (n = 4182, divergent throughout the lifetime) genes to test whether the convergence pattern is a result of the regression towards the mean. (a) Density plots of Spearman’s correlation coefficients (x-axis) between heterogeneity and age for DiCo and DiDi genes in each tissue. Heterogeneity was calculated as the absolute residuals of the linear regression between age (log2 scale) and expression (see Materials and methods). Only in muscle tissue the two-sided Kolmogorov–Smirnov (KS) test result was marginally significant in the direction of higher heterogeneity change for DiDi genes (p=0.0496). (b) Density plots of chi-square test statistics (x-axis) from Breusch–Pagan test (from ‘car’ package in R) between expression level and age (log2 scale) for DiCo and DiDi genes in each tissue. Only in muscle tissue the two-sided KS test result was significant in the direction of higher heterogeneity change for DiDi genes (p=0.0423). p-Values of KS test results between DiCo and DiDi genes are given within each plot.
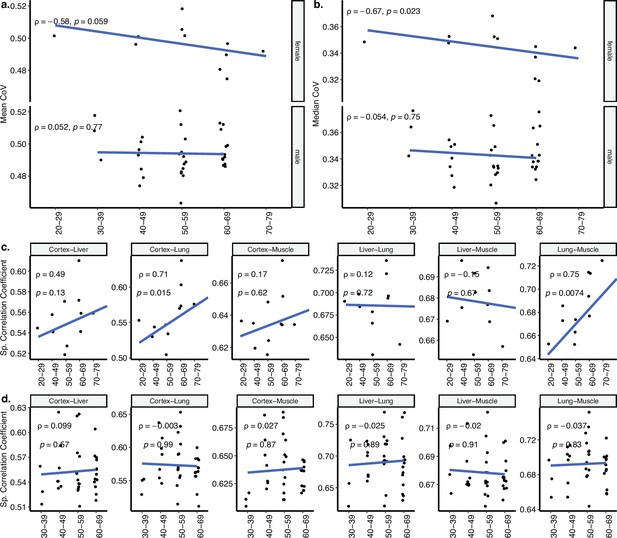
Sex effect on coefficient of variation (CoV) analysis using GTEx.
(a, b) Transcriptome-wide mean (a) and median (b) CoV change with age across four tissues (cortex, liver, lung, muscle) in GTEx for female (n = 11) and male (n = 36) individuals separately. Each point represents the mean or median CoV value of all protein-coding genes (16,197) for each individual. Spearman’s correlation coefficients and p-values are also presented in the plots. (c, d) The change in pairwise Spearman’s correlation coefficient between gene expression values of the same individual (y-axis) for (c) females (n = 11) and (d) males (n = 36), across ages (x-axis). Spearman’s correlation coefficient and p-values between the pairwise tissue correlations and age are also presented in each plot.
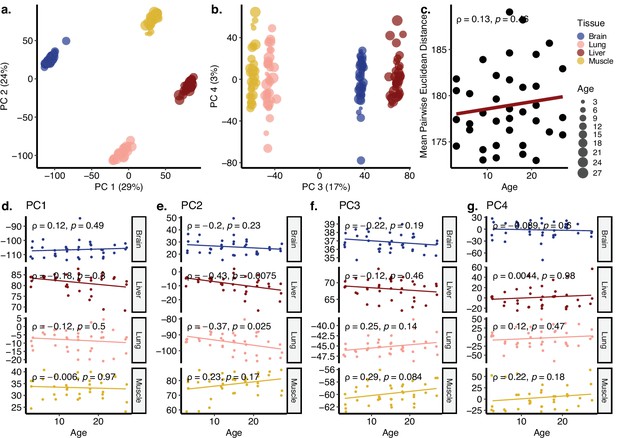
Principal components analysis (PCA) of Schaum dataset covering cortex, liver, lung, and muscle tissues.
(a, b) PCA of expression values of 16,806 genes across four tissues (cortex, liver, lung, muscle) of 37 individuals in the Schaum dataset. Values in parentheses show the variance explained by each PC. (c) The change in mean pairwise Euclidean distance between the PC values for the tissues of the same individuals (y-axis) with age (x-axis, in months). (d–g) Association between the first four PCs (y-axis) and age (x-axis, in months). The tissue and age of the samples are indicated by the colour and size of the points, respectively. Spearman’s correlation test results are indicated in each plot.
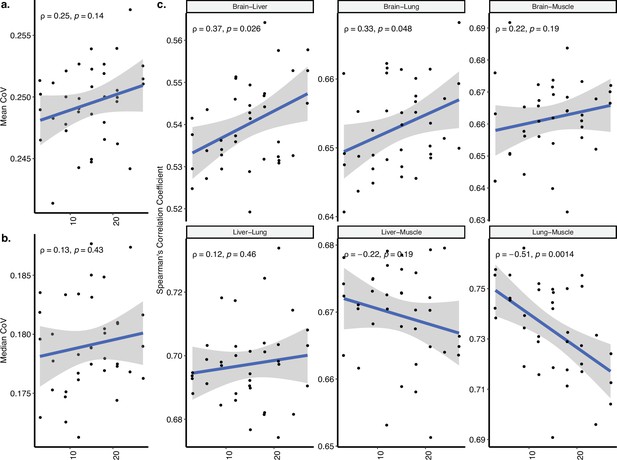
Coefficient of variation (CoV) and pairwise correlation analysis of Schaum dataset covering cortex, liver, lung, and muscle tissues.
