Structural insights into the activation of human calcium-sensing receptor
Figures
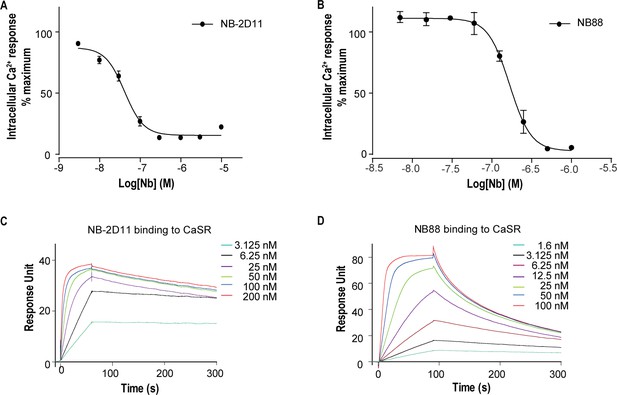
The function and binding affinity of NB-2D11 and NB88.
(A) Dose-dependent NB-2D11-inhibited intracellular Ca2+ mobilization in response to Ca2+ ions. N = 3, data represent mean ± SEM (Figure 1—source data 1). (B) Dose-dependent NB-88-inhibited intracellular Ca2+ mobilization in response to Ca2+ ions. N = 3, data represent mean ± SEM (Figure 1—source data 1). (C) SPR sensorgram showing that NB-2D11 bound to CaSR with 0.24 nM affinity (Figure 1—source data 2). (D) SPR sensorgram showing that NB88 bound to CaSR with 3.9 nM affinity (Figure 1—source data 2).
-
Figure 1—source data 1
Intracellular Ca2+ flux assay on CaSR-NB-2D11 and CaSR-NB88 complex.
- https://cdn.elifesciences.org/articles/68578/elife-68578-fig1-data1-v2.xlsx
-
Figure 1—source data 2
SPR sensorgram of NB-2D11 and NB88 binding affinity.
- https://cdn.elifesciences.org/articles/68578/elife-68578-fig1-data2-v2.xlsx
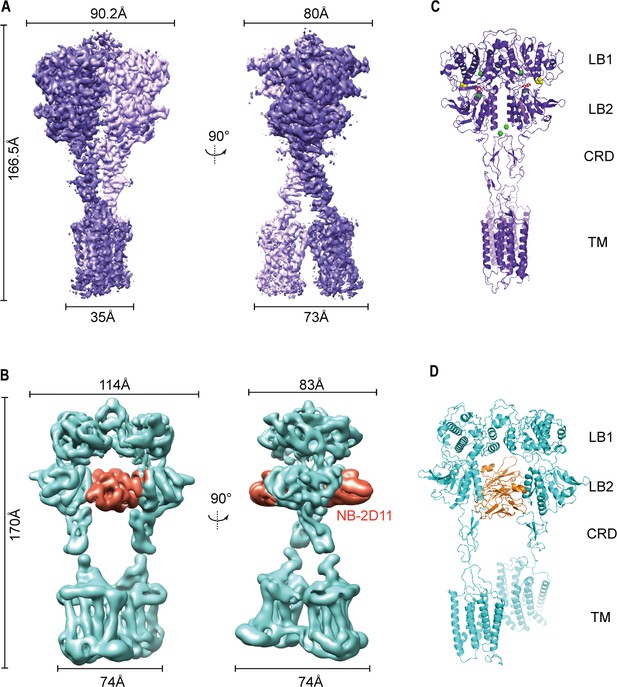
Cryo-EM maps and models of full-length CaSR.
(A) Left panel shows the view of CaSR in the active conformation (purple) from front view, and the right panel shows the view after a 90° rotation as indicated. (B) Left panel shows the view of CaSR in the inactive conformation (cyan) bound to NB-2D11 (orange) from front view, and the right panel shows the view after a 90° rotation as indicated. (C) Model (Ribbon representation) of CaSR shows the structure of the active state (purple) bound to TNCA (red) and Ca2+ ion (green) viewed from the side. (D) Model (Ribbon representation) of CaSR shows the structure of the inactive state (cyan) bind with NB-2D11 (orange).
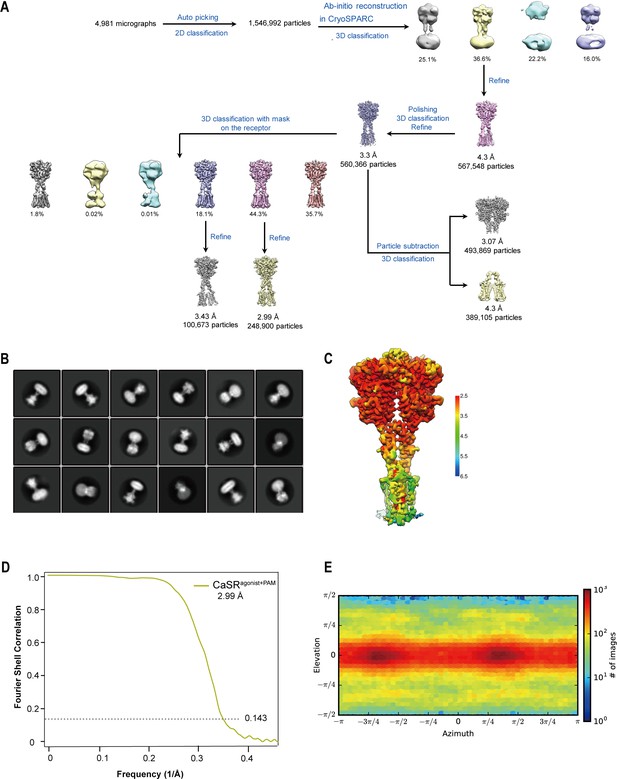
Cryo-EM processing workflow of CaSR bound to agonist+PAM.
