Ferroptotic stress promotes the accumulation of pro-inflammatory proximal tubular cells in maladaptive renal repair
Figures
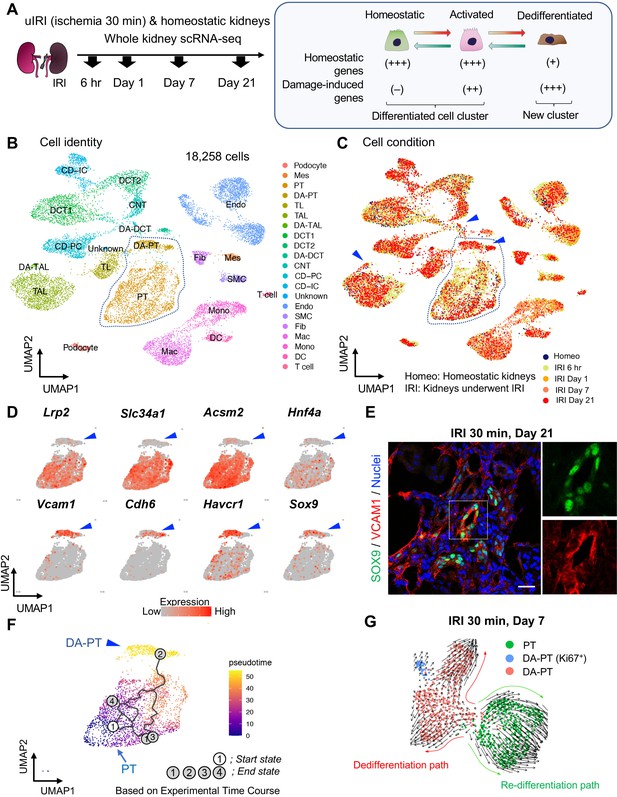
Single-cell RNA sequencing (scRNA-seq) identifies dynamic cellular state transitions of tubular epithelial cells after severe IRI.
(A) Drop-seq strategy. uIRI, unilateral IRI. A schematic illustration of epithelial cell states is shown. (B) and (C) Integrated single-cell transcriptome map. Unsupervised clustering identified 21 distinct clusters in the UMAP plot. Arrowheads indicate damage-associated tubular epithelial cells. The dotted area (PT cell clusters; PT and DA-PT) was used for the downstream analyses in (D)–(G). (D) UMAP plots showing the expression of indicated genes in PT cell clusters (PT and DA-PT in (B)). Differentiated/mature PT cell markers: Lrp2 (megalin), Slc34a1 (sodium-dependent phosphate transporter 2a, NaPi2a), Acsm2 (acyl-coenzyme A synthetase), and Hnf4a (hepatocyte nuclear factor 4α); and damage-induced genes: Vcam1 (vascular adhesion molecule 1), Cdh6 (cadherin 6), Havcr1 (kidney injury molecule-1, KIM1), Sox9 (Sry-box 9). Arrowheads; DA-PT. (E) Immunostaining for SOX9 and VCAM1 using post-severe IRI kidneys on day 21. Scale bar: 20 μm. (F) Pseudotime trajectory analysis of proximal tubular cells (PT and DA-PT clusters) that underwent IRI. A region occupied with cells from 6 hr after post-IRI was set as a starting state. (G) RNA velocity analysis of PT clusters (PT and DA-PT) from post-IRI kidneys on day 7. Cells in PT clusters from IRI day 7 dataset was extracted for the analysis. The arrows indicate predicted lineage trajectories. PT, proximal tubule; DA-PT, damage-associated PT; TL, thin limb; TAL, thick ascending limb; DA-TAL, damage-associated TAL; DCT, distal convoluted tubule; DA-DCT, damage-associated DCT; CNT, connecting tubule; CD, collecting duct (P, principal cells, IC, intercalated cells); Mes, mesangial cells; Endo, endothelial cells; SMC, smooth muscle cells; Fib, fibroblasts; Mac, macrophages; Mono, monocytes; DC, dendritic cells.

Characterization of severe and mild unilateral IRI models.