(a, b) Transcriptome-wide mean (a) and median (b) CoV change with age (in months) across four tissues (cortex, liver, lung, muscle) in Schaum dataset. Each point represents the mean or median CoV value of all protein-coding genes (16,806) for each individual (n = 37). Spearman’s correlation coefficients and p-values are also presented in the plot. (c) The change in pairwise Spearman’s correlation coefficient between gene expression values of the same individual (y-axis) with age (x-axis, in months). Spearman’s correlation coefficient and p-values between the pairwise tissue correlations and age are also presented in each plot.
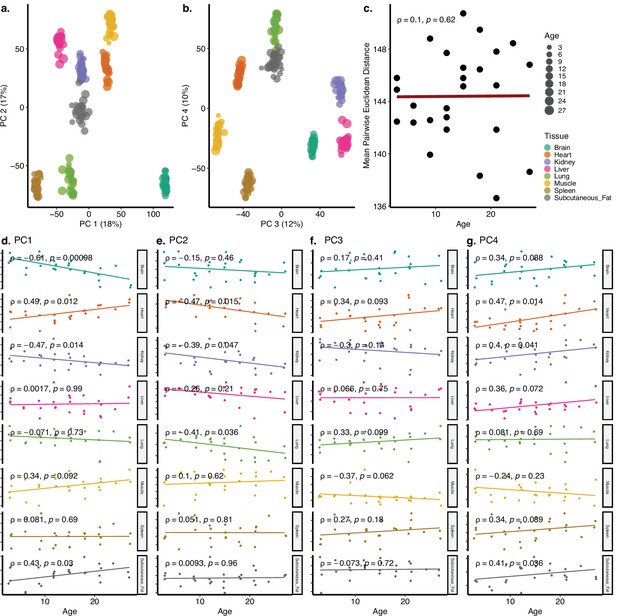
Principal components analysis (PCA) of Schaum dataset with eight tissues.
(a, b) PCA of expression values of 17,619 genes across eight tissues of 26 individuals in the Schaum dataset. Values in parentheses show the variance explained by each PC. (c) The change in mean pairwise Euclidean distance between the PC values for the tissues of the same individuals (y-axis) with age (x-axis, in months). (d–g) Association between the first four PCs (y-axis) and age (x-axis, in months). The tissue and age of the samples are indicated by the colour and size of the points, respectively.
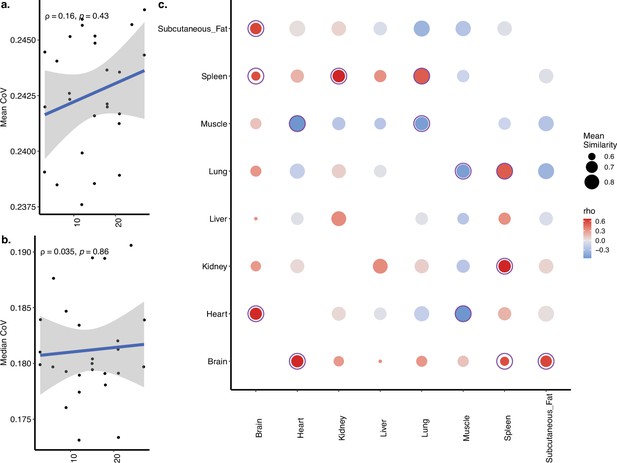
Coefficient of variation (CoV) and pairwise correlation analysis of Schaum dataset with eight tissues.
(a, b) Transcriptome-wide mean (a) and median (b) CoV change with age (in months) across eight tissues (brain [cortex], heart, kidney, liver, lung, muscle, spleen, subcutaneous fat) in Schaum dataset. Each point represents the mean or median CoV value of all protein-coding genes (17,619) for each individual (n = 26). Spearman’s correlation coefficients and p-values are also presented in the plot. (c) Age-related changes in pairwise Spearman’s correlation coefficient between gene expression values of the same individual. The colour of points shows the correlations between age and pairwise correlations, where darker red colour indicates an increased correlation with age and darker blue indicates a decreased correlation. The size of points shows the mean similarity (correlation) between tissues using all ages. Significant correlations are indicated with circles around the points after multiple testing correction using ‘Benjamini–Hochberg (BH) (5/7 of significant correlations were positive).