(A) The flow chart displaying the cryo-EM processing of agonist+PAM bound CaSR in GDN. (B) Cryo-EM class averages of agonist+PAM bound CaSR in GDN micelles. (C) Local resolution map of agonist+PAM bound CaSR. (D) Gold standard Fourier shell correlation (FSC) curve indicates that the map for agonist+PAM bound CaSR reaches nominal resolutions of 2.99 Å at FSC = 0.143. (E) Particle angular distribution of the final cryo-EM reconstruction of agonist+PAM bound CaSR.
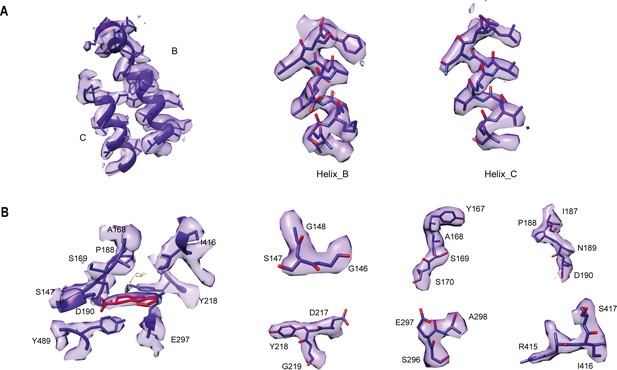
Agreement between the cryo-EM map of CaSR bound to agonist+PAM and the model.
(A) The cryo-EM densities and fitted atomic models of B and C helices of CaSR in agonist+PAM bound conformation. (B) The cryo-EM densities and fitted atomic models of TNCA and related amino acids.
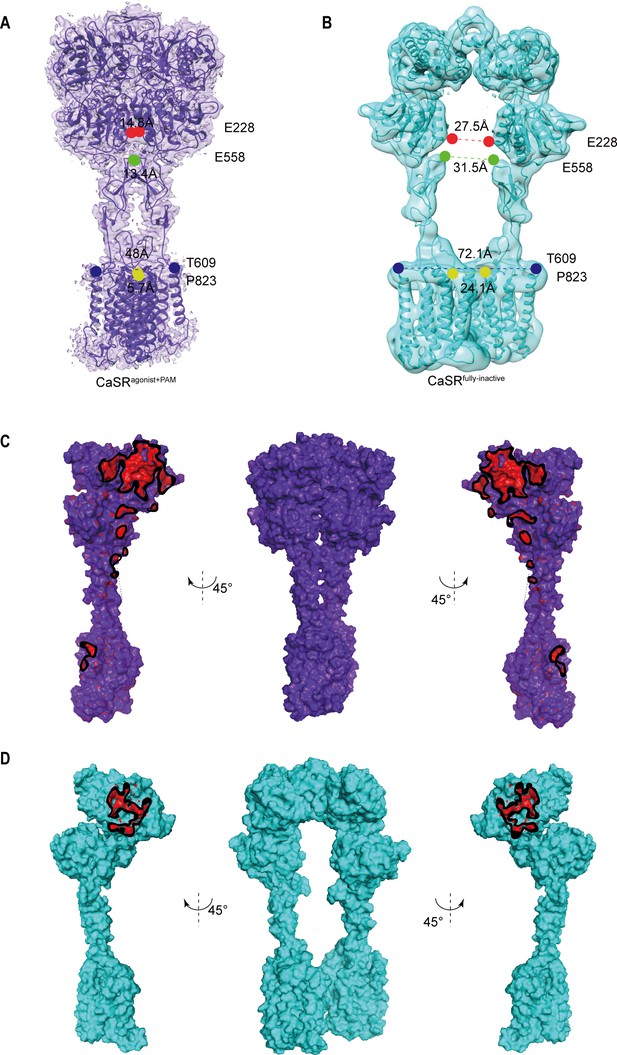
Cryo-EM maps and models of CaSR.
(A, B) The full-length CaSR cryo-EM maps and models present view from front view in the agonist+ PAM bound state (purple) (A), and the fully inactive state (cyan) with deletion of NB-2D11 for clarity (B). Positions in the VFT (red, E228), CRD (green, E558), CRD/7TMD interface (blue, T609), and 7TM domain (yellow, P823) show that the active state is characterized by smaller intersubunit distances. The P823 position in the active model (yellow) shows the contact of 7TMDs. (C, D) Comparison of the intersubunit interfaces in active and inactive CaSR are shown for active (C) and inactive (D) CaSR. Contact regions (red) show residues within 4 Å of the opposite subunit. It should be noted that there is only one LB1–LB1 interaction interface for the inactive CaSR, and its total buried surface area is much smaller than that of the active CaSR with four different contact interfaces.