(A) Experimental workflow for the mild and severe IRI models. Left kidneys from wild-type C57BL/6J mice were subjected to mild (20 min) and severe (30 min) ischemia. Contralateral kidneys (CLK) were used as controls. uIRI, unilateral IRI. Kidneys were harvested on day 21 post-IRI. (B) Severe IRI results in renal atrophy. Relative size of post-IRI kidney compared to contralateral kidney (CLK) was quantified. N = 4–5. (C) Hematoxylin-eosin staining of IRI kidneys on day 21 (D21). Note that severe-IRI resulted in tubular dilatation, flattening of tubular epithelial cells, cast formation, and inflammatory cell infiltration. (D) Immunofluorescence for KIM1, NGAL, F4/80, and αSMA. Severe IRI resulted in persistent expression of proximal and distal tubular injury markers. Kidney injury molecule 1, KIM1 (encoded by Havcr1) is a marker for proximal tubular injury. Neutrophil gelatinase-associated lipocalin, NGAL (encoded by Lcn2) is a marker for distal tubular injury. Note that post-severe IRI kidneys exhibit accumulation of F4/80+ cells (mostly macrophages) and αSMA+ myofibroblasts in renal interstitium, both are classical features of failed renal repair. (E) Real-time PCR analyses of indicated gene expression. Acta2 gene encodes αSMA. N = 4–5. Scale bars: 100 μm in stitched images of (C); 20 μm in higher magnifications of (C); 50 μm in (D). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Unpaired Student’s t-test for (B) and one-way ANOVA with post hoc multiple comparisons test for (E).
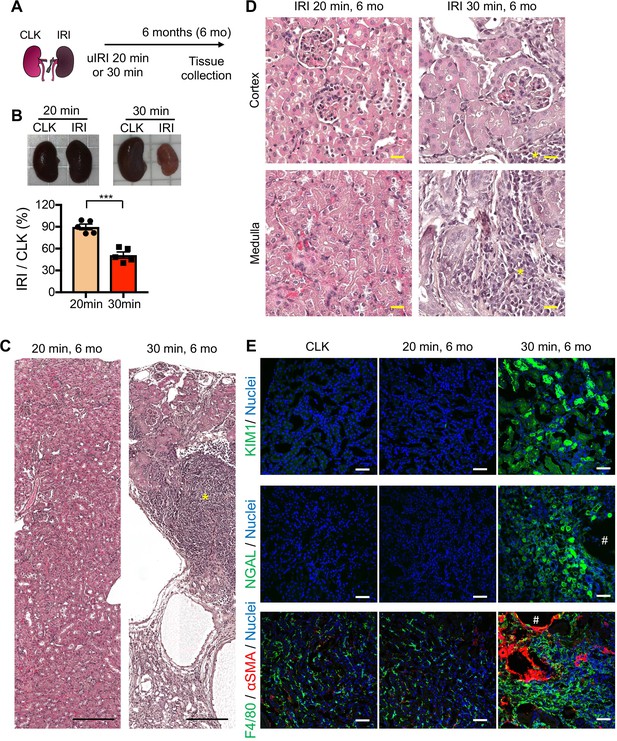
Severe IRI leads to cystic and atrophic kidneys 6 months after severe IRI.
(A) Experimental workflow for mild and severe IRI models with long-term observation. Kidneys were harvested on 6 months post-IRI. (B) Severe IRI (30 min ischemia) results in marked renal atrophy on 6 months post-IRI. Relative size of post-IRI kidney compared to contralateral kidney (CLK) was quantified. N = 5. Unpaired Student’s t-test. (C) and (D) Hematoxylin-eosin staining of IRI kidneys. Note that severe-IRI resulted in flattened and necrotic tubular epithelial cells with massive infiltration of inflammatory cells, which occupied the renal parenchyma. * indicates clusters of inflammatory cells, occupying renal parenchyma. (E) Immunofluorescence for KIM1, NGAL, F4/80, and αSMA. Severe IRI resulted in persistent expression of proximal and distal tubular injury markers (KIM1 and NGAL, respectively). Note that post-severe IRI kidneys exhibit accumulation of F4/80+ myeloid cells (mostly macrophages) and αSMA+ myofibroblasts in renal interstitium. Small cystic structures (#) were encircled by myofibroblasts. Scale bars: 20 μm in (C), 200 μm in (D) and 50 μm in (E).
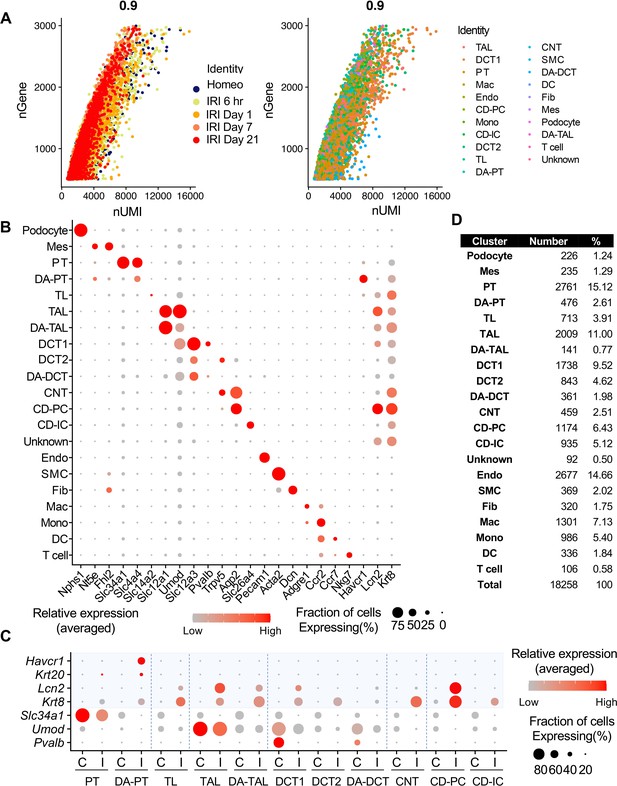
scRNA-seq identifies major cell types in homeostatic and post-IRI kidneys.
