Heparin-binding motif mutations of human diamine oxidase allow the development of a first-in-class histamine-degrading biopharmaceutical
Figures
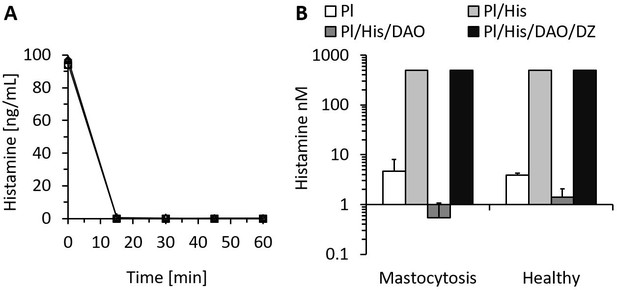
Recombinant human diamine oxidase (rhDAO) efficiently degrades histamine in phosphate buffered saline (PBS) and human plasma samples from mastocytosis patients and healthy volunteers.
(A) 6 nM purified rhDAO-WT with and without 100× high molecular weight heparin (HMWH; 600 nM) were incubated at 37°C for 60 min with 100 ng/mL histamine in 1% human serum albumin-phosphate buffered saline (HSA-PBS) and EDTA plasma of a healthy volunteer. Histamine concentrations were determined in duplicate. Plasma: △; plasma with 100× HMWH: ◇; HSA-PBS: ○; HSA-PBS with 100× HMWH: □. (B) Plasma of mastocytosis patients (n = 7) and healthy volunteers (n = 3) was spiked with 6 nM rhDAO-WT, 545 nM histamine, and 20 µM diminazene aceturate (DZ, potent DAO inhibitor) and incubated for 60 min at 37°C. Mean histamine concentrations with standard deviation (SD) as applicable are shown; column Pl (plasma): no histamine and no DAO spiking but with endogenous histamine; column Pl/His/DAO: spiking of exogenous histamine and exogenous DAO; column Pl/His: spiking of exogenous histamine; column Pl/His/DAO/DZ: spiking of exogenous histamine, DAO, and DIMAZ; t-test p-value comparing Pl with Pl/His/DAO data was 0.015 for mastocytosis patients and 0.011 for healthy volunteers; Histamine concentrations in Pl/His and Pl/His/DAO/DZ samples were >500 nM and the SD could not be calculated. Pl: plasma; His: histamine.
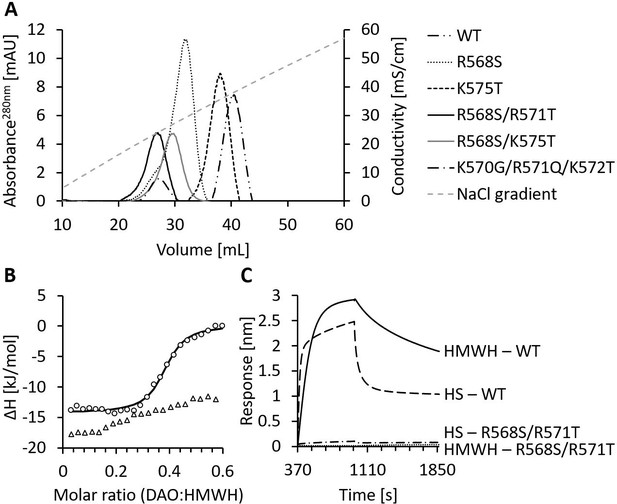
Mutations of the heparin-binding motif of recombinant human diamine oxidase (rhDAO) significantly reduce binding to heparin and heparan sulfate (HS).
(A) Purified rhDAO-WT and various heparin-binding motif (HBM) mutants were loaded onto a heparin-sepharose column and eluted with a linear gradient of increasing NaCl concentration. (B) Isothermal titration calorimetry was performed by titrating 25 × 1.5 µL high molecular weight heparin (HMWH) at a concentration of 120 µM into 36.7 µM rhDAO-WT (○) or rhDAO-R568S/R571T (△). The graph represents single experiments for each protein-ligand pair. A summary of all experiments is shown in Figure 2—source data 1. (C) Biolayer interferometry. Streptavidin sensors loaded with biotinylated HMWH or HS were incubated for 10 min with 88 nM or 471 nM rhDAO, respectively. Dissociation was measured for 15 min. The graphs represent one of three individual measurements and show the association and dissociation curves after subtraction of the negative control (no DAO added). The data of rhDAO-WT have already been published (Gludovacz et al., 2020) but are added for better presentation.
-
Figure 2—source data 1
All raw plots and integrated heat plots of the isothermal titration calorimetry (ITC) analyses of the heparin-binding motif (HBM) mutant are presented in Figure 2B.