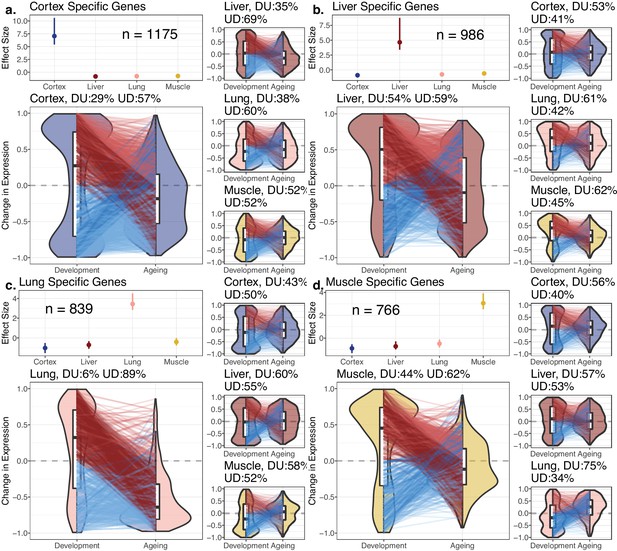
Reversal patterns among tissue-specific genes.
Age-related expression changes of the tissue-specific genes. In each panel (a–d), the upper-left subpanels show effect size (ES) calculated with the Cohen’s d formula, using expression levels of each gene among tissues (Materials and methods). The IQR (line range) and median (point) ES for each tissue are shown. The number of tissue-specific genes is indicated inside each subpanel. The lower-left subpanels show violin plots of the distribution of age-related expression change values (Materials and methods) among tissue-specific genes in development and in ageing. Each quadrant represents the plots for each tissue-specific gene group. The red and blue lines connect gene expression changes for the same genes in development and ageing. DU: percentage of down-up reversal genes among downregulated, tissue-specific genes in development; UD: percentage of up-down reversal genes among upregulated, tissue-specific genes in development. Tissue-specific genes are enriched among UD reversal genes except in the liver (Fisher’s exact test; ORcortex = 1.65, ORlung = 6.52, ORliver = 0.87, ORmuscle = 1.26, p<0.05 for each test except in liver).
-
Figure 3—source data 1
Effect sizes for determination of tissue-specific genes, enrichment of divergence-convergence (DiCo), and reversal genes within tissue-specific genes.
- https://cdn.elifesciences.org/articles/68048/elife-68048-fig3-data1-v4.xlsx
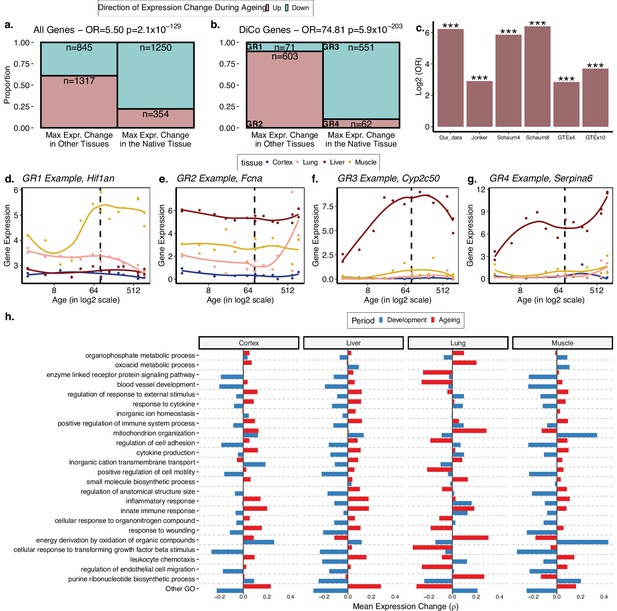
The loss of tissue-specific expression during ageing and functional enrichment of divergence-convergence (DiCo) genes.
(a) Mosaic plot showing the association between maximal expression change in native vs. non-native tissues (x-axis) vs. down- (cyan) or upregulation (pink) during ageing across all tissue-specific genes (n = 3766). The highly significant odds ratio indicates that genes native to a tissue tend to be downregulated during ageing in that native tissue if they show maximal expression change during ageing in that tissue. Conversely, if they show maximal expression change during ageing in non-native tissue, those genes are upregulated during ageing. Consequently, tissue-specific expression patterns established during development will tend to be lost during ageing. (b) The same as (a) but using only the tissue-specific genes that show the DiCo pattern (n = 1287). (c) Summary of the association tests for ‘direction of maximal expression change in native vs. non-native tissues’ across all datasets analysed. The y-axis shows log2-transformed odds ratio (OR) for each dataset (x-axis) – Schaum4: using the same four tissues as our dataset. Schaum8: using eight tissues. GTEx4: using the same four tissues as our dataset. GTEx10: using 10 tissues. ***False discovery rate (FDR)-corrected p-value<10–87. p-Values are given in Table 2. The four groups are annotated as GR1–4 and gene expression changes for each group in our dataset is exemplified in (d–g). (h) Trends of expression change with age of genes (x-axis) in categories enriched in DiCo (Gene Set Enrichment Analysis [GSEA]). Enriched categories (n = 184) are summarised into representatives (y-axis) using hierarchical clustering and Jaccard similarities (Materials and methods). Categories are ordered by the number of genes they contain from highest (bottom, n = 290) to lowest (top, n = 26). The most distant cluster with low within-cluster similarity in the hierarchical clustering (other Gene Ontology [GO]) was clustered separately and given in Figure 4—figure supplement 1.
-
Figure 4—source data 1
Gene Set Enrichment Analysis (GSEA) result of divergence-convergence (DiCo) genes, DiCo enrichment with tissue-specific expression loss, age-related expression change correlations, and convergence overlaps among datasets.