(A) Pearson correlation plot showing the linear relationship between the number of genes (nGene) and unique molecular identifiers (nUMI). Experimental conditions and cell types are color-coded. (B) Dot plot shows the gene expression patterns of cluster-enriched canonical markers. Note that damage-associated tubular epithelial cells (DA-PT, DA-TAL, and DA-DCT) have reduced expression of canonical cellular markers. (C) Tubular injury marker gene expressions are selectively observed in damaged kidneys (IRI) but not in homeostatic control kidneys. ‘C’ indicates cells from the control homeostatic kidneys. ‘I’ indicates cells from IRI kidneys from all time points. Proximal tubule-specific injury markers (Havcr1, Krt20), distal tubule-specific injury markers (Lcn2), and pan-tubular injury marker (Krt8) are shown. Both homeostatic and activated cells show high canonical cell type marker gene expressions, such as Slc34a1 in PT, but damage-associated clusters (DA-PT, DA-TAL, DA-DCT) show reduced homeostatic gene expressions and increased damage-induced gene expressions (See Figure 1A). (D) Our single-cell preparation resulted in high yields of podocytes (1.24%) and endothelial cells (14.66%). Abbrev: PT, proximal tubule; TL, thin limb; TAL, thick ascending limb; DCT, distal convoluted tubule (DCT1 and DCT2); CNT, connecting tubule; CD, collecting duct (P, principal cells, IC, intercalated cells); Mes, mesangial cells; Endo, endothelial cells; SMC, smooth muscle cells; Fib, fibroblasts; Mac, macrophages; Mono, monocytes; DC, dendritic cells; DA-PT, damage-associated PT; DA-TAL, damage-associated TAL; DA-DCT, damage-associated DCT.
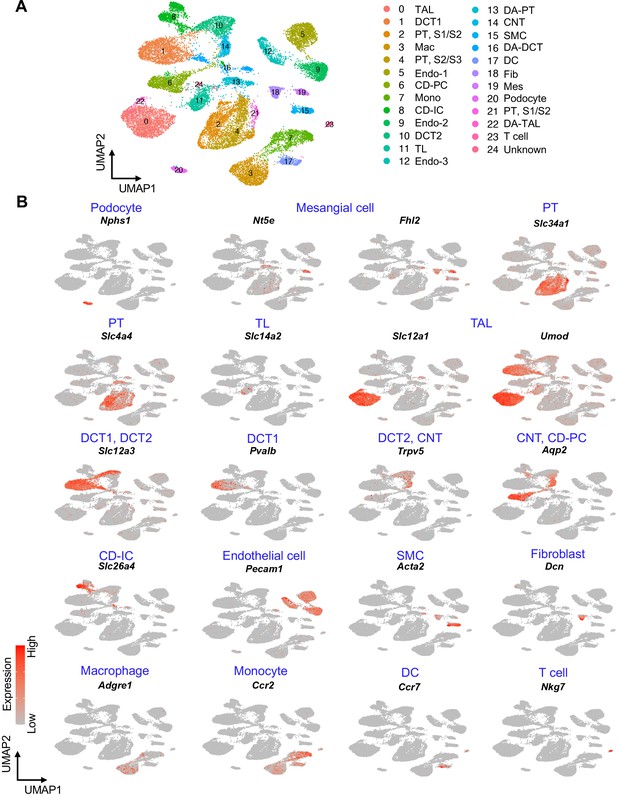
UMAP plots show the expression pattern of anchor genes in homeostatic and post-IRI kidneys.
(A) UMAP plots show the identified cell clusters (resolution was set as 1.0). (B) UMAP plots show the expression pattern of indicated canonical marker genes (anchor genes) of each cluster. We manually combined three clusters of differentiated/mature proximal tubular cells (PT, S1/S2 and PT, S2/S3) into one cluster (PT) to generate a more coarse-grained cell-type annotation and data visualization in other figures. We also combined three clusters of endothelial cells (Endo-1, Endo-2, and Endo-3) into one cluster (Endo) for data visualization in other figures. S1, S1 segment; S2, S2 segment 2; S3, S3 segment of proximal tubule.
