APOE4 is associated with elevated blood lipids and lower levels of innate immune biomarkers in a tropical Amerindian subsistence population
Figures
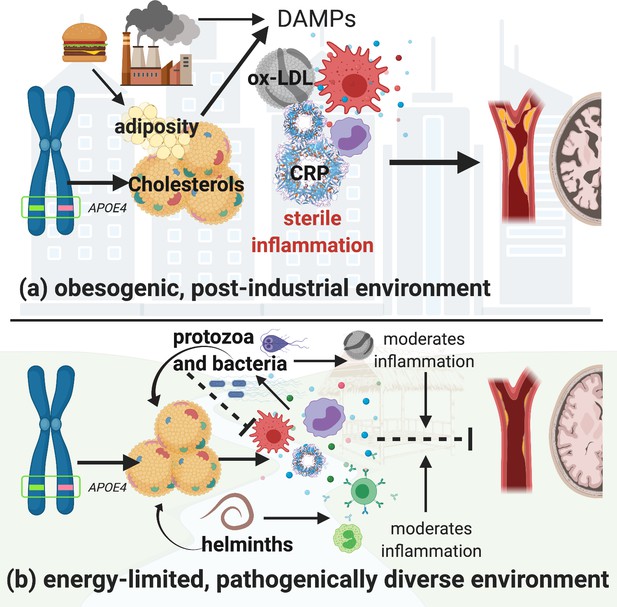
Hypothetical pathways through which the apolipoprotein E4 (APOE4) allele influences lipid processing, immune regulation and disease risk in post-industrialized and non-industrialized contexts.
In both contexts, the APOE4 allele leads to increased levels of circulating lipids; however, in post-industrialized contexts (a), lipid levels can reach dangerously high levels due to obesogenic diets, and an absence of moderation by parasites and pathogen-driven immune activation. Immune activation by non-pathogenic elements triggers damage-associated molecular pattern pathways, which generates a proinflammatory ‘sterile’ immune response. Obesity and hyperlipidemia can simultaneously fuel sterile inflammation and promote oxidization of cholesterols, which, due to their lack of function, cause further tissue damage associated with cardiovascular and neurodegenerative disease risk. In energetically limited, pathogenically diverse contexts (b), the pathway between APOE4 and disease risk is considerably more complex. Briefly, immune responses to parasites and microbes require cholesterol, and there are both direct and indirect effects of different species of parasites which further regulate cholesterol production and utilization. In addition, anti-inflammatory immune responses are generated by ox-LDL (e.g. in response to bacteria and protozoal infections), and helminthic parasites, which balance the immune system’s overall response. It is possible that in contexts where there is higher pathogen diversity, an APOE4 phenotype may be less harmful because it minimizes the damage caused by upregulated innate immune function, while also maintaining higher cholesterol levels which would buffer the cost of innate immune activation due to infection. Whereas in high-calorie, low-pathogen environments, the utility of having an APOE4 allele may be muted, and the costs more severe. Image created with BioRender.com.
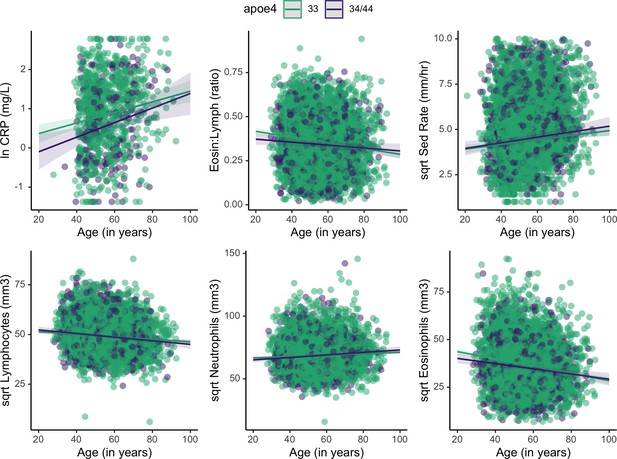
Plots showing estimated change in immune markers across age, split by apolipoprotein E4 (APOE) genotype.
Slopes were taken from mixed effects linear regression models and represent estimates of the interactive effects of APOE genotype and age on each immune marker. Models adjust for sex, season, and current illness (Supplementary file 1). Erythrocyte sedimentation rate is abbreviated as ‘Sed Rate’.