- https://cdn.elifesciences.org/articles/68542/elife-68542-fig2-data1-v2.docx
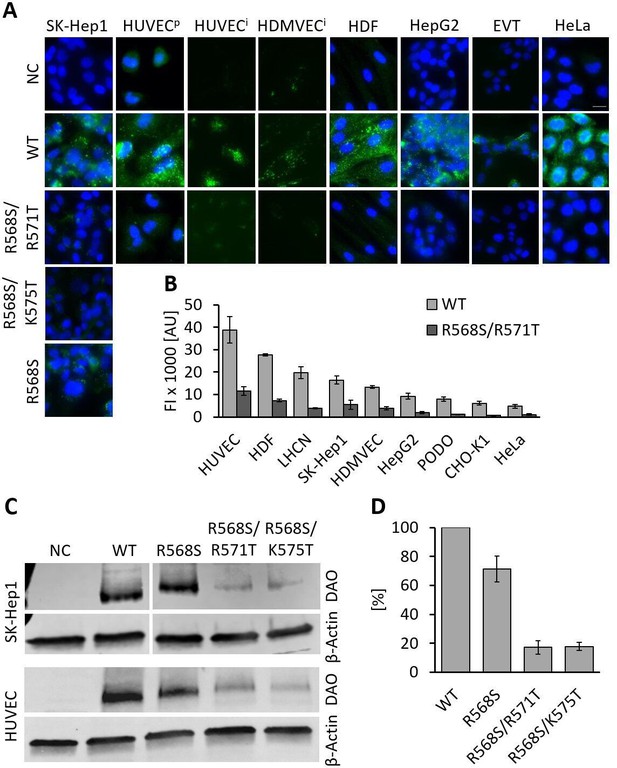
Recombinant human diamine oxidase (rhDAO) heparin-binding motif (HBM) double mutants show significantly reduced uptake into various cell lines.
(A) SK-Hep1, primary human umbilical vein endothelial cells ( = HUVECp), human dermal fibroblasts (HDF), HepG2, and HeLa cells were incubated with 120 nM unlabeled purified rhDAO-WT and various HBM mutants and detected with rabbit anti-ABP1 antibodies and Alexa Fluor 488 donkey anti-rabbit antibodies after fixation and permeabilization. Immortalized HUVEC/TERT2 ( = HUVECi) and human dermal microvascular endothelial cells (HDMVEC/TERT164-B = HDMVECi) were incubated with 120 nM and extravillous trophoblasts (EVT) with 60 nM Alexa488-labeled rhDAO-WT and rhDAO-R568S/R571T; Scale bar = 20 μm. (B) Cells were incubated with 30 nM Alexa488-labeled rhDAO-WT and rhDAO-R568S/R571T mutant (no DAO added = negative control), washed, and analyzed flow cytometrically (500 cells per sample). Background corrected mean median values ± standard error of the mean (SEM) are shown (n = 4 biological replicates, two individual experiments in duplicate). (C) 106 SK-Hep1 and 5 × 105 HUVEC/TERT2 cells were incubated with 120 nM and 60 nM rhDAO-WT and three HBM mutants. The cell lysates were analyzed for DAO uptake using western blotting. β-Actin was used as an internal standard. (D) After background subtraction, rhDAO band intensities were corrected against β-Actin and normalized against the rhDAO-WT band. The mean ± SEM of two individual experiments with SK-Hep1 cells and one experiment with HUVEC/TERT2 cells are shown. (A–D) Incubations were performed for 60 min at 37°C. FI: fluorescence intensity; AU: arbitrary units.
-
Figure 3—source data 1
Statistical evaluation of flow cytometry data is summarized in Figure 3B.
- https://cdn.elifesciences.org/articles/68542/elife-68542-fig3-data1-v2.docx
-
Figure 3—source data 2
Raw unedited western blots are presented in Figure 3C.
- https://cdn.elifesciences.org/articles/68542/elife-68542-fig3-data2-v2.docx
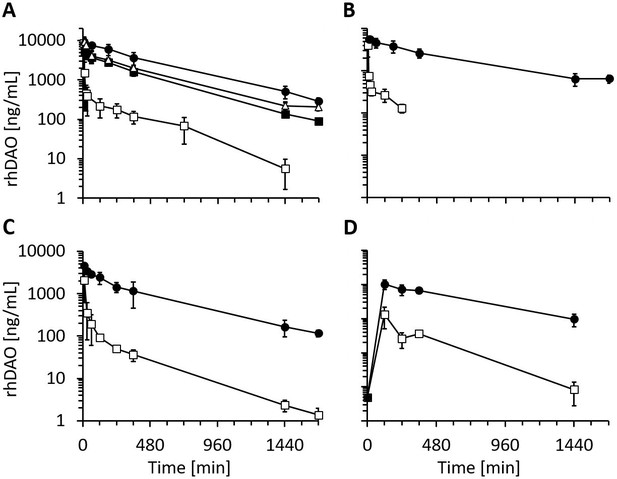
Recombinant human diamine oxidase (rhDAO) heparin-binding motif (HBM) mutants show strongly reduced clearance compared to the wildtype protein in rats and mice.