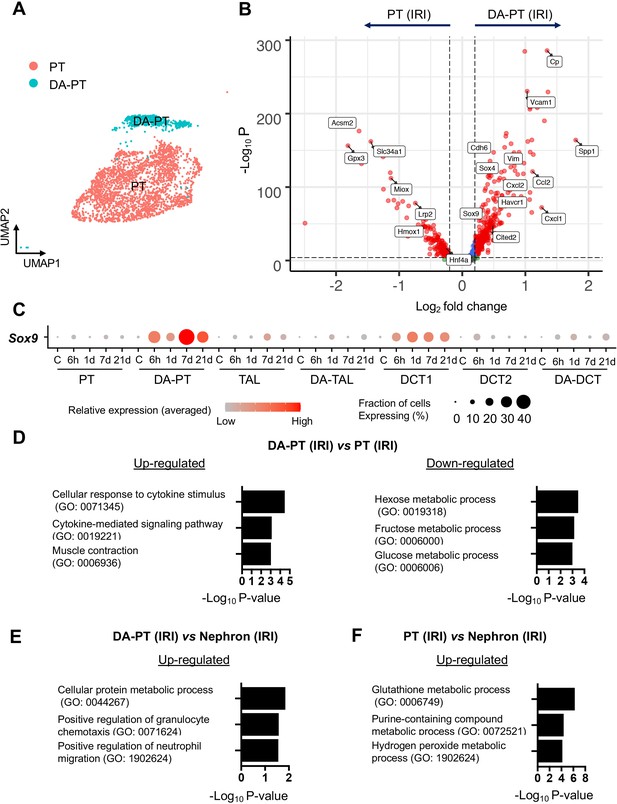
Damage-associated PT cells show an inflammatory transcriptional signature.
(A) UMAP of PT clusters (PT, differentiated proximal tubular cell cluster; DA-PT, damage-associated proximal tubular cell cluster). See Figure 1B. (B) Volcano plot showing a distinct transcriptional profile of damage-associated PT cells (PT cells in DA-PT cluster). A Wilcoxon rank-sum test was used for the statistical analysis comparing cells in PT cluster from IRI kidneys and cells in DA-PT cluster from IRI kidneys. Blue and gray data points indicate transcripts that fall below the set threshold for fold change (Log2 fold change, a threshold value of 0.25) and p value (a cutoff value of 10e-5). Note that cells in the DA-PT cluster show reduced expression of homeostatic marker genes (Acsm2, Slc34a1, and Lrp2), oxidative stress defense genes (Hmox, Miox, and Gpx3), and a gene encodes transcription factor for PT maturation in renal development (Hnf4a). The cells in the DA-PT cluster also show enrichment of developmental genes (Sox4, Sox9, Cited2, and Cdh6), damage-induced genes (Havcr1 and Vcam1) and inflammatory cytokines and chemokines (Spp1, Ccl2, Cxcl1, and Cxcl2). (C) Dot plots show the expression of Sox9 gene is highly enriched in damage-associated PT (DA-PT) cell population. (D) and (E) Gene ontology enrichment analyses identify that DA-PT cluster is enriched for immune response gene ontology. The cells in the DA-PT cluster from IRI kidneys (all time points) were compared with the cells in the PT cluster or cells in all other nephron segments and collecting duct (IRI kidneys, all time points) in (D) and (E), respectively. (F) Gene Ontology enrichment analyses identify the PT cluster is enriched for glutathione-mediated anti-oxidative stress responses. The cells in the PT cluster from IRI kidneys (all time points) were compared with the cells from all other nephron segments and collecting duct from IRI kidneys (all time points).

Severe IRI reduces expressions of proximal tubular differentiation markers.
(A) UMAP plots showing the expression of indicated genes. Note that damage-associated PT cell state (cells in DA-PT cluster) are enriched with damage-induced genes (Havcr1, Krt20, Vcam1, and Vim) and exhibit less differentiated signature (upregulation of Cdh6 and downregulation of Hnf4a). Arrow, PT cells; arrowheads; DA-PT cells. (B) and (C) Comparative analyses of kidneys underwent severe IRI (ischemic time 30 min) and mild IRI (ischemic time 20 min). Kidneys were harvested on day 21 post-IRI. Severe IRI reduces LTLhigh proximal tubular cells. Lotus tetragonolobus lectin (LTL) binds fully differentiated proximal tubular cells. LTLhigh areas from kidneys on day 21 were quantified. Wild-type C57BL/6J mice were used. Scale bars: 50 μm. N = 4–7. (D) Real-time PCR analyses of Slc34a1. mRNA expression of Slc34a1 was reduced after severe IRI (ischemic time 30 min on day 21). Whole kidney lysates from post-IRI kidneys on day 21 were used. Contralateral kidneys (CLK) were used as controls. N = 4–5. **p < 0.01; ****p < 0.0001, one-way ANOVA with post hoc multiple comparisons test.