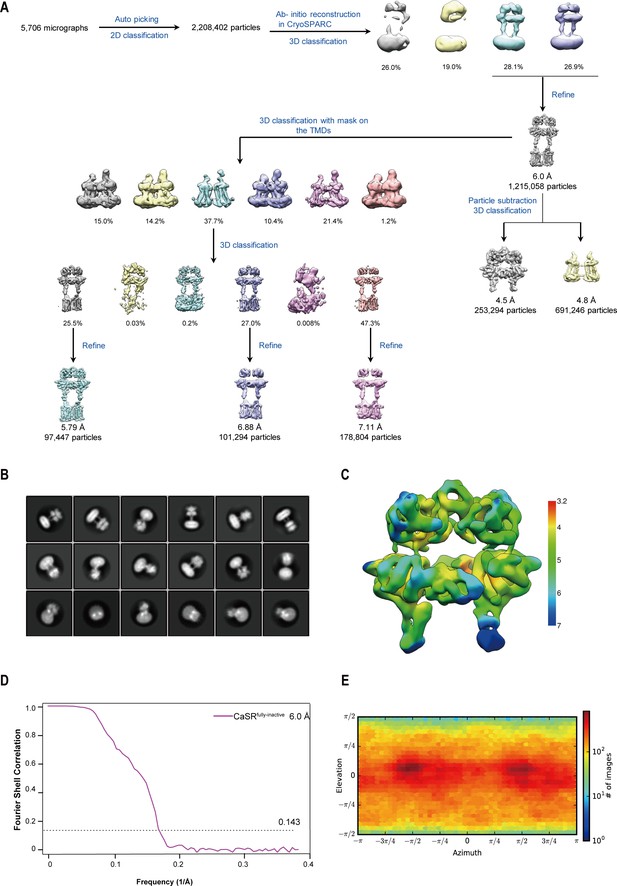
Cryo-EM processing workflow of inactive CaSR bound to NB-2D11 in GDN.
(A) The flow chart displaying the cryo-EM processing of inactive CaSR bound to NB-2D11 in GDN. (B) Cryo-EM class averages of inactive CaSR bound to NB-2D11 in GDN micelles. (C) Local resolution map of inactive CaSR bound to NB-2D11. (D) Gold standard Fourier shell correlation (FSC) curve indicates that the overall map for inactive CaSR bound to NB-2D11 reaches nominal resolutions of 6.0 Å at FSC = 0.143. (E) Particle angular distribution of the final cryo-EM reconstruction of inactive CaSR bound to NB-2D11.
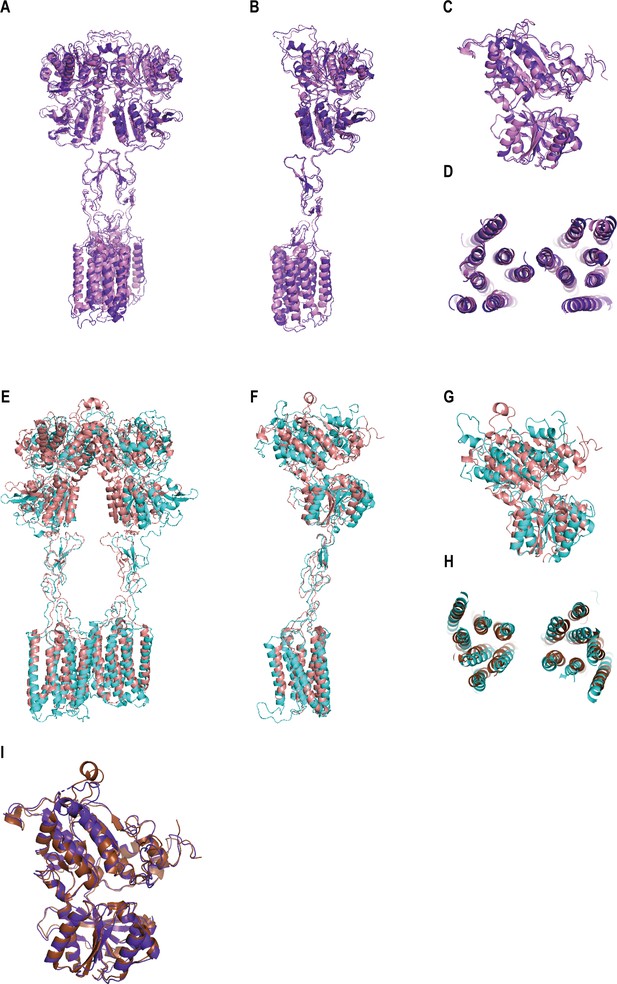
Comparisons of the structures of CaSR in different conformations.
(A) The superimposition of CaSRagonist+PAM (purple) and CaSRAcc (lavender, PDB:7DTV) displays the similar structures with the r.m.s.d of 1.299 Å. (B–D) The superimpositions of CaSRagonist+PAM (purple) and CaSRAcc (lavender, PDB:7DTV) in intersubunit (B), the VFT (C), and 7TMD (D) regions. (E) The superimposition of CaSRfully inactive (cyan) and CaSRIcc (brown, PDB:7DTW) presents a significant difference with the r.m.s.d of 8.007 Å. (F–H) The alignment of CaSRfully inactive (cyan) and CaSRIcc (brown, PDB:7DTW) in intersubunit (F), the VFT (G), and 7TMD (H) regions. (H) The superimpositions of CaSRagonist+PAM (purple), CaSRAcc (lavender, PDB:7DTV), and CaSRIcc (brown, PDB:7DTW) for VFT domain.