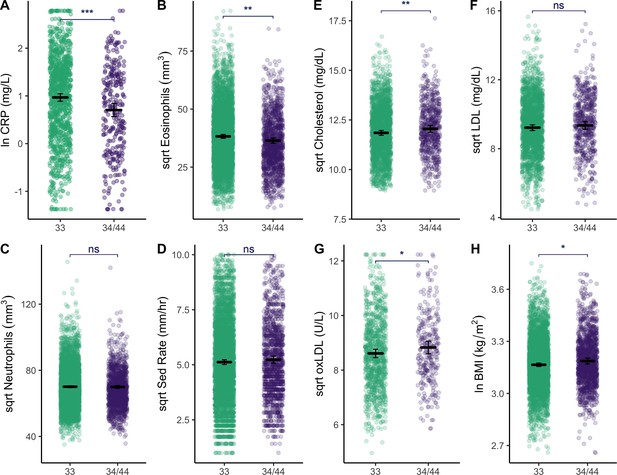
Plots show distributions of biomarker data (raw) grouped by APOE genotype (homozygous APOE 33 versus those that have at least one copy of the E4 allele).
All plots include estimated means (horizontal lines) and standard errors (crossbars) per genotype, derived from mixed effects linear regression models that adjust for age, sex, season, and current illness. For full models with covariates, see Supplementary file 1b, c and e. Erythrocyte sedimentation rate is abbreviated as ‘Sed Rate’. See Figure 3—figure supplement 1 for additional plots comparing of biomarkers by APOE genotype. Statistical significance is denoted as: *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
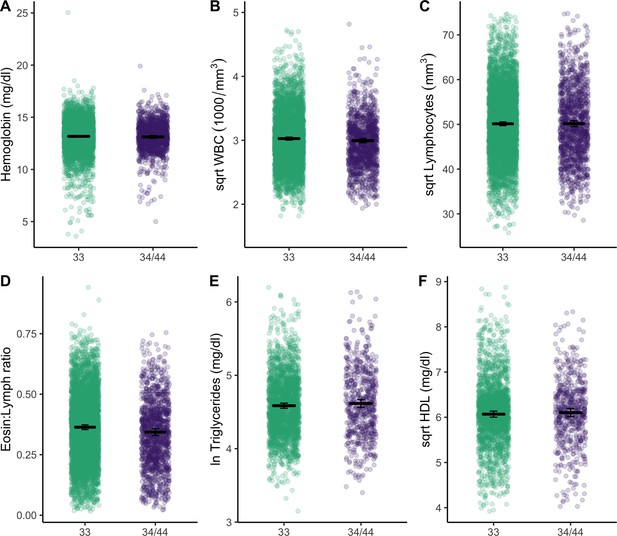
Plots show distributions of biomarker data (raw) grouped by APOE genotype (homozygous APOE 33 versus those that have at least one copy of the E4 allele).
All plots include estimated means (horizontal lines) and standard errors (crossbars) per genotype, derived from mixed effects linear regression models that adjust for age, sex, season, and current illness. For full models with covariates, see Supplementary file 1b, c and e.
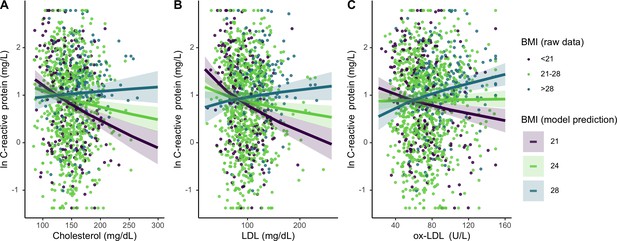
Differing influence of cholesterols on C-reactive protein (CRP), based on three levels of body mass index (BMI) (mean, ± 1 standard deviation).
Panels A-C show interaction effects between BMI and (A) total cholesterol, (B) LDL, and (C) oxidized LDL. For total and LDL cholesterol, among those with low (purple line) and mean (lime green line) BMI, cholesterol is negatively associated with CRP. Oxidized LDL is only associated with higher CRP among individuals with high BMI (turquoise line). Lines are predicted from mixed effects linear regressions that adjust for age, sex, seasonality, with random effects for individual and community residence (Supplementary file 1f-h). Data points are raw values, color-coded based on BMI level. Variables are transformed and centered. See Figure 4—figure supplement 1 for independent associations between BMI and cholesterols with markers of inflammation.