Comparative analyses of damage-associated PT cells and neonatal proximal tubular cells.
(A) and (B) Characterization of mouse neonatal kidney single-cell RNA-seq data. UMAP plots show mouse neonatal kidney cells from GSM2473317 (4693 cells; post-natal day 1) in (A). Dot plots show the gene expression patterns of cluster-enriched canonical markers in (B). (C) UMAP rendering of Top 100 genes characterizing mature and immature early proximal tubular (PT) cells. We obtained the top 100 genes representing mature PT and immature early PT cells by performing differential gene expression analysis using the ‘FindMarkers’ command in Seurat. As expected, the mature PT Top 100 genes are enriched in mature PT cluster. The early PT Top 100 genes are enriched in immature early PT and nephron progenitor clusters. (D) UMAP rendering of mature and early PT genes on adult PT clusters from our dataset (PT and DA-PT). Note that the early PT Top 100 genes are highly enriched in damage-associated PT (DA-PT) population, indicating they are in a less differentiated state. (E) UMAP plots showing gene expressions of early PT genes (Cd24a, Jag1, and Aldh1a2) are enriched in DA-PT cluster.

Trajectory analyses predict lineage hierarchy from differentiated mature PT cells to damage-associated PT cells.
(A) UMAP plots showing mature and damage-associated PT cells (PT and DA-PT clusters) underwent IRI. (B) Pseudotime trajectory analysis using Monocle 3. A region occupied with Slc34a1high cells was set as a starting state. Note that predicted trajectory starting from PT to DA-PT state. See Figure 1F for the analysis with a different starting ‘root’ setting. The earliest time point of injury was used as a starting root in Figure 1F. Both analyses resulted in a similar predicted trajectory. (C) UMAP plots showing proximal tubular cells from PT and DA-PT clusters on IRI day 7 (D7) . Cells from post-IRI kidneys on day seven are shown and used for RNA velocity analysis to investigate potential cellular plasticity at this stage (single data point). See Figure 1G for RNA velocities (trajectory). UMAP plots in the right panels show the indicated genes; homeostatic genes (Lrp2, Acsm2, and Slc34a1), damage-induced genes (Vcam1, Cdh6, and Sox9), and cellular proliferation (Mki67 and Top2a). Fig. S8A and S8B are supporting data for Figure 1F. Fig. S8C is supporting data for Figure 1G.
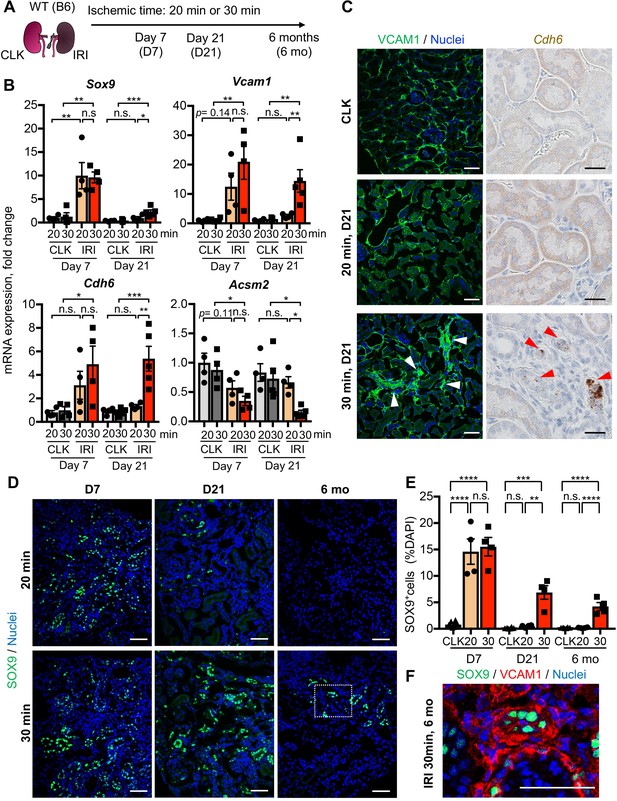
Damage-associated PT cells emerge transiently after mild injury but persist after severe injury.