- https://cdn.elifesciences.org/articles/68048/elife-68048-fig4-data1-v4.xlsx
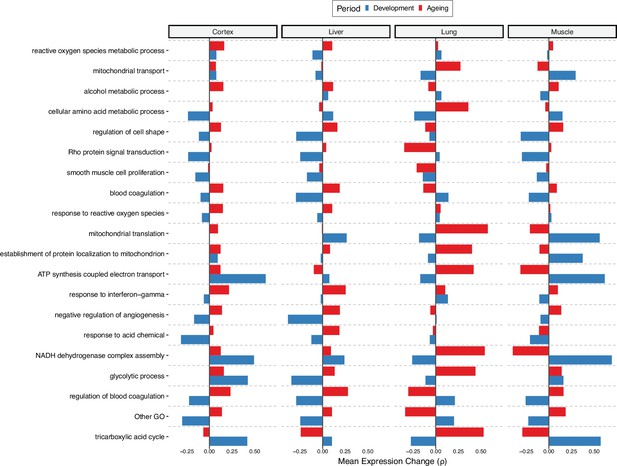
Age-related expression change trends in divergence-convergence (DiCo)-enriched categories denoted as ‘Other GO’ in the first clustering.
Age-related expression change trends of genes (x-axis) in categories enriched in DiCo (Gene Set Enrichment Analysis [GSEA]) that were grouped into one cluster ‘Other GO’ in Figure 4g. These categories (n = 69) were again summarised into representatives (y-axis) using hierarchical clustering and Jaccard similarities (see Materials and methods). Categories are ordered by the number of genes they contain from highest (bottom, n = 97) to lowest (top, n = 21). One cluster containing unrelated categories (n = 17) was again denoted as ‘Other GO’.
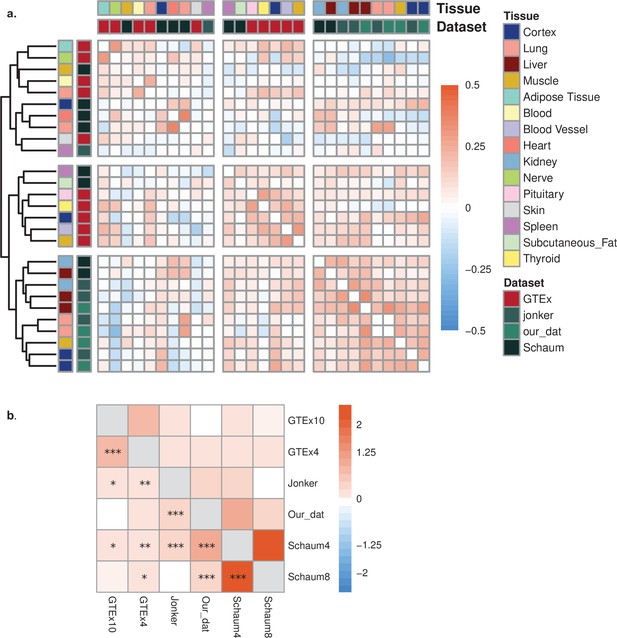
Comparison of datasets.
(a) Heatmap using Spearman’s correlation coefficients among expression trajectories (Spearman’s correlation coefficients between expression and age) across datasets during ageing. As the pairwise tissue correlations range between –0.2 and 0.52, the colour palette was restricted to –0.52 to 0.52 range. The same tissues of our dataset and Jonker dataset were clustered together (cortex, lung, liver) in the lower-right corner. (b) Enrichment of convergent genes among datasets during ageing. GTEx10 and GTEx4: coefficient of variation (CoV) calculation was performed with 10 tissues and with the same 4 tissues as our dataset in GTEx. Schaum8 and Schaum4: CoV calculation was performed with eight tissues and with the same four tissues as our dataset in Schaum dataset. ***False discovery rate (FDR)-corrected p-value<0.001; **FDR-corrected p-value<0.01; *FDR-corrected p-value<0.1. All log2(OR) values were positive except for our data vs. GTEx10 (log2(OR) = –0.04) and Jonker vs. Schaum8 (log2(OR) = –0.06), both of which were non-significant.
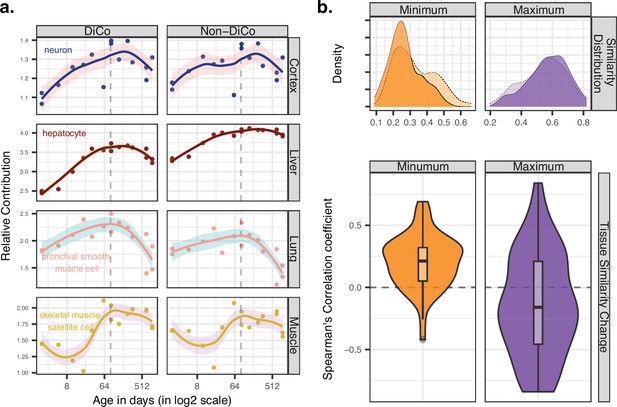
Contribution of tissue composition and cell-autonomous changes to the divergence-convergence (DiCo) pattern.