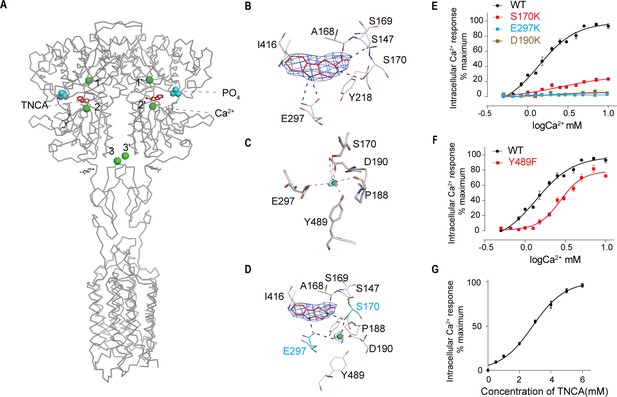
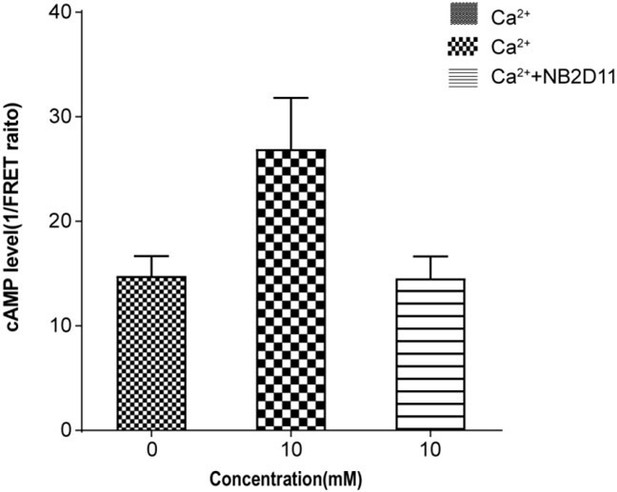
Ca2+ and TNCA as a composite agonist activate the full-length CaSR dimer directly.
(A) Ribbon representation of the active CaSR (gray), showing the location of the Ca2+-binding sites (green sphere) and TNCA (red). (B) Specific contacts between CaSR (gray) and TNCA (red space-filling model), mesh represents the final density map contoured at 17σ surrounding. (C) Specific interactions between CaSR and newly identified Ca2+ ion (green sphere), the mesh represents the cryo-EM density map contoured at 6.0σ surrounding Ca2+. (D) Highlighting the newly identified Ca2+ and TNCA sharing two common binding residues S170 and E297 (cyan space-filling model). (E) Dose-dependent intracellular Ca2+ mobilization expressing WT (black dots), mutant S170K (red dots), E297K (cyan dots), and D190K (brown dots) of CaSR. The single mutations were designed based on Ca2+ binding sites. N = 4, data represent mean ± SEM (Figure 3—source data 1). (F) Dose-dependent intracellular Ca2+ mobilization expressing WT (black dots), mutant Y489K (red dots) of CaSR. The single mutation was designed based on Ca2+ binding sites. N = 4, data represent mean ± SEM (Figure 3—source data 1). (G) Dose-dependent TNCA-activated intracellular Ca2+ mobilization in response to 0.5 mM Ca2+ ions. N = 3, data represent mean ± SEM (Figure 3—source data 2).
-
Figure 3—source data 1
Intracellular Ca2+ flux assay on CaSR mutations.
- https://cdn.elifesciences.org/articles/68578/elife-68578-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Intracellular Ca2+ flux assay on CaSR-TNCA complex.
- https://cdn.elifesciences.org/articles/68578/elife-68578-fig3-data2-v2.xlsx
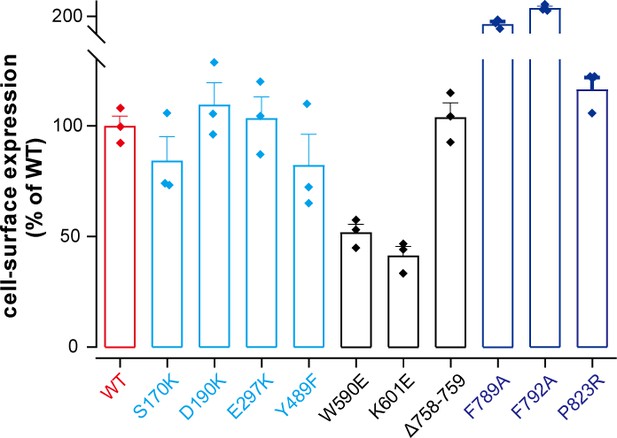
Cell surface expression.
Cell surface expression of indicated CaSR mutants. N = 3, data represent mean ± SEM.
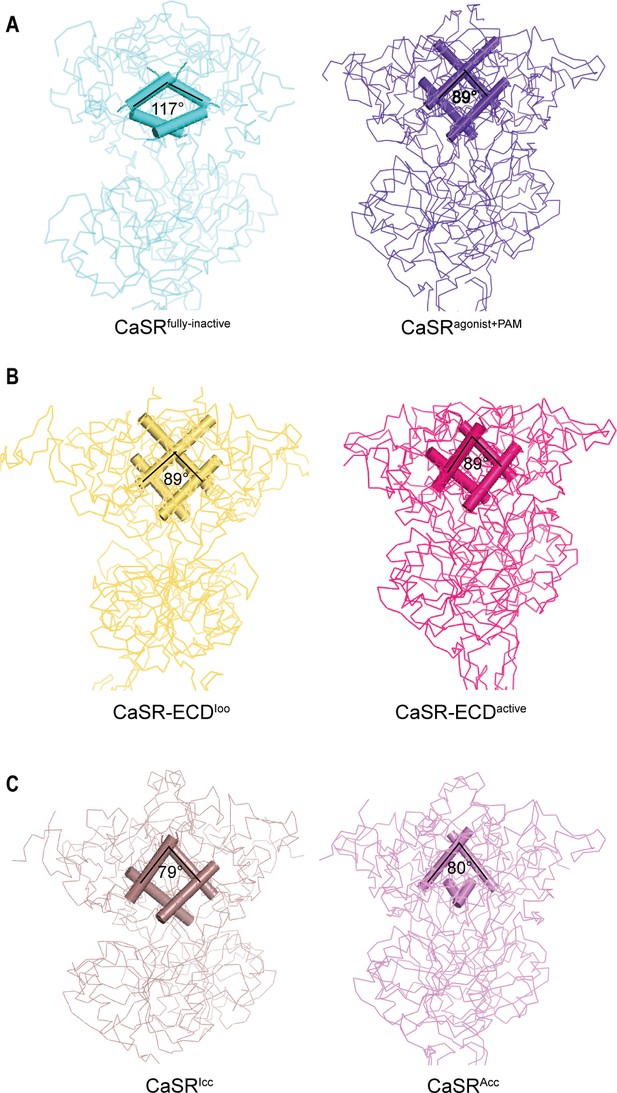
Comparisons of intersubunit LB1 domains interfaces in the inactive and active states of CaSR.