(A, B) Reduced clearance of HBM mutants in rats. Means of the measured values ± standard deviation (SD) after administration of 1 mg/kg purified rhDAO: (A) WT (n = 9; □), -R568S (n = 4; ■), -R568S/R571T (n = 5; ●), -R568S/K575T (n = 4; △); (B) rhFcDAO (n = 6; □), Fc-R568S/R571T (n = 4; ●). (C, D) rhDAO-R568S/R571T mutant increases the area under the curve (AUC) more than 15-fold compared to wildtype rhDAO after intravenous and intraperitoneal injection into mice. (C) rhDAO-WT (□) and rhDAO-R568S/R571T (●) were injected at 1 mg/kg into the tail vein of C57BL6 mice with a body weight of about 20 g. The mean (n = 3–4 mice per time point) ± SD are shown. (D) rhDAO-WT and rhDAO-R568S/R571T mutant were injected intraperitoneally at 1 mg/kg in mice with a body weight of 21 g. Each time point represents the mean of 3 mice, and therefore in total 15 mice with WT and 15 mice with rhDAO-R568S/R571T were used. The means ± SD are shown. n: number of animals.
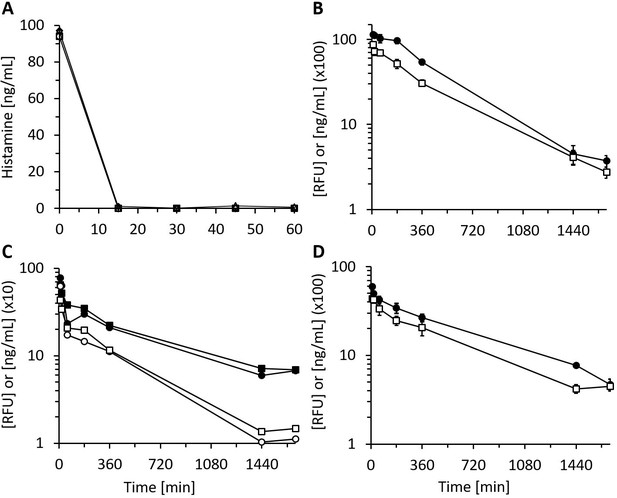
Recombinant human diamine oxidase (rhDAO) heparin-binding motif (HBM) mutants are enzymatically fully active in vitro and in vivo.
(A) Purified rhDAO-R568S/R571T mutant rapidly and completely degrades histamine. 6 nM rhDAO-R568S/R571T with and without 100× high molecular weight heparin (HMWH) (600 nM) were incubated with 100 ng/mL histamine in 1% human serum albumin-phosphate buffered saline (HSA-PBS) and EDTA plasma of a healthy volunteer for 60 min at 37°C. Histamine concentrations were determined in duplicate. Plasma: △; plasma with 100× HMWH: ◇; HSA-PBS: ○; HSA-PBS with 100× HMWH: □. (B–D) rhDAO HBM mutants are enzymatically active in rats. Means of the measured values ± standard error of the mean (SEM) using 1 mg/kg rhDAO-R568S/R571T (B, n = 3; R2 = 94.8% with p-value<0.001, linear regression) and rhFcDAO-R568S/R571T (D, n = 3; R2 = 98% with p-value<0.001, linear regression) and two individually plotted measurements of rhDAO-R568S/K575T (C); rhDAO concentration (ng/mL; □ and ○) and activity (relative fluorescence units [RFU]; ■ and ●); RFU data downscaled by a factor 50 in panels (B) and (C) and by 100 in (D) for easier presentation. n: number of animals.
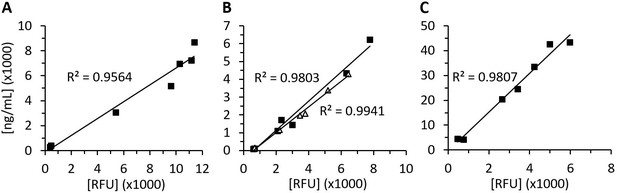
Recombinant human diamine oxidase (rhDAO) antigen concentrations and activity are highly correlated in rat plasma.