(A) Experimental workflow for the mild and severe IRI models. Left kidneys from wild-type (WT) C57BL/6J (B6) mice were subjected to mild (20 min) and severe (30 min) ischemia (unilateral IRI, uIRI). Contralateral kidneys (CLK) were used as controls. (B) Real-time PCR analyses of indicated gene expression. Whole kidney lysates were used. N = 4–5. (C) Expression analyses of VCAM1 and Cdh6 using post-IRI kidneys on day 21. Immunostaining for VCAM1 revealed clusters of VCAM1high tubular epithelial cells. In situ hybridization (ISH) was used to detect Cdh6 gene expression on kidney sections. (D) Immunostaining for SOX9 in mild (20 min) and severe (30 min) IRI kidneys collected at indicated time points (day 7, day 21, and 6 months after IRI). (E) Quantification of SOX9+ cells over the DAPI+ area. Note that SOX9+ cells persist after severe IRI up to 6 months after IRI (30 for 30 min ischemia). In contrast, they disappear after a transient appearance in post-mild IRI kidneys (20 for 20 min ischemia). N = 4–8. (F) Immunostaining for SOX9 and VCAM1 (6 months post-severe IRI kidneys, dotted area in D). Scale bars, 20 μm in (C, Cdh6), and 50 μm (C, VCAM1, D and F). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, one-way ANOVA with post hoc multiple comparisons test. n.s., not significant.
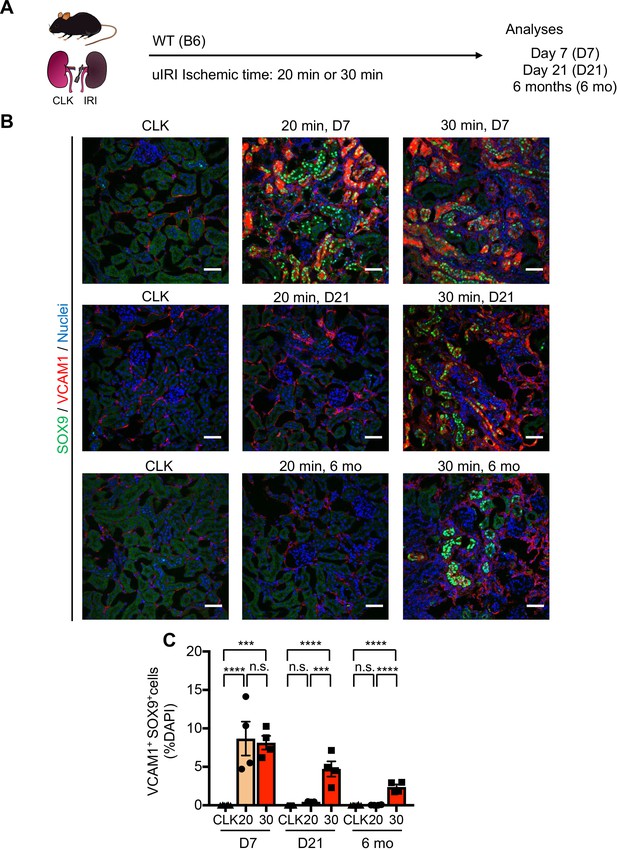
Comparative analyses of mild and severe IRI identify distinct temporal dynamics of damage-associated PT cells.
(A) Experimental workflow. Wild type C57BL/6J mice were used. Mild IRI (20 min ischemia) and severe IRI (30 min ischemia). Kidneys were harvested on day 7, day 21 and 6 months post-IRI. (B) Immunostaining for SOX9 and VCAM1. Note that double-positive cells (indicative for damage-associated PT cell state) emerge after mild and severe IRI similarly on day 7, but they are differentially regulated subsequently. SOX9+VCAM1+ cells disappear after mild IRI (20 min ischemia) on day 21, but they persist at least for 6 months after severe IRI (30 min ischemia). Scale bars: 50 μm. * indicates cystic lesions. (C) Quantification of double-positive cells over DAPI+ area in (B). N = 3–8. ***p < 0.001, ****p < 0.0001, one-way ANOVA with post hoc multiple comparisons test. n.s., not significant.

Lineage-tracing identifies the cellular plasticity of damage-associated PT cells.