(A) Left panel: The Cα trace of VFT module of CaSRfully inactive cryo-EM structure (cyan). The B-C Helix angle is 117°. Right panel: The Cα trace of VFT module of CaSRagonist+PAM cryo-EM structure (purple). The B-Helix angle is 89°. (B) Left panel: The Cα trace of VFT module of crystal structure of CaSR-ECDIoo (yellow) (PDB:5K5T). The B-Helix angle is 89°. Right panel: The Cα trace of VFT module of CaSR-ECDactive crystal structure (red) (PDB:5K5S). The B-Helix angle is 89°. (C) Left panel: The Cα trace of VFT module of CaSRIcc cryo-EM structure (brown) (PDB:7DTW). The B-Helix angle is 79°. Right panel: The Cα trace of VFT module of CaSRAcc cryo-EM structure (lavender) (PDB:7DTV). The B-Helix angle is 80°.
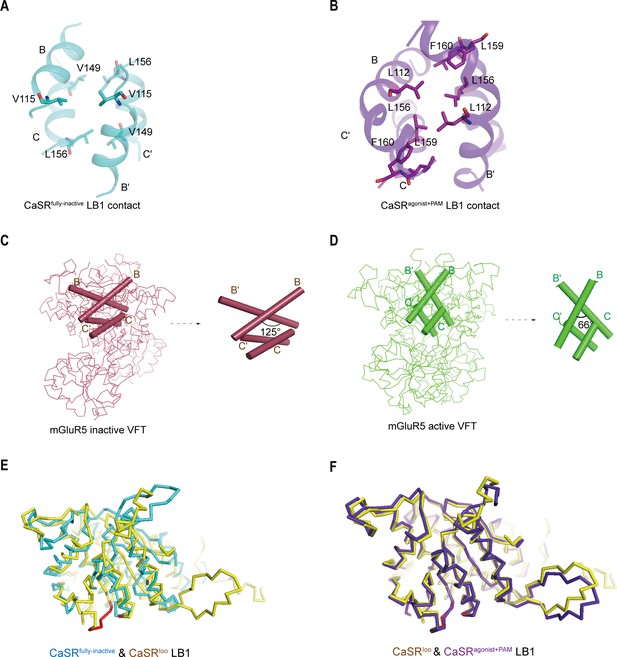
Comparisons of intersubunit LB1 domains interfaces in the inactive and active states of CaSR.
(A, B) The interface is a hydrophobic patch between residues on the B and C helices of each protomer. In the inactive conformation, it is an interface involving V115, V149, and L156 residues (A, cyan), whereas LB1 interface of the agonist+PAM bound state (B, purple) is packed with residues L112, L156, L159, and F160. (C) Left panel: The Cα trace of VFT module of inactive mGluR cryo-EM structure (raspberry) (PDB: 6N52). Right panel: The B-Helix angle is 125°. (D) Left panel: The Cα trace of VFT module of active mGluR cryo-EM structure (green) (PDB: 6N51). Right panel: The B-Helix angle is 66°. (E, F) Alignment of LB1 domain of CaSR in three conformations: fully inactive (cyan), intermediate (yellow, PDB:5K5T), and agonist+PAM bound (purple) states, ligand-binding residues (red). (E) Fully inactive and intermediate LB1 domain, the conformation of the ligand-binding region in LB1 domain is significantly different in two states. (F) intermediate and agonist+PAM LB1 domain, showing a well superposition.
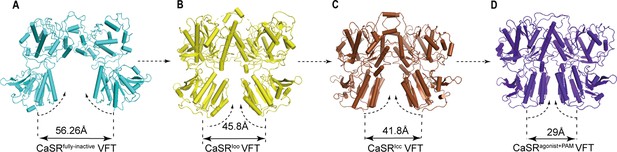
The conformational changes of LB2 domains in three states.
(A) The CaSRfully inactive (cyan) conformation of VFT module. The distance between C termini of the two LB2 domains is 56.26 Å. (B) CaSR-ECDIoo (yellow) (PDB:5K5T) conformation of VFT module. The distance between C termini of the two LB2 domains is 45.8 Å. (C) CaSRIcc (brown) conformation of VFT module (PDB:7DTW). The distance between C termini of the two LB2 domains is 41.8 Å. (D) CaSRagonist+PAM (purple) conformation of VFT module. The distance between C termini of the two LB2 domains is 29 Å.