(a) Deconvolution analysis of our mouse dataset with the 3-month-old scRNA-seq data (Tabula Muris Senis) using DiCo (n = [4007, 4106]) and non-DiCo (n = [8485, 8743]) genes. Only the cell types with the highest relative contributions to each tissue bulk transcriptome are shown (cell-type names are given within each plot). Contributions of all cell types to bulk tissue transcriptomes are shown in Figure 5—figure supplement 1. (b) Distribution of correlations for minimally (left) and maximally (right) correlated cell-type pairs among tissues (n = 54 pairs). For each cell type of a given tissue, one minimally (or maximally) correlated cell type is chosen from other tissues among the 3-month age group of the Tabula Muris Senis dataset (density plots with solid line edges). Dashed lines show the correlation distributions in 24-month age of minimally or maximally correlated cell-type pairs identified in the 3-month age group. Bottom panel shows age-related expression similarity (ρ) changes of minimally (left) and maximally (right) correlated cell-type pairs. The correlation between age and tissue similarity (expression correlations) was calculated for each pair of cell types identified in the 3-month age group. All pairwise cell-type correlations and their age-related changes are given in Figure 5—source data 1.
-
Figure 5—source data 1
Cell-type proportion estimation and cell-autonomous changes using the Tabula Muris Senis dataset.
- https://cdn.elifesciences.org/articles/68048/elife-68048-fig5-data1-v4.xlsx
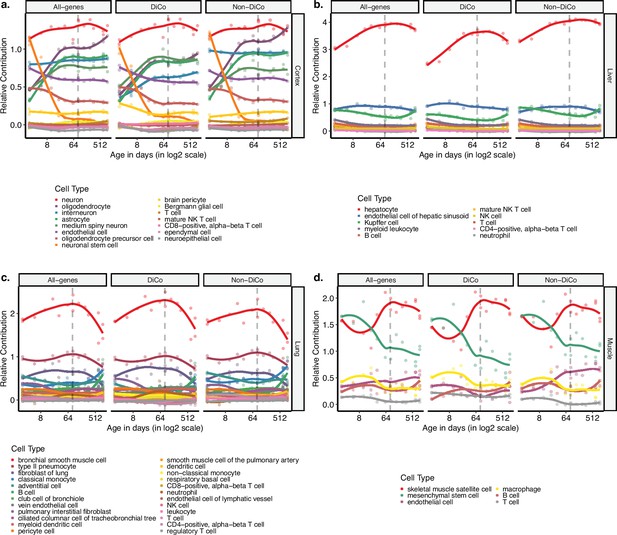
Age-related changes in cell-type proportions calculated using divergence-convergence (DiCo) and non-DiCo genes.
Deconvolution of bulk tissue expression profiles of the mice in our dataset with regression analysis using the single-cell expression profile of the 3-month-old mice in the Tabula Muris Senis dataset for cortex (a), liver (b), lung (c) and muscle (d). Contribution of each cell type was measured using three gene sets; all genes (n = [12,492, 12,849]), DiCo (n = [4007, 4106]), and non-DiCo genes (n = [8485, 8743]). Age-related changes of the relative contribution of each cell type in each tissue are given in Figure 5—source data 1.
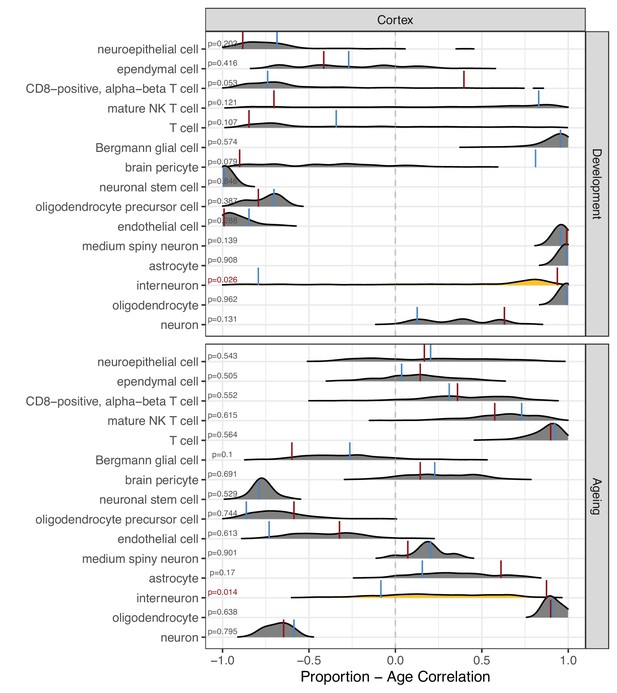
Permutation-based comparison between divergence-convergence (DiCo) and non-DiCo-related cell-type proportion changes with age in the cortex.