Means of the measured values using 1 mg/kg rhDAO-R568S/R571T (A, n = 3) and Fc-rhDAO-R568S/R571T (C, n = 3) and two individually plotted measurements of rhDAO-R568S/K575T (B); relative fluorescence units (RFU) data downscaled by factor 50 in panels (A) and (B) and by factor 100 in (C). n: number of animals.
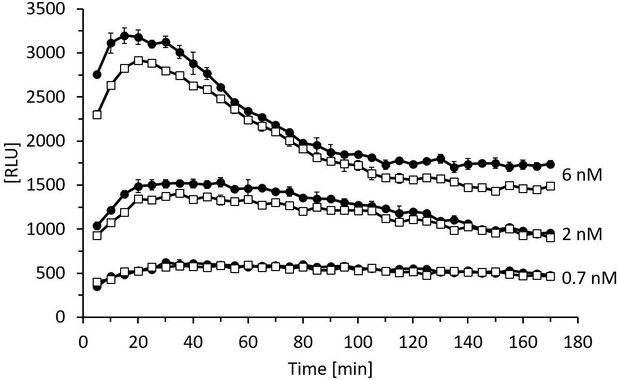
The enzymatic activity of rhDAO-R568S/R571T is comparable to rhDAO-WT.
Enzymatic activity of rhDAO-WT (□) and rhDAO-R568S/R571T (●) at 0.7 nM, 2 nM, and 6 nM was measured as published (Boehm et al., 2020; Boehm et al., 2017). Means ± standard deviation (SD) of duplicate measurements are shown. RLU: relative light units; rhDAO: recombinant human diamine oxidase.
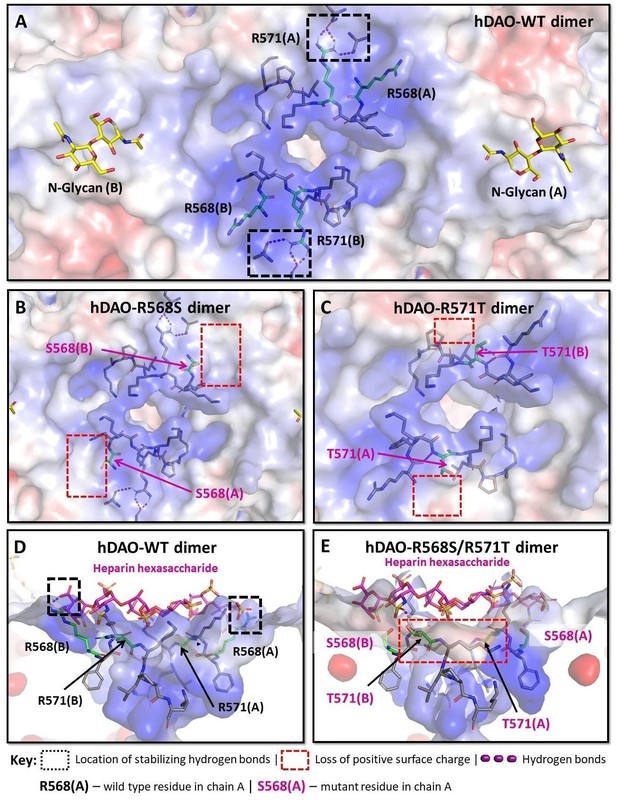
Structural analysis of the heparin-binding motif (HBM) in the 3D models for the R568S, R571T, and R568S/R571T mutants compared with the crystal structure of hDAO-WT.
(A) Top view of the HBM in hDAO-WT. R568 and R571 (green sticks) are the key residues in the positively charged HBM formed by arginines and lysines from both chains in the hDAO-WT dimer. Intramolecular interactions formed by R571 stabilize the HBM. (B) Top view of the HBM in hDAO-R568S dimer. The positively charged area is reduced around S568s in the R568S mutant compared to hDAO-WT (A). (C) Top view of the HBM in hDAO-R571T. The stabilizing interactions formed by R571 in hDAO-WT (A) are lost, and positive patches around T571s are reduced in hDAO-R571T. (D) Complex of hDAO-WT dimer with heparin hexasaccharide, sliced side view. R568s form hydrogen bonds with heparin hexasaccharide. (E) Complex of hDAO-R568S/R571T dimer with heparin hexasaccharide, sliced side view. In the R568S/R571T mutant, the positively charged surface patches and the stabilizing interactions are lost. Red color corresponds to the negatively charged surface, blue color indicates positively charged regions. hDAO: human diamine oxidase.