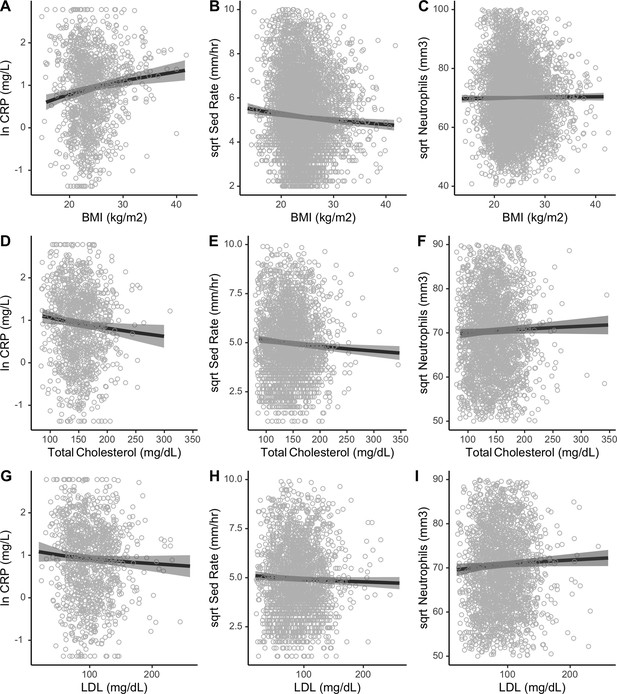
This figure shows independent associations between body mass index (BMI) and cholesterols with markers of inflammation from mixed effects models, adjusting for age, sex, seasonality, and current illness with random effects for community and individual ID.
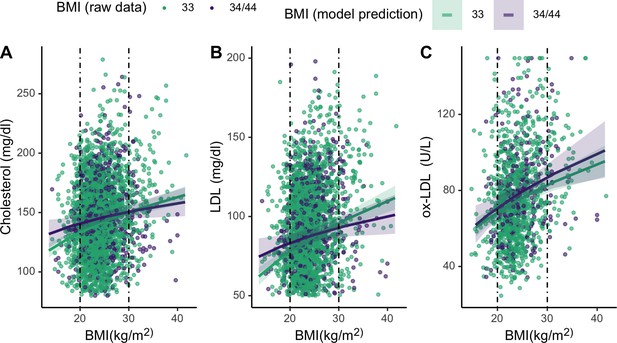
Plots showing moderating effects of APOE genotype on associations between body mass index (BMI) and cholesterols (see Table 2 in the manuscript for models).
Dotted vertical lines represent cutoffs for low (<21) and high (>28) BMI. For the primary cholesterols utilized during an immune response to pathogens – total cholesterol (A) and LDL (B) – individuals with an E4 allele maintain slightly higher levels of those cholesterols at low BMIs, compared to homozygous E3. Plotted lines are derived from mixed effects linear regressions that include age, sex, season, and current infection as covariates, as well as random effects for individual and community residence. Data points are raw values, color-coded by APOE genotype.
Tables
Description of immune and lipid measures for homozygous APOE3/3 and APOE4+ carriers for whom age and sex are available.
Values are reported as mean (standard deviation). Linear mixed effects models fit by REML were used to test for differences between groups, controlling for age, sex, current immune activation, and seasonality, with random effects for community and individual ID. Model to test age differences includes only a random effect for individual ID. Multiple test correction was conducted for models used for hypothesis-testing; False Discovery Rate (FDR) adjusted p-values are reported for these models. No. of observations is reported as: total number of observations for the main biomarker (number of unique individuals) in each model. T-statistics use Satterthwaite's method. Due to skewness of biomarkers, statistical models use transformed and scaled data for normalization, as such, estimates are reported as standardized betas (b) with standard errors (SE). Full models with covariates and 95% confidence intervals can be found in Tables 2-3 in the Supplement. See methods for transformations for each marker.