The difference between DiCo (4106) and non-DiCo (8743)-related cell-type proportion changes with age was tested in the cortex tissue. The x-axis is the Spearman’s correlation coefficient between age and relative contribution of a given cell type. The red vertical lines show the cell-type proportion changes calculated with DiCo genes (observed value), and the blue vertical lines indicate the same but with non-DiCo genes. Overlapping DiCo and non-DiCo values are indicated with blue. Null distributions for non-DiCo genes (density plots) were created with resampling among all genes (n = 12,849) (Materials and methods). Significant results are represented with yellow density plots, and the nominal p-values for permutation tests are indicated on the left side of the density plots. Permutation test results are also provided in Figure 5—source data 1.
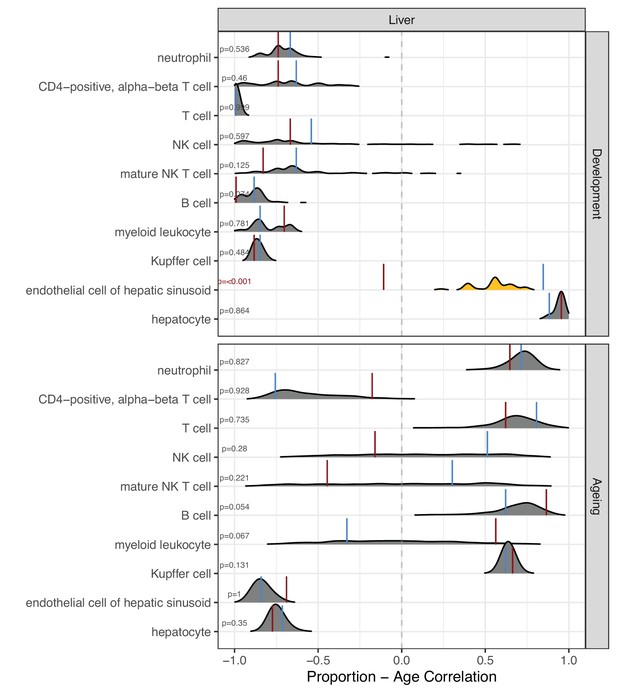
Permutation-based comparison between divergence-convergence (DiCo) and non-DiCo-related cell-type proportion changes with age in the liver.
The difference between DiCo (4007) and non-DiCo (8485)-related cell-type proportion changes with age was tested in the liver tissue. The x-axis is the Spearman’s correlation coefficient between age and relative contribution of a given cell type. The red vertical lines show the cell-type proportion changes calculated with DiCo genes, and the blue vertical lines indicate the same but with non-DiCo genes. Overlapping DiCo and non-DiCo values are indicated with blue. Null distributions for non-DiCo genes (density plots) were created with resampling among all genes (n = 12,492) (see Materials and methods). Significant results are represented with yellow density plots, and the nominal p-values for permutation tests are indicated on the left side of the density plots. Permutation test results are provided in Figure 5—source data 1.
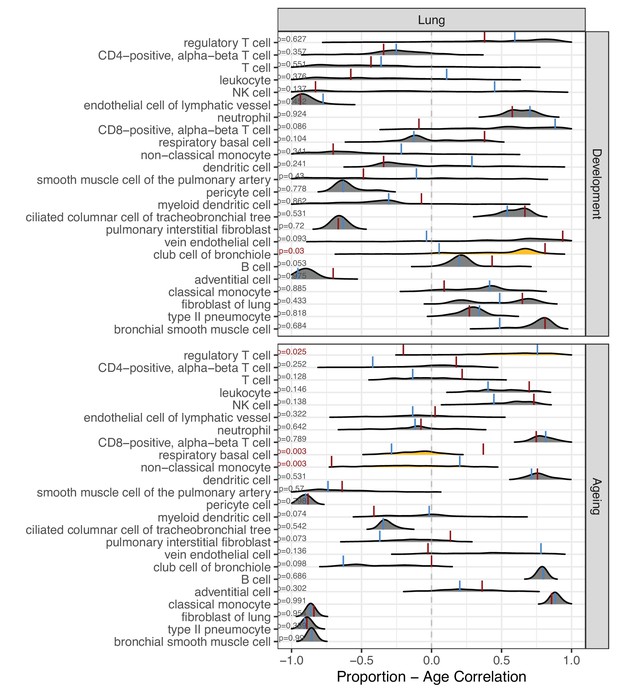
Permutation-based comparison between divergence-convergence (DiCo) and non-DiCo-related cell-type proportion changes with age in the lung.
The difference between DiCo (4084) and non-DiCo (8670)-related cell-type proportion changes with age was tested in the lung tissue. The x-axis is the Spearman’s correlation coefficient between age and relative contribution of a given cell type. The red vertical lines show the cell-type proportion changes calculated with DiCo genes, and the blue vertical lines indicate the same but with non-DiCo genes. Overlapping DiCo and non-DiCo values are indicated with blue. Null distributions for non-DiCo genes (density plots) were created with resampling among all genes (n = 12,754) (see Materials and methods). Significant results are represented with yellow density plots, and the nominal p-values for permutation tests are indicated on the left side of the density plots. Permutation test results are provided in Figure 5—source data 1.