(A) Schematic of fate-mapping strategy using Sox9IRES-CreERT2; Rosa26tdTomato mice. Tamoxifen was administered three times on alternate days. Contralateral kidneys (CLK) were used as controls. (B) Distribution of tdTomato-expressing cells (Sox9-lineage cells) in contralateral (CLK), mild (20 min) and severe (30 min) IRI kidneys on day 21 (D21). (C) Quantification of tdTomato+ area relative to DAPI+ area in (B). DAPI was used for nuclear staining. N = 4–5. (D) Immunostaining for VCAM1 in Sox9-lineage-tagged kidneys (post-IRI, day 21). Sox9-lineage cells express native tdTomato red fluorescence (TdT). Insets: individual fluorescence channels. (E) Quantification of double-positive cells in total tdTomato+ cells in (D). N = 3–4. Note that more Sox9-lineage cells express VCAM1 after severe IRI (30 min) compared to mild IRI (20 min) on day 21. One-way ANOVA with post hoc multiple comparisons test and unpaired Student’s t-test were used for (C) and (E), respectively. Scale bars, 200 μm in (B); and 50 μm in (D). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. (F) Schematic illustration of PT cell state dynamics. Differentiated/mature PT cells are activated, transit into a molecularly distinct PT cell state (damage-associated PT cells in DA-PT cluster), and redifferentiate into their original state after mild injury (left). Severe injury prevents the redifferentiation of damage-associated PT cells into normal PT cell state, leading to the accumulation and persistence of damage-associated PT cells (right).

Damage-associated PT cells create a proinflammatory milieu with myeloid cells.
(A) Schematic model of intercellular communications between damage-associated PT cells and macrophages. NicheNet was used to predict intercellular interactions using our integrated single-cell map of failed renal repair. (B) Predicted ligands from damage-associated PT cells and receptors in macrophages. (C) Schematic model of intercellular communications between damage-associated PT cells and monocytes. (D) Predicted ligands from damage-associated PT cells and receptors in monocytes. (E) UMAP plots showing the expression of indicated genes. Our integrated single-cell map of mouse failed renal repair is shown (See Figure 1, B and C). Arrowheads indicate damage-associated PT cells (DA-PT cluster). Arrows indicate differentiated PT cells (PT cluster). (F) Real-time PCR analyses of indicated gene expression. Post-IRI kidneys on day 21 that underwent mild (20 min) or severe (30 min) ischemia were used. N = 4–5. *p < 0.05; **p < 0.01; ***p < 0.001, one-way ANOVA with post hoc multiple comparisons test.
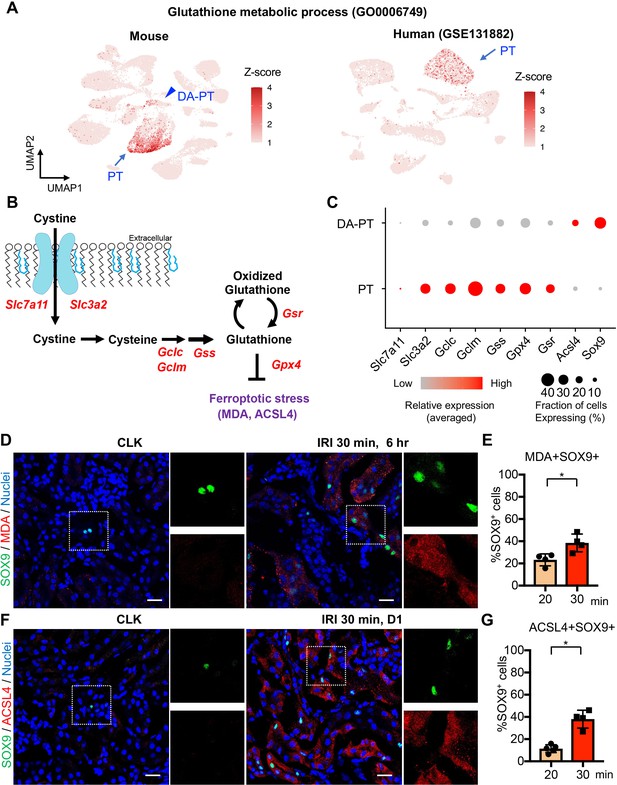
Damage-associated PT cells undergo high ferroptotic stress after severe IRI.
(A) UMAP rendering of glutathione metabolic process in mouse and human kidneys. (B) A scheme showing glutathione-glutathione peroxidase 4 (GPX4) anti-ferroptotic defense pathway. Slc7a11 and Slc3a2 (system xc-); Gclc and Gclm (glutamate-cysteine ligase); Gss (glutathione synthetase); Gsr (glutathione reductase): and Gpx4. MDA (malondialdehyde, a lipid peroxidation product) and ACSL4 (acyl-CoA synthetase long-chain family member 4) are markers for ferroptotic stress. (C) Dot plots show the expression of genes for glutathione-GPX4 axis, Sox9, and Acsl4. (D) Immunostaining for SOX9 and MDA (6 hr post-IRI), and (E) quantification of double-positive cells in total SOX9+ cells. N = 4. (F) Immunostaining for SOX9 and ACSL4 (1 day post-IRI), and (G) quantification of double-positive cells in total SOX9+ cells. N = 4. Insets: individual fluorescence channels of the dotted box area. Note that severe ischemia (30 min) induces more ferroptotic stress markers (MDA and ACSL4) in SOX9+ cells in damaged kidneys than mild ischemia (20 min). Wild-type C57BL/6J mice were used for (D) to (G). Scale bars, 20 μm in (D) and (F). *p < 0.05. unpaired Student’s t-test.
