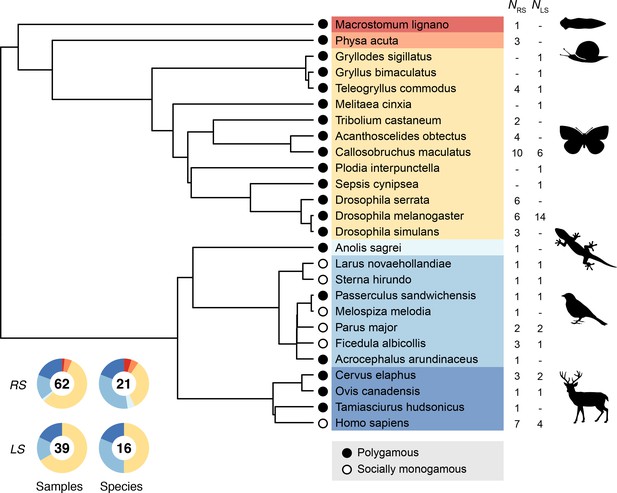
Stronger net selection on males across animals
Figures
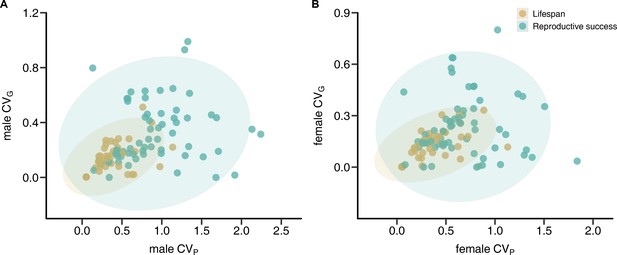
Correlations between phenotypic and genetic coefficients of variation (CVP and CVG, respectively) for lifespan and reproductive success.
Scatterplots show relationships between CVP and CVG for male (A) and female (B) reproductive success (green) and lifespan (brown). Shaded areas indicate the 95% confidence ellipses.
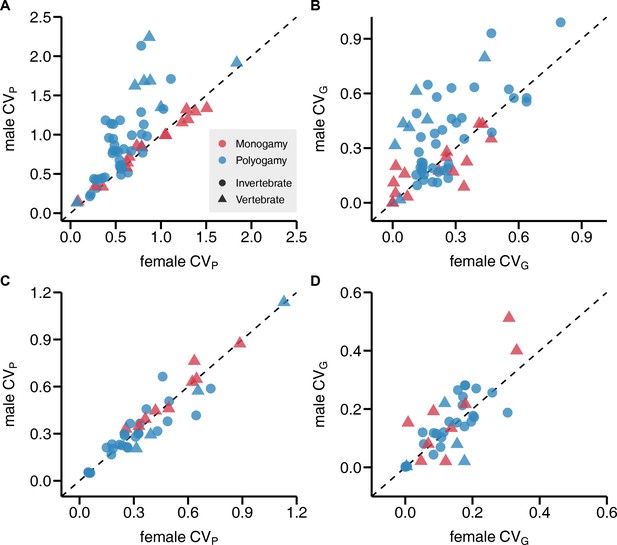
Sex bias in phenotypic and genetic variances in reproductive success and lifespan.
Scatterplots show the coefficient of phenotypic variation CVP (A, C) and genetic variation CVG (B, D) for reproductive success (A, B) and lifespan (C, D). Monogamous species are represented in red, polygamous species in blue. All data points above the diagonals indicate a male bias.
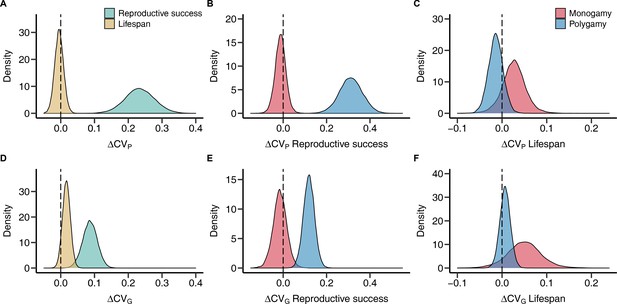
Sex differences in phenotypic and genetic coefficients of variation for reproductive success and lifespan.