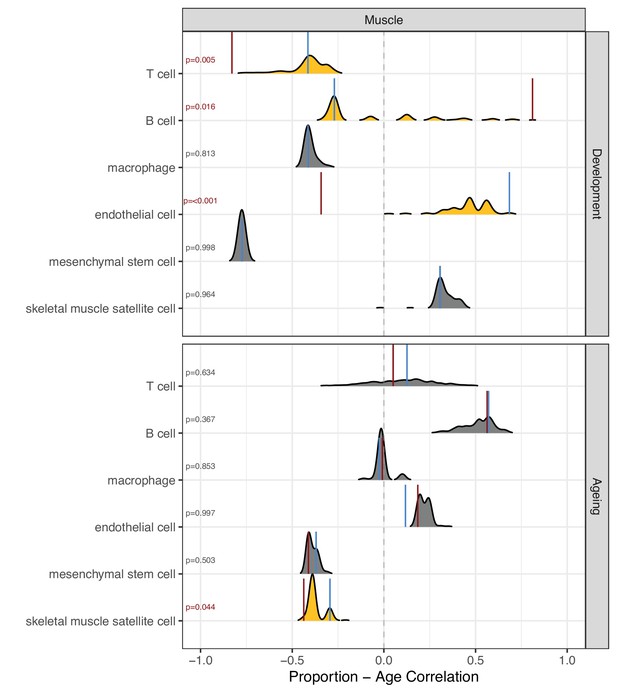
Permutation-based comparison between divergence-convergence (DiCo) and non-DiCo-related cell-type proportion changes with age in the muscle.
The difference between DiCo (4055) and non-DiCo (8568)-related cell-type proportion changes with age was tested in the muscle tissue. The x-axis is the Spearman’s correlation coefficient between age and relative contribution of a given cell type. The red vertical lines show the cell-type proportion changes calculated with DiCo genes, and the blue vertical lines indicate the same but with non-DiCo genes. Overlapping DiCo and non-DiCo values are indicated with blue. Null distributions for non-DiCo genes (density plots) were created with resampling among all genes (n = 12,623) (see Materials and methods). Significant results are represented with yellow density plots, and the nominal p-values for permutation tests are indicated on the left side of the density plots. Permutation test results are provided in Figure 5—source data 1.
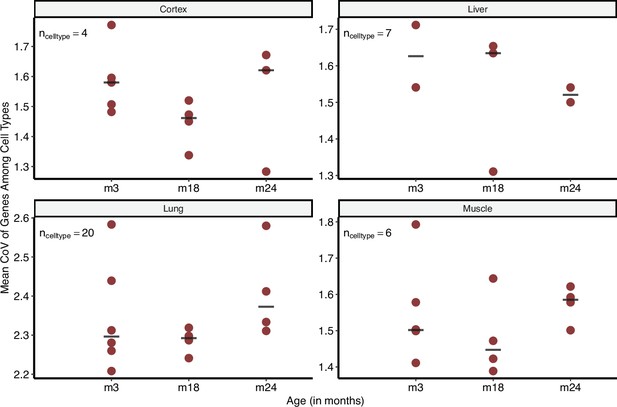
Intra-tissue coefficient of variation (CoV) changes between cell types using the Tabula Muris Senis dataset.
Intra-tissue CoV: CoV is calculated among cell types within each tissue for each individual mouse and in three age groups. Y-axis shows the mean CoV value of genes for each individual. The horizontal line on each age group shows the median of points. Cell types found in at least two individuals at every time point were considered.
Tables
Dataset characteristics summarising species, tissues, number of individuals, age range, sex, and platform used for measuring gene expression values.
| Dataset | Species | Tissues | N | Age range | Sex | Method |
|---|---|---|---|---|---|---|
| Izgi et al.,four tissues | Mice | Brain, lung, liver, muscle | 8 | 3–30 months | Male | RNA-seq |
| Jonker et al.,five tissues | Mice | Brain, lung, liver, kidney, spleen | 18 | 3–30 months | Female | Microarray |
| Schaum et al.,four tissues | Mice | Brain, lung, liver, muscle | 37 | 3–27 months | Male (n = 26)Female (n = 11) | RNA-seq |
| Schaum et al., eight tissues | Mice | Brain, lung, liver, muscle, subcutaneous fat, kidney, heart, spleen | 26 | 3–27 months | Male (n = 20)Female (n = 6) | RNA-seq |
| GTEx,four tissues | Humans | Brain, lung, liver, muscle | 47 | 20–75 years | Male (n = 36)Female (n = 11) | RNA-seq |
| GTEx,10 tissues | Humans | Adipose, tibial artery, cerebellum, lung, skeletal muscle, tibial nerve, pituitary, sun-exposed skin, thyroid, whole blood | 35 | 20–75 years | Male (n = 27)Female (n = 8) | RNA-seq |
Result summary of the all datasets analysed.