| Variables | APOE 3/3 | Apoe4+ | No. observations per model | β | SE | t-value | p-value | FDR adj p-value |
|---|---|---|---|---|---|---|---|---|
| N | 998 | 268 | – | – | – | – | – | – |
| % female | 51% | 48% | – | – | – | – | – | – |
| Age (in years) | 54 (11.5) | 54 (11.2) | 6615 (1268) | 0.007 | 0.07 | 0.106 | 0.915 | – |
| BMI (kg/m2) | 23.9 (3.4) | 24.6 (3.7) | 6224 (1263) | 0.15 | 0.06 | 2.421 | 0.016* | – |
| C-reactive protein (mg/L) | 3.7 (3.3) | 2.9 (2.6) | 1032 (907) | -0.29 | 0.08 | -3.731 | <0.001*** | <0.001*** |
| Eosinophil (mm3) | 1617 (1060) | 1375 (910) | 6129 (1259) | -0.16 | 0.04 | -3.871 | <0.001*** | <0.001*** |
| Neutrophil (mm3) | 5044 (1803) | 4816 (1697) | 6144 (1259) | -0.03 | 0.03 | -0.816 | 0.415 | 0.435 |
| Sed Rate (mm/hr) | 29.4 (19.5) | 29.7 (19.2) | 5987 (1253) | 0.06 | 0.04 | 1.334 | 0.182 | 0.217 |
| Eosin : Lymph ratio | 0.36 (0.14) | 0.33 (0.14) | 6121 (1260) | -0.14 | 0.04 | -3.197 | 0.001** | –--- |
| Leukocytes (1000/mm3) | 9.3 (2.5) | 8.8 (2.4) | 6229 (1266) | -0.08 | 0.03 | -2.281 | 0.023* | – |
| Lymphocytes (mm3) | 2590 (832) | 2565 (852) | 6136 (1260) | 0 | 0.04 | 0.104 | 0.917 | – |
| Hemoglobin (g/dL) | 13.2 (1.5) | 13.2 (1.4) | 6195 (1263) | -0.04 | 0.05 | -0.805 | 0.421 | – |
| Triglycerides (mg/dL) | 107 (51) | 114 (61) | 2633 (1174) | 0.07 | 0.06 | 1.157 | 0.248 | – |
| Total cholesterol (mg/dL) | 144 (32) | 148 (33) | 2581 (1168) | 0.15 | 0.06 | 2.594 | 0.010** | 0.021 |
| HDL cholesterol (mg/dL) | 38 (9) | 38 (9) | 2477 (1167) | 0.05 | 0.06 | 0.83 | 0.407 | – |
| LDL cholesterol (mg/dL) | 90 (32) | 93 (32) | 2390 (1154) | 0.08 | 0.06 | 1.307 | 0.192 | 0.217 |
| Oxidized LDL (U/L) | 76 (24) | 79 (23) | 1033 (907) | 0.16 | 0.08 | 1.947 | 0.052* | 0.090t |
-
Statistical significance is denoted as: t p<0.10; * p<0.05; ** p<0.01; *** p<0.001.
Models evaluating the moderating effects of APOE genotype on associations between BMI and cholesterols.
Results are fixed effects estimates from mixed effects linear regressions, which include random effects for ID and community residence. In addition to age, sex, and season, a dummy variable was used as a proxy for current illness (leukocytes > 12 mm3). Results are reported as standardized betas; CI is the 95% confidence interval. All dependent variables were transformed and centered prior to analyses. APOE genotype is coded as a categorical variable, binned as individuals that are homozygous E3 (E3) versus those that have at least one copy of the E4 allele (E4).
| Predictors | Total cholesterol | LDL | Oxidized LDL | ||||||
|---|---|---|---|---|---|---|---|---|---|
| β | CI | p | β | CI | p | β | CI | p | |
| E4 * BMI | −0.08 | −0.18–0.02 | 0.111 | −0.09 | −0.19–0.00 | 0.061 | 0.02 | −0.13–0.17 | 0.786 |
| APOE [E4] | 0.14 | 0.03–0.26 | 0.013 | 0.07 | −0.04–0.18 | 0.221 | 0.13 | −0.03–0.29 | 0.112 |
| BMI | 0.18 | 0.14–0.23 | <0.001 | 0.2 | 0.15–0.25 | <0.001 | 0.21 | 0.14–0.28 | <0.001 |
| Sex [women] | 0.15 | 0.06–0.24 | 0.001 | 0.2 | 0.11–0.28 | <0.001 | 0.09 | −0.03–0.21 | 0.158 |
| Age [in years] | 0.01 | 0.01–0.01 | <0.001 | 0.01 | 0.01–0.02 | <0.001 | 0 | −0.01–0.01 | 0.869 |
| Currently ill | 0.02 | −0.13–0.17 | 0.773 | −0.03 | −0.18–0.13 | 0.744 | 0.15 | −0.09–0.39 | 0.229 |
| Season [wet] | −0.12 | −0.20–−0.04 | 0.004 | −0.29 | −0.38–−0.21 | <0.001 | -0.28 | −0.44–−0.13 | <0.001 |
| (Intercept) | −0.7 | −0.94–−0.46 | <0.001 | −0.71 | −0.95–−0.47 | <0.001 | 0.09 | −0.28–0.46 | 0.626 |
| Observations | 2581 | 2390 | 1032 | ||||||
| Marginal R2 | 0.049 | 0.074 | 0.07 | ||||||
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68231/elife-68231-transrepform1-v1.docx
-
Supplementary file 1
Full models with covariates described in analyses and figures presented in main article.
- https://cdn.elifesciences.org/articles/68231/elife-68231-supp1-v1.docx
-
Supplementary file 2
Spanish-language abstract.
Link to the Spanish version of the article: https://tsimane.anth.ucsb.edu/results.html#pubs.
- https://cdn.elifesciences.org/articles/68231/elife-68231-supp2-v1.docx