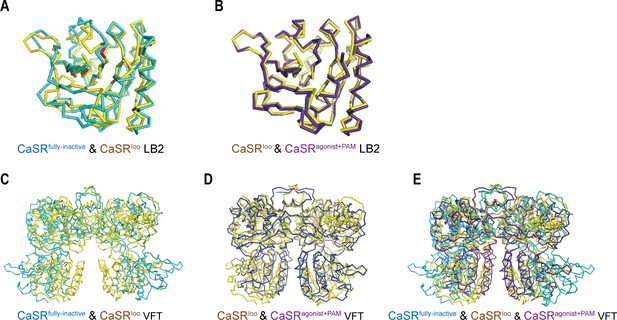
Superposition of LB1 and VFT domains of CaSR.
(A) Inactive and intermediate LB2 domains, (B) intermediate (PDB:5K5T) and active LB2 domains, which indicate the three conformations of LB2 domain have no distinguishing differences, suggesting the ligand-binding residues in LB2 domain do not change during approach of LB2 domain. (C–E) Superpositions of VFT module based on LB1 domains in the inactive and intermediate (C), intermediate and active (D), and inactive, intermediate, and active states (E) of CaSR.
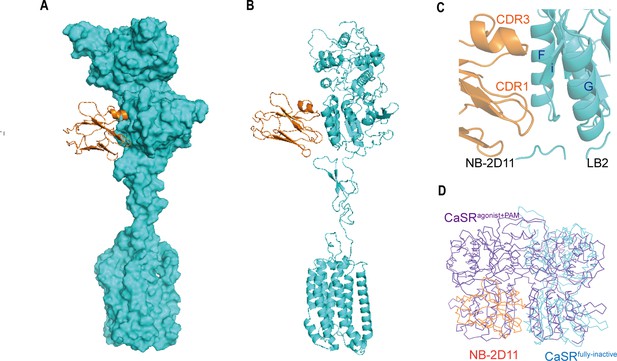
The NB-2D11 blocks the interaction of LB2 domains.
(A) Structure of the inactive CaSR protomer (surface representation, cyan) with NB-2D11 (ribbon diagram, orange) from front view. (B) The NB-2D11 (orange) binds the left lateral of the LB2 (cyan) from the front view of the protomer. (C) NB-2D11 binds the LB2 domain through a series of polar interactions through CDR1 and CDR3 of the nanobody and the Helix F and Strand I of the CaSR. (D) Superposition of NB-2D11 (orange) binding inactive conformations (cyan) and active (purple) conformations based on the LB2 domain of VFT module, showing the whole NB-2D11 in the inactive state crashes with the LB2 domain of another VFT module in the active state.
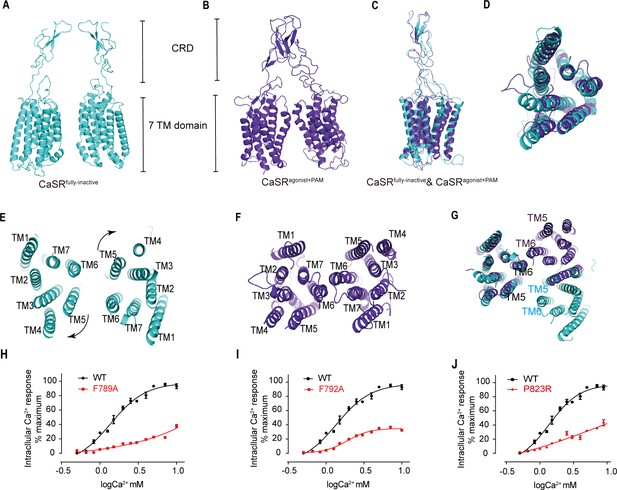
The closure of VFT leading to the rearrangement of inter-7TMDs.
(A) Front view of CaSRfully inactive CRDs and 7TMDs (cyan). (B) Front view of CaSRagonist+PAM CRDs and 7TMDs (purple). (C) The alignment of the part of CRD and 7TMDs in both fully inactive and agonist+PAM bound CaSR. (D) The alignment of inactive and agonist+PAM bound 7TMDs from top view. (E–G) The 7TMDs interface in the fully inactive state of CaSR is mediated by TM5 and TM6 (cyan) from top view and that of the agonist+PAM state is driven by TM6 from top view. Superposition of 7TMD of the inactive (cyan) and agonist+PAM bound CaSR (purple) show the rotation of 7TMDs. (H–J) Dose-dependent intracellular Ca2+ mobilization expressing WT (black dots) and mutant (red dots) CaSR (Figure 7—source data 1). The single mutations of F789A (H), F792A (I), and P823R (J) were designed based on the inactive density map. For (H–J), N = 4, data represent mean ± SEM.
-
Figure 7—source data 1
Intracellular Ca2+ flux assay on various CaSR mutations.
- https://cdn.elifesciences.org/articles/68578/elife-68578-fig7-data1-v2.xlsx
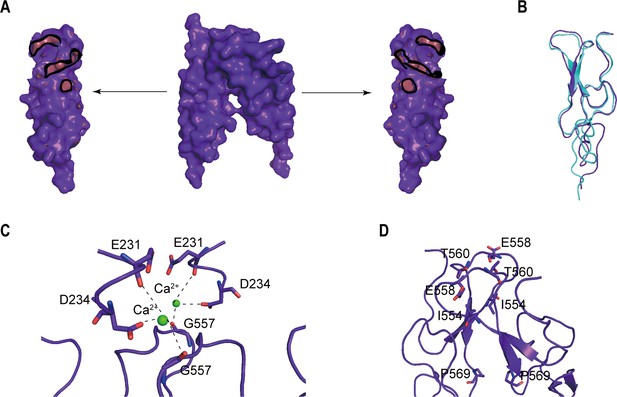
The homodimer interface of the CRDs in the active state of CaSR.