Plots show posterior distributions for the sex difference of the phenotypic (A–C) and genetic (D–F) coefficient of variation (ΔCVP and ΔCVG, respectively) obtained from phylogenetic general linear mixed-effects models (PGLMMs; see Methods). Positive values indicate a male, negative values a female bias. Density plots contrast fitness components pooled across mating systems (A and D) or compare socially monogamous and polygamous species separately for reproductive success (B and E) and lifespan (C and F).
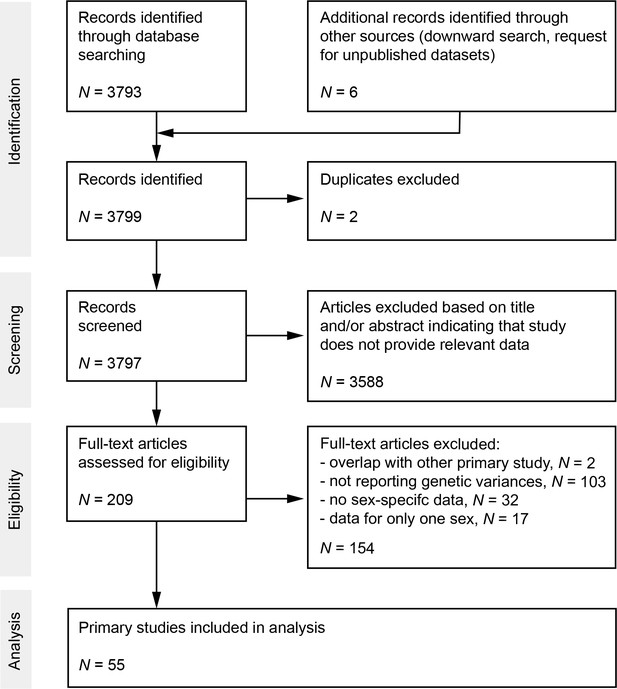
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) diagram.
Flow chart maps the number of records identified during the different phases of the systematic literature search.
Tables
Results of phylogenetic general linear mixed-effect models testing for an effect of sex on phenotypic (CVP) and genetic (CVG) coefficient of variation.
Results shown for reproductive success (RS) and lifespan (LS) for models ran across mating systems and when ran separately for socially monogamous and polygamous species. Estimates are shown as posterior means with 95% highest posterior density (HPD) intervals, with positive values indicating a male bias. PMCMC is the probability of the posteriors including zero. The variance explained by sex is given as the marginal R2 and the phylogenetic signal is reported as H2.