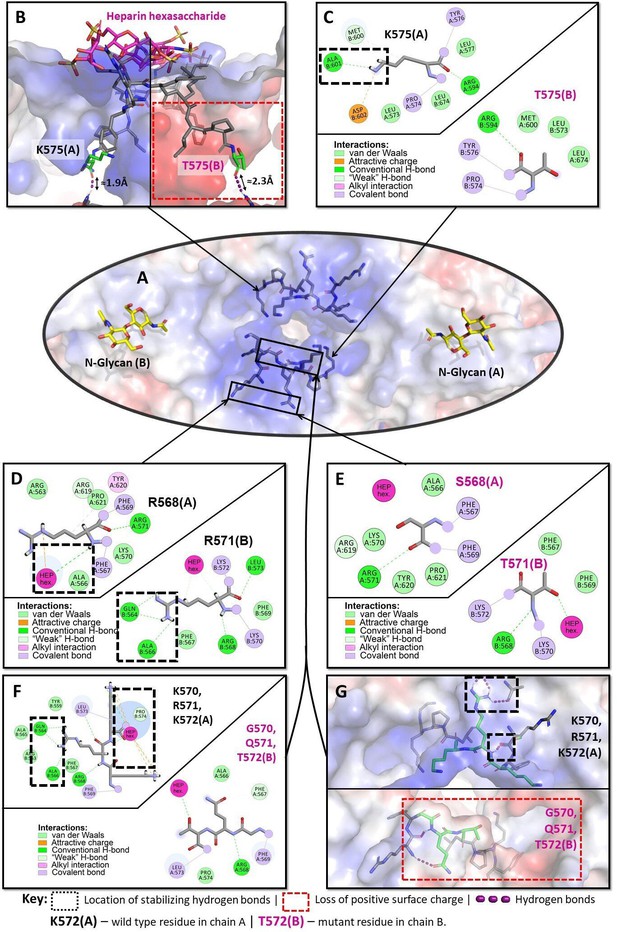
Structural analysis of the heparin-binding motif (HBM) in 3D models of the K575T, K570G/R571Q/K572T, and R568S/R571T mutants versus the crystal structure of hDAO-WT.
(A) HBM in hDAO-WT, top view. (B) Complex of hDAO-WT (left) and hDAO-K575T (right) dimers with heparin hexasaccharide, sliced side view. Change of lysine to threonine leads to a dramatic decrease in the positively charged surface inside the HBM. (C) Local interaction networks of K575 (chain A) in hDAO-WT and the corresponding site with T575 (chain B) of hDAO-K575T show that the mutation eliminates stabilizing hydrogen bonds. (D) Local interaction network of R568 is shown in chain A and R571 – in chain B of hDAO-WT. R568 interacts with heparin hexasaccharide, and R571 establishes multiple interactions with other residues stabilizing the HBM. (E) Local interaction networks are shown for S568 in chain A and T571 – in chain B of hDAO-R568S/R571T. The double mutation leads to an evident disruption of the hydrogen bonding network due to R571T mutation and loss of interactions with the heparin hexasaccharide due to R568S mutation. (F) Local interaction networks of K570, R571, and K572 (chain A) in hDAO-WT compared to the corresponding site in hDAO-K570G/R571Q/K572T (chain B). The triple mutation reduces the number of favorable interactions with other binding site residues and heparin hexasaccharide. (G) Top view of hDAO-WT compared to the K570G/R571Q/K572T triple mutant (chain B). The surface charge of the HBM changes from positive to negative charge in the triple mutant. Red color corresponds to the negatively charged surface, blue color indicates positively charged regions. hDAO: human diamine oxidase.
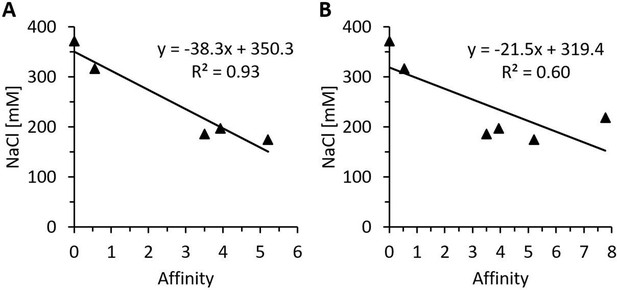
Regression analysis of in silico affinity change estimations and measured heparin-sepharose salt elution concentrations of various heparin-binding motif (HBM) mutants.