First column shows the names of datasets analysed. Numbers in parentheses show the sample sizes. ‘Among all genes’ column refers to the analyses performed using all genes relevant to those analyses (subcolumns) without a significance cutoff. ‘Within significant CoV changes’: genes show significant CoV change with age with FDR-corrected p-value<0.1. In the ‘DiCo vs. tissue specificity (Di- as background)’ column, divergent genes in development (Di-) were chosen as background. ‘Co vs. expression change in native tissue association (Figure 4b)’ column refers to the analysis performed in Figure 4b for each dataset, and the results are presented in Figure 4c. The association tests were performed among convergent genes in ageing except in our dataset, which was performed with DiCo genes. Significant test results are indicated with italic fonts. Bold fonts show the results that support convergence or tissue-specific expression loss in ageing whether as a significant result or as a trend. Unsupportive test results and inapplicable tests are written in normal font.
| Among all genes | Within significant CoV changes | ||||||
|---|---|---|---|---|---|---|---|
| PCA change in Euclidean distance | Mean CoV change | Median CoV change | Pairwise tissue correlations | DiCo vs. tissue specificity(Di- as background) | Co vs. expression change in native tissue association (Figure 4b) | Co vs. Di proportions | |
| Izgi2022 | ρ = −0.87, p=0.0026 | ρ = −0.5, p=0.2 | rho = −0.48, P = 0.23 | 4/6 positive, none significant* | OR = 1.56, p=1.3 × 10–18 | OR = 74.81,p=5.9 × 10–203(among 1287 DiCo genes) | 68% convergence(among 62 significant genes*) |
| Jonker 2013five tissues, two different than ours (n = 18) | ρ=−0.57, p=0.014 | ρ=−0.48, p=0.044 | ρ = −0.03, p=0.91 | 7/10 positive, none significant* | Di- background missing | OR = 7.52, p=6.5 × 10–109(among 2967 convergent genes) | 66% convergence(among 1735 significant genes*) |
| Schaum 2020,same four tissues (n = 37) | ρ = 0.13, p=0.46 | ρ = 0.25, p=0.14 | ρ = 0.13, p=0.43 | 4/6 positive,two significant* | OR = 1.33, p=1.07 × 10–8 | OR = 58.03, p=1.5 × 10–197(among 2124 convergent genes) | 53% convergence(among 319 significant genes*) |
| Schaum 2020,eight tissues (n = 26) | ρ = 0.1, p=0.62 | ρ = 0.16, p=0.43 | ρ = 0.04, p=0.86 | 16/28 positive,five significant* | Di- background missing | OR = 84.2, p=9.7 × 10–96(among 2380 convergent genes) | 54% convergence(among 244 significant genes*) |
| GTEx,same four tissues | ρ = −0.23, p=0.12 | ρ = −0.12, p=0.42 | ρ = −0.18, p=0.23 | 5/6 positive, none significant* | Di- background missing | OR = 7.21, p=7 × 10–87(among 2407 convergent genes) | (no significant CoV changes) |
| GTEx,10 tissues | ρ = −0.26, p=0.13 | ρ = −0.14, p=0.44 | ρ = −0.3, p=0.08 | 29/45 positive, none significant* | Di- background missing | OR = 13.01, p=5.7 × 10–114(among 2195 convergent genes) | (all three significant genes were convergent) |
-
ρ = Spearman’s correlation coefficient; OR = odds ratio; FDR = false discovery rate; CoV = coefficient of variation; DiCo = divergence-convergence; PCA = principal components analysis.
-
* FDR-corrected p-value<0.1.
Additional files
-
Supplementary file 1
Gene set over-representation analysis (GORA) of age-related genes in tissues.
Tissue-specific age-related gene expression changes and functional enrichment test results, performed with GORA using ‘topGO’ package.
- https://cdn.elifesciences.org/articles/68048/elife-68048-supp1-v4.xlsx
-
Supplementary file 2
Gene set over-representation analysis (GORA) of shared age-related genes among tissues.
Functional enrichment for shared genes across tissues. The same GORA that was performed for Supplementary file 1 was used to test the enrichment of shared up-/downregulated genes in development among the background genes which are chosen as the all-significant age-related genes across tissues in development. We did not apply the test for the ageing period as there were no shared ageing-related expression changes.
- https://cdn.elifesciences.org/articles/68048/elife-68048-supp2-v4.xlsx
-
Supplementary file 3
Gene set over-representation analysis (GORA) of reversal patterns.
Functional enrichment for gene expression reversals. GORA was performed with the same criteria as explained above. Up-down reversal genes were tested against up-up genes, and down-up reversal genes were tested against down-down genes in each tissue.
- https://cdn.elifesciences.org/articles/68048/elife-68048-supp3-v4.xlsx
-
Supplementary file 4
Gene set over-representation analysis (GORA) of divergence-convergence (DiCo) gene clusters determined with coefficient of variation (CoV) values.
Functional enrichment of DiCo genes clustered with k-means algorithm according to their CoV values. GORA was performed using gene sets in each cluster (Figure 2—figure supplement 2) which were tested among all DiCo genes.
- https://cdn.elifesciences.org/articles/68048/elife-68048-supp4-v4.xlsx
-
Supplementary file 5
Gene set over-representation analysis (GORA) of divergence-convergence (DiCo) gene clusters determined with expression levels.
Functional enrichment of DiCo genes clustered with k-means algorithm according to their expression levels. GORA was performed using gene sets in each cluster (Figure 2—figure supplement 3) which are tested among all DiCo genes.
- https://cdn.elifesciences.org/articles/68048/elife-68048-supp5-v4.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68048/elife-68048-transrepform1-v4.docx