| Response | Variance component | Sex effect estimate | PMCMC | Marginal R2 | Phylogenetic H2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Across mating systems | |||||||||||
| RS | CVP | 0.234 | (0.149 ,0.322) | <0.001 | 0.07 | (0.02, | 0.12) | 0.12 | (0.00, | 0.32) | |
| CVG | 0.086 | (0.043, 0.128) | <0.001 | 0.04 | (0.00, | 0.08) | 0.28 | (0.03, | 0.58) | ||
| LS | CVP | −0.005 | (−0.031, 0.021) | 0.704 | 0.00 | (0.00, | 0.00) | 0.41 | (0.06, | 0.81) | |
| CVG | 0.017 | (−0.006, 0.040) | 0.149 | 0.01 | (0.00, | 0.02) | 0.46 | (0.12, | 0.81) | ||
| Monogamy | |||||||||||
| RS | CVP | −0.012 | (−0.063, | 0.035) | 0.602 | 0.00 | (0.00, | 0.00) | 0.37 | (0.01, | 0.96) |
| CVG | −0.015 | (−0.080, | 0.046) | 0.628 | 0.01 | (0.00, | 0.02) | 0.43 | (0.05, | 0.92) | |
| LS | CVP | 0.026 | (−0.028, | 0.079) | 0.281 | 0.00 | (0.00, | 0.02) | 0.37 | (0.01, | 0.87) |
| CVG | 0.050 | (−0.030, | 0.125) | 0.179 | 0.02 | (0.00, | 0.07) | 0.46 | (0.05, | 0.89) | |
| Polygamy | |||||||||||
| RS | CVP | 0.312 | (0.207, | 0.417) | <0.001 | 0.11 | (0.03, | 0.18) | 0.20 | (0.01, | 0.47) |
| CVG | 0.119 | (0.068, | 0.170) | <0.001 | 0.07 | (0.01, | 0.13) | 0.25 | (0.02, | 0.56) | |
| LS | CVP | −0.014 | (−0.046, | 0.018) | 0.373 | 0.00 | (0.00, | 0.00) | 0.69 | (0.25, | 0.97) |
| CVG | 0.007 | (−0.016, | 0.031) | 0.554 | 0.00 | (0.00, | 0.01) | 0.44 | (0.12, | 0.78) | |
Additional files
-
Supplementary file 1
Results of phylogenetic general linear mixed-effects models (PGLMMs) testing for sex by mating system interaction on phenotypic (CVP) and genetic (CVG) coefficient of variation.
- https://cdn.elifesciences.org/articles/68316/elife-68316-supp1-v1.docx
-
Supplementary file 2
Phylogenetic general linear mixed-effect models testing for an effect of sex on phenotypic (CVP) and genetic (CVG) coefficient of variation for a reduced dataset including only vertebrates.
- https://cdn.elifesciences.org/articles/68316/elife-68316-supp2-v1.docx
-
Supplementary file 3
Results of phylogenetic general linear mixed-effects models (PGLMMs) testing for sex by mating system interaction on phenotypic (CVP) and genetic (CVG) coefficient of variation obtained from a reduced dataset including only vertebrates.
- https://cdn.elifesciences.org/articles/68316/elife-68316-supp3-v1.docx
-
Supplementary file 4
Results of phylogenetic general linear mixed-effects models (PGLMMs) testing for the effect of sex, study type (lab versus field studies) and their interaction on phenotypic (CVP) and genetic (CVG) coefficients of variation.
- https://cdn.elifesciences.org/articles/68316/elife-68316-supp4-v1.docx
-
Supplementary file 5
Results of phylogenetic general linear mixed-effects models (PGLMMs) testing for the effect of sex, VG estimate (additive versus total) and their interaction on genetic (CVG) coefficient of variation.
- https://cdn.elifesciences.org/articles/68316/elife-68316-supp5-v1.docx
-
Supplementary file 6
Results of phylogenetic general linear mixed-effects models (PGLMMs) testing for the effect of sex, RS estimate (temporal versus lifetime reproductive success) and their interaction on phenotypic (CVP) and genetic (CVG) coefficients of variation.
- https://cdn.elifesciences.org/articles/68316/elife-68316-supp6-v1.docx
-
Supplementary file 7
Analysis of cross-sex genetic correlations.
- https://cdn.elifesciences.org/articles/68316/elife-68316-supp7-v1.docx
-
Supplementary file 8
Search terms and list of primary studies.
- https://cdn.elifesciences.org/articles/68316/elife-68316-supp8-v1.docx
-
Supplementary file 9
Methodological characteristics of the 55 primary studies.
- https://cdn.elifesciences.org/articles/68316/elife-68316-supp9-v1.docx
-
Supplementary file 10
Mating system classification of the 26 sampled species.
- https://cdn.elifesciences.org/articles/68316/elife-68316-supp10-v1.docx
-
Supplementary file 11
Phylogenetically independent meta-analysis of lnCVR of phenotypic variation.
- https://cdn.elifesciences.org/articles/68316/elife-68316-supp11-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68316/elife-68316-transrepform1-v1.docx