Regression analysis of predicted free energy changes in heparin affinity and the NaCl concentrations necessary for the elution from heparin-sepharose of the HBM mutants rhDAO-R568S, rhDAO-K575T, rhDAO-R568S/R571T, and rhDAO-R568S/K575T versus rhDAO-WT (0 affinity; 372 mM NaCl). The rhDAO-K570G/R571Q/K572T triple mutant corresponding to the HBM in guinea pig, dog, rat, mouse, and Chinese hamster proteins was excluded (A, p-value 0.0088), or included (B, p-value 0.08) in the calculations.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
| Gene (Homo sapiens) | AOC1 | GenBank | HGNC: 80 | |
| Recombinant DNA reagent | pRMCE-hDAO (plasmid) | Gludovacz et al., 2016 | hDAO-WT expression vector | |
| Recombinant DNA reagent | pRMCE-hDAO-R568S (plasmid) | This paper | R568S mutant of hDAO | |
| Recombinant DNA reagent | pRMCE-hDAO-R571T (plasmid) | This paper | R571T mutant of hDAO | |
| Recombinant DNA reagent | pRMCE-hDAO-K575T (plasmid) | This paper | K575T mutant of hDAO | |
| Recombinant DNA reagent | pRMCE-hDAO-R568S/R571T (plasmid) | This paper | R568S/R571T mutant of hDAO | |
| Recombinant DNA reagent | pRMCE-hDAO-R568S/K575T (plasmid) | This paper | R568S/K575T mutant of hDAO | |
| Recombinant DNA reagent | pRMCE-hDAO-R571T/K575T (plasmid) | This paper | R571T/K575T mutant of hDAO | |
| Recombinant DNA reagent | pRMCE-hDAO- K570G/R571Q/K572T (plasmid) | This paper | K570G/R571Q/K572T mutant of hDAO | |
| Recombinant DNA reagent | pRMCE-hFcDAO (plasmid) | This paper | Fusion of human IgG Fc and hDAO | |
| Recombinant DNA reagent | pRMCE-hFcDAO-R568S/R571T (plasmid) | This paper | Fusion of human IgG Fc and R568S/R571T mutant of hDAO | |
| Sequence-based reagent | R568S-FP | This paper | PCR primers | ttcaaaaggaagctgccc |
| Sequence-based reagent | R568S-RP | This paper | PCR primers | gctgaaggccgcctggc |
| Sequence-based reagent | R571T-FP | This paper | PCR primers | acgaagctgcccaagtacc |
| Sequence-based reagent | R571T-RP | This paper | PCR primers | tttgaagcggaaggc |
| Sequence-based reagent | K575T-FP | This paper | PCR primers | acgtacctgctctttaccagcc |
| Sequence-based reagent | K575T-RP | This paper | PCR primers | gggcagcttccttttga |
| Sequence-based reagent | R568S/R571T-FP | This paper | PCR primers | aaaacgaagctgcccaagtacctg |
| Sequence-based reagent | R568S/R571T-RP | This paper | PCR primers | gaagctgaaggccgcctgg |
| Sequence-based reagent | R571T/K575T-FP | This paper | PCR primers | gcccacgtacctgctctttaccagccc |
| Sequence-based reagent | R571T/K575T-RP | This paper | PCR primers | agcttcgttttgaagcggaaggc |
| Sequence-based reagent | K570G/R571Q/K572T-FP | This paper | PCR primers | cagacgctgcccaagtacctgct |
| Sequence-based reagent | K570G/R571Q/K572T-RP | This paper | PCR primers | tccgaagcggaaggccg |
| Peptide, recombinant protein | rhDAO-WT | Gludovacz et al., 2016 | Recombinant human diamine oxidase, wildtype | |
| Peptide, recombinant protein | rhDAO-R568S | This paper | R568S mutant of rhDAO-WT | |
| Peptide, recombinant protein | rhDAO-K575T | This paper | K575T mutant of rhDAO-WT | |
| Peptide, recombinant protein | rhDAO-R568S/R571T | This paper | R568S/R571T mutant of rhDAO-WT | |
| Peptide, recombinant protein | rhDAO-R568S/K575T | This paper | R568S/K575T mutant of rhDAO-WT | |
| Peptide, recombinant protein | rhDAO- K570G/R571Q/K572T | This paper | K570G/R571Q/K572T mutant of rhDAO-WT | |
| Peptide, recombinant protein | rhFcDAO | This paper | Human IgG Fc-rhDAO fusion protein | |
| Peptide, recombinant protein | rhFcDAO-R568S/R571T | This paper | Human IgG Fc-rhDAO-R568S/R571T fusion protein | |
| Cell line (Cricetulus griseus) | CHO-K1_rhDAO-WT | Gludovacz et al., 2016 | CHO-K1 stably expressing rhDAO-WT | |
| Cell line (C. griseus) | CHO-K1_rhDAO-R568S | This paper | CHO-K1 stably expressing rhDAO-R568S | |