(A) The homodimer interface of the CRDs (surface representation, purple). Contact regions (red) show residues within 4 Å of the opposite. It covers approximately 1079.09 Å2 of solvent accessible surface area. (B) There is a bound Ca2+ coordinated by carboxylate group of D234 and carbonyl oxygen of E231 and G557, holding the interface that is required to activate the receptor. (C) The CR–CR contacts were maintained through the cross-subunit hydrogen bonds between T560 and E558, and hydrophobic interaction of I554 and P569.
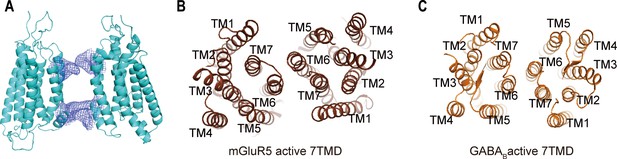
Conformational change of the 7TMDs interface during activation.
(A) The 7TMD configuration in the inactive state from front view. There are pairwise symmetrical undefined maps that link the extracellular and intracellular part of TM5 and TM6 in the 7TM dimer interface, blocking the association of 7TMDs could regulate the function of CaSR. (B, C) The 7TMDs interface of the active state of mGluR5 (C, PDB:6N51) and GABAB (D, PDB:7C7Q) have the interface contact with TM6.
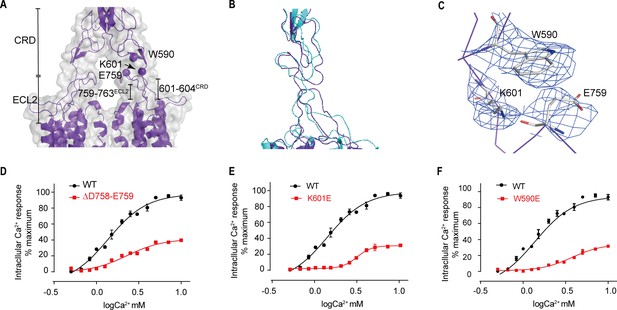
Upward movement of LB2 is converted into the intra-7TM conformational rearrangement through ECL2.
(A) Model in CaSRagonist+PAM state (purple) and cryo-EM map (gray) showing the contact between the CRD and the ECL2 of the 7TMDs. Critical residues at this interface are shown as spheres at their Cα positions. (B) Superposition of the interface between the CRD and the ECL2 of the 7TMD between both fully inactive and agonist+PAM bound conformations. (C) Specific contacts between the loop of CR domain and the loop of ECL2 to shift the ECL2 up. (D–F) Deletion of residues D758 and E759 (D), the single mutation of K601E (E) and W590E (F) significantly reduced Ca2+-induced receptor activity. (WT in black dots and mutant in red dots). For (D–F), N = 4, data represent mean ± SEM (Figure 8—source data 1).
-
Figure 8—source data 1
Intracellular Ca2+ flux assay on CaSR mutations.
- https://cdn.elifesciences.org/articles/68578/elife-68578-fig8-data1-v2.xlsx
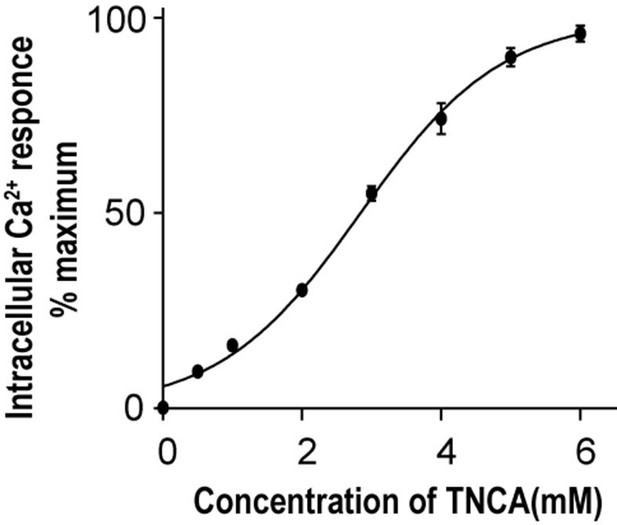
Does-response curves of TNCA-induced intracellular Ca2+ mobilization in presence of 0.
5mM extracellular Ca2+ ion.
Tables
Cryo-EM data collection, refinement, and validation statistics.