| Cell line (C. griseus) | CHO-K1_rhDAO-K575T | This paper | CHO-K1 stably expressing rhDAO-K575T | |
| Cell line (C. griseus) | CHO-K1_rhDAO-R568S/R571T | This paper | CHO-K1 stably expressing rhDAO-R568S/R571T | |
| Cell line (C. griseus) | CHO-K1_rhDAO-R567S/K575T | This paper | CHO-K1 stably expressing rhDAO- R567S/K575T | |
| Cell line (C. griseus) | CHO-K1_rhDAO- K570G/R571Q/K572T | This paper | CHO-K1 stably expressing rhDAO- K570G/R571Q/K572T | |
| Cell line (C. griseus) | CHO-K1_rhFcDAO | This paper | CHO-K1 stably expressing rhFcDAO | |
| Cell line (C. griseus) | CHO-K1_ rhFcDAO-R568S/R571T | This paper | CHO-K1 stably expressing rhFcDAO-R568S/R571T | |
| Cell line (C. griseus) | ExpiCHO-S | Thermo Fisher Scientific | Cat#: A29133RRID:CVCL_5J31 | |
| Cell line (C. griseus) | CHO-K1 | ATCC | Cat#: CCL-61RRID:CVCL_0214 | |
| Cell line (H. sapiens) | SK-Hep1 | Sigma-Aldrich | Cat#: 91091816RRID:CVCL_0525 | |
| Cell line (H. sapiens) | HUVEC67 | Evercyte | Cat#: CPT-006-0067 | |
| Cell line (H. sapiens) | HUVEC/TERT2 | Evercyte | Cat#: CHT-006-0008RRID:CVCL_9Q53 | |
| Cell line (H. sapiens) | HDMVEC/TERT164-B | Evercyte | Cat#: CHT-013-0164-B | |
| Cell line (H. sapiens) | HDF76 | Evercyte | Cat#: CPT-008-0076 | |
| Cell line (H. sapiens) | PODO/TERT256 | Evercyte | Cat#: CHT-033-0256RRID:CVCL_JL76 | |
| Cell line (H. sapiens) | LHCN-M2 | Evercyte | Cat#: CkHT-040-231-2RRID:CVCL_8890 | |
| Cell line (H. sapiens) | HepG2 | ATCC | Cat#: HB-8065RRID:CVCL_0027 | |
| Cell line (H. sapiens) | HeLa | Ellmeier Lab – Medical University of Vienna | ||
| Cell line (H. sapiens) | EVT | Velicky et al., 2018 | ||
| Antibody | Anti-ABP1 (rabbit polyclonal) | Sigma-Aldrich | Cat#: SAB1410491-100UG | IF (1:500) |
| Antibody | Alexa Fluor 488 anti-rabbit (H + L) (donkey polyclonal) | Jackson Research | Cat#: 711-545-152RRID:AB_2313584 | IF (1:500) |
| Antibody | IgG serum fraction (rabbit polyclonal) | Boehm et al., 2017 | WB (1:1000) | |
| Antibody | β-actin mAB (AC-15) (mouse monoclonal) | Invitrogen | Cat#: AM4302RRID:AB_2536382 | WB (1:5000) |
| Antibody | IRDye 800CW anti-rabbit IgG (H + L) (goat polyclonal) | Li-Cor | Cat#: 926-32211RRID:AB_621843 | WB (1:5000) |
| Antibody | IRDye 680RD anti-mouse IgG (H + L) (goat polyclonal) | Li-Cor | Cat#: 925-68070RRID:AB_2651128 | WB (1:5000) |
| Chemical compound, drug | Histamine | Sigma-Aldrich | Cat#: 53300 | |
| Chemical compound, drug | Heparin | Gilvasan | 1000 IU/mL | |
| Chemical compound, drug | Heparin (sodium salt from intestinal mucosa) | Sigma-Aldrich | Cat#: H3149 | |
| Chemical compound, drug | Diminazene aceturate | Sigma-Aldrich | Cat#: D7770 | |
| Commercial assay or kit | Cisbio HTRF histamine 500 test kit | Biomedica | Cat#: 62HTMDPET | |
| Commercial assay or kit | Immunotech histamine ELISA | Beckman Coulter | Cat#: IM2562 | |
| Commercial assay or kit | ExpiCHO Expression System Kit | Thermo Fisher Scientific | Cat#: A29133 | |
| Software, algorithm | R (version 3.6.3) | R Development Core Team, 2020 | ||
| Software, algorithm | Kaluza Flow Cytometry Analysis Software (version 2.1) | Beckman Coulter | ||
| Software, algorithm | ImageJ/Fiji | Schneider et al., 2012 | ||
| Software, algorithm | Pymol (version 2.0) | The PyMOL Molecular Graphics System, version 2.0 Schrödinger, LLC | ||
| Software, algorithm | Chimera | UCSF | AmberTools GUI incorporated | |
| Software, algorithm | BIOVIA Discovery Studio 2019 | Dassault Systemes | Modeller 9.2 GUI incorporated | |
| Software, algorithm | FoldX 4.0 | Schymkowitz et al., 2005 | ||
| Other | DAPI stain | Invitrogen | Cat#: D1306 | (80 ng/mL) |
Mutuations in the heparin-binding motif of recombinant human diamine oxidase (rhDAO) reduce binding to heparin-sepharose.
| rhDAO | NaCl | KCl | Mean (SD) | ||
|---|---|---|---|---|---|
| Concentration (mM) | Reduction (mM) | Concentration (mM) | Reduction (mM) | Reduction (mM) | |
| WT | 372 | 0 | 310 | 0 | 0 (na) |
| R568S | 186 | 186 | 172 | 138 | 162 (24) |
| K575T | 317 | 55 | 233 | 77 | 66 (11) |
| R568S/R571T | 175 | 197 | 138 | 172 | 185 (13) |
| R568S/K575T | 197 | 175 | 164 | 146 | 161 (15) |
| K570G/R571Q/K572T* | 219 | 153 | - | - | - |
-
Purified rhDAO-WT and various HBM mutants were loaded onto a heparin-sepharose column and eluted with linear gradients of 0–1 M NaCl or KCl. The salt concentrations necessary for rhDAO elution and the reduction thereof compared to the wildtype protein are shown. The physiological salt concentration in blood plasma and interstitial fluid is approximately 145 mM.
-
*
Corresponds to the HBM in guinea pig, dog, rat, mouse, and Chinese hamster.
-
HBM = heparin-binding motif; SD = standard deviation; na = not applicable.
Recombinant human diamine oxidase (rhDAO) heparin-binding motif mutants eliminate the fast α-distribution half-life and strongly reduce clearance in rats and mice.
| rhDAO | Rats IV | |||||||
|---|---|---|---|---|---|---|---|---|
| AUC (µg/mL/min)*† | Fold increase | t1/2 α (min) | t1/2 β (min) | |||||
| WT | 116 | 1.0 | 3.7 | 250 | ||||
| R568S | 1531 | 13.2 | na | 294 | ||||
| R568S/R571T | 3788 | 32.6 | na | 353 | ||||
| R568S/K575T | 2113 | 18.2 | 42 | 361 | ||||
| Fc-WT | 69 | 1.0 | 1.7 | 120 | ||||
| Fc-R568S/R571T | 2723 | 39.5 | na | 268 | ||||
| Mice IV | Mice IP | |||||||
| AUC (µg/mL/min)† | Fold increase | t1/2 (min) ‡ ‡ | AUC (µg/mL/min) § ‡ | Fold increase | t1/2 (min) ‡* § | |||
| WT | 76 | 1.0 | 76.1 | 41.2 | 1.0 | 200 | ||
| R568S/R571T | 1468 | 19.4 | 192.1 | 666.3 | 16.2 | 394 | ||
-
*
Calculated from 5 to 1440 min.
-
†
Calculated from 10 to 1680 min.
-
‡
Calculated from 60 to 1680 min.
-
§
Calculated from 0 to 1440 min.
-
AUC = area under the curve; IV = intravenous; IP = intraperitoneal; na = not applicable.
Predicted free energy changes in the human diamine oxidase (hDAO) dimer by mutation of the heparin-binding motif (HBM).
| rhDAO mutant variant | Stability | Affinity | ||
|---|---|---|---|---|
| ∆∆G (kcal/mol) | ∆∆G (kcal/mol) | |||
| R568S | +0.29 | +3.50 | ||
| R571T | +0.73 | +1.96 | ||
| K575T | +2.13 | +0.55 | ||
| R568S/R571T | +0.60 | +5.21 | ||
| R568S/K575T | +2.42 | +3.93 | ||
| R571T/K575T | +2.86 | +2.52 | ||
| K570G/R571Q/K572T* | +1.53 | +7.77 | ||
-
The plus sign (+) in the ‘Stability’ column indicates the destabilizing effect on the 3D protein structure, while the plus sign (+) in the ‘Affinity’ column means reduced affinity to heparin hexasaccharide compared to wildtype hDAO. A higher value indicates higher effect on stability or affinity.
-
*
Corresponds to the HBM in guinea pig, dog, rat, mouse, and Chinese hamster.
Additional files
-
Supplementary file 1
The evolutionary conservation analysis of the diamine oxidase (DAO) heparin-binding motif is summarized in Supplementary file 1.
- https://cdn.elifesciences.org/articles/68542/elife-68542-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68542/elife-68542-transrepform1-v2.docx