| CaSR | #1 inactive(EMD-30997)(PDB 7E6U) | #2 agonist+PAM(EMD-30996)(PDB 7E6T) |
|---|---|---|
| Data collection and processing | ||
| Magnification | 81,000× | 81,000× |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 70 | 70 |
| Defocus range (μm) | –1.5 to –2.5 | –1.5 to –2.5 |
| Pixel size (Å) | 1.071 | 1.071 |
| Symmetry imposed | C2 | C2 |
| Initial particle images (no.) | 2,208,402 | 1,546,992 |
| Final particle images (no.) | 1,215,058 | 560,366 |
| Map resolution (Å)FSC threshold | 6.00.143 | 3.30.143 |
| Map resolution range (Å) | 3.2–7.0 | 2.5–6.5 |
| Refinement | ||
| Initial model used (PDB code) | 5k5s, 6n51 | 5k5s, 6n51 |
| Model resolution (Å)FSC threshold | 4.3/5.9/8.00/0.143/0.5 | 3.3/3.4/3.70/0.143/0.5 |
| Model resolution range (Å) | 4.3–8.0 | 3.3–3.7 |
| Map sharpening B factor (Å2) | –217 | –115 |
| Model composition | ||
| Non-hydrogen atoms | 14,214 | 12,751 |
| Protein residues | 1796 | 1592 |
| Ligands | 0 | PO43-: 2 |
| Ca2+: 6 | ||
| NAG: 4 | ||
| TNCA: 2 | ||
| B factors (Å2) | ||
| Protein | 102.59/530.90/286.91 | 61.44/302.84/157.67 |
| Ligand | N/A | 91.52/151.96/105.80 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.002 | 0.002 |
| Bond angles (°) | 0.559 | 0.602 |
| Validation | ||
| MolProbity score | 2.5 | 1.49 |
| Clashscore | 14 | 5 |
| Poor rotamers (%) | 0 | 0 |
| Ramachandran plot | ||
| Favored (%) | 94 | 97 |
| Allowed (%) | 6 | 3 |
| Disallowed (%) | 0 | 0 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | CaSR | NCBI | NM_000388.4 | |
| Strain, strain background (Escherichia coli) | BL21(DE3) | New England Biolabs | Cat#: C2527I | E. coli strain for expression of the nanobody |
| Strain, strain background (Escherichia coli) | TG1 | Lucigen | Cat#: 60,502 | Electrocompetent cells |
| Strain, strain background (Escherichia coli) | TOP10F' | Huayueyang Biotech | WXR15-100S | E. coli strain for expression of the nanobody |
| Cell line (Homo sapiens) | (HEK) 293 S GnTI- cells | ATCC | Cat# CRL-3022RRID: CVCL_A785 | Mycoplasma negative |
| Cell line (Homo sapiens) | HEK 293T/17 | ATCC | Cat# CRL-11268RRID: CVCL_1926 | Mycoplasma negative |
| Antibody | HA-Tag, (Mouse monoclonal) | Yeasen | 30,701ES60 | Dilution: (1/2000) |
| Antibody | Flag-Tag (DYKDDDDK), (Mouse monoclonal) | Yeasen | 30,503ES20 | Dilution: (1/2000) |
| antibody | Peroxidase AffiniPure Goat Anti-Mouse IgG (H + L)(Goat monoclonal) | Yeasen | 33,201ES60 | Dilution: (1/2500) |
| Recombinant DNA reagent | AxyPrep Plasmid Miniprep Kit | CORNING LIFE SCIENCES | Cat#:220 | |
| Recombinant DNA reagent | pMECS vector | BioVector NTCC | pMECS | Phage display vector |
| Recombinant DNA reagent | pEG BacMam vector | Addgene | Cat#:160,451 | Vector |
| Recombinant DNA reagent | pCMV-HA | Addgene | Cat#:631,604 | Vector |
| Recombinant DNA reagent | pcDNA3.1 | Addgene | Cat#:128,034 | Vector |
| Peptide, recombinant protein | Flag peptide | Genscript | DYKDDDDK | |
| Peptide, recombinant protein | NB88(camel nanobody) | This study | Isolated from phage display library of immunized cammel with hCaSR | |
| Peptide, recombinant protein | NB-2D11 (camel nanobody) | This study | Isolated from phage display library of immunized cammel with hCaSR | |
| Commercial assay or kit | Luciferase assay kit | Promega | E152A | For signaling assay |
| Commercial assay or kit | SuperSignal ELISA Femto Substrate | Thermo Scientific | Cat#: 37,075 | Protein Assays and Analysis |
| Commercial assay or kit | Fluo-4, AM, Cell Permeant | YEASEN | 40,704ES50 | |
| Chemical compound, drug | TNCA | aladdin | 42438-90-4 | |
| Chemical compound, drug | NPS-2143 | aladdin | 284035-33-2 | |
| Chemical compound, drug | cinacalcet | aladdin | 364782-34-3 | |
| Chemical compound, drug | Lauryl Maltose Neopentyl Glycol (LMNG) | Anatrace | NG310 | Membrane protein purification |
| Chemical compound, drug | Glyco-Diosgenin (GDN) | Anatrace | GDN101 | Membrane protein purification |
| Chemical compound, drug | Cholesterol Hemisuccinate tris Salt (CHS) | Anatrace | CH210 | Membrane protein purification |
| Chemical compound, drug | TMB substrate | Thermo Fisher Scientific | 34,021 | Protein Assays and Analysis |
| Software, algorithm | cryoSPARC | https://cryosparc.com | Version 3.0.0RRID:SCR_016501 | Cryo-EM data processing |
| Software, algorithm | PHENIX | http://www.phenix-online.org/ | Version 1.19.2RRID:SCR_014224 | Structure refinement |
| Software, algorithm | Coot | Coot (cam.ac.uk) | Version 0.9.4 RRID:SCR_014222 | Structure refinement |
| Software, algorithm | MolProbity | DOI:10.1107/S0907444909042073 | RRID:SCR_014226 | Structure verification |
| Software, algorithm | UCSF Chimera | https://wwwcgl.ucsf.edu/chimera/ (PMID:15264254) | Version 1.15 RRID:SCR_004097 | Initial homology model docking |
| Software, algorithm | PyMol | Schrodinger | Version 2.5 RRID:SCR_000305 | Structural visualization/figure preparation |
| Software, algorithm | GraphPad Prism 7 | GraphPad | RRID:SCR_002798 | Analysis of signaling data |
| Other | Lipofectamine 2000 | Invitrogen | 11668030 | Transfection reagent for signaling assay |