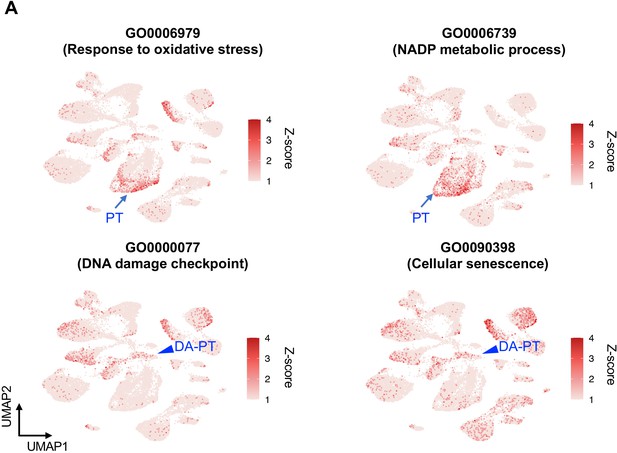
Gene ontology analyses identify enrichment of anti-oxidative stress defense genes in differentiated/mature PT cells.
UMAP rendering of signaling pathways. Upper panels show the pathways enriched in differentiated PT cells (PT cluster). Lower panels show the pathways enriched in damage-associated PT cells (DA-PT cluster). Arrows indicate differentiated PT cell cluster (PT). Arrowheads indicate DA-PT cell cluster.

Characterization of human normal kidney single-nucleus RNA-seq data.
(A) UMAP plots showing human normal kidney cells from GSE131882 (7,631 cells). Two normal kidney datasets were integrated and analyzed. (B) Dot plot showing the gene expression patterns of cluster-enriched canonical markers. (C) UMAP rendering of signaling pathways. Note that the signaling pathways for anti-oxidative stress, which are enriched in differentiated PT cells in mouse kidneys, are also enriched in normal differentiated PT cells in humans. Blue arrowheads: PT cells, green arrowheads, VCAM1+ PT cells.
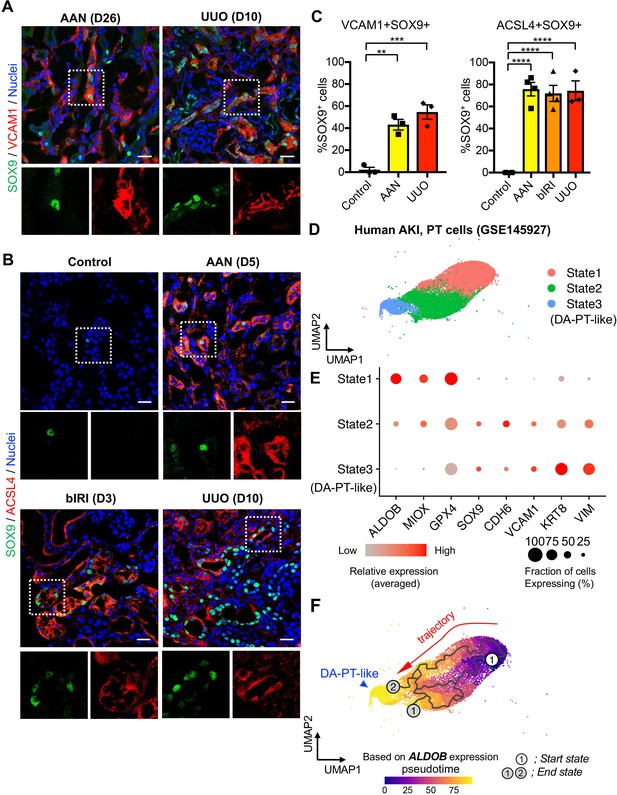
Damage-associated PT cells emerge after injury in mouse and human kidneys.
(A) Immunostaining for SOX9 and VCAM1. Aristolochic acid nephropathy (AAN) and unilateral ureteral obstruction (UUO) models were used. Kidneys from wild-type C57BL/6J mice were harvested on day 26 (D26) for AAN and day 10 (D10) for UUO. Insets: individual fluorescence channels of the dotted box area. (B) Immunostaining for SOX9 and ACSL4. bIRI, bilateral IRI model. Kidneys were harvested on day 3 (D3) for bIRI, day 5 (D5) for AAN, and day 10 (D10) for UUO. Insets: individual fluorescence channels of the dotted box area. (C) Quantification of double-positive cells in total SOX9+ cells from panel (A) and (B). Scale bars, 20 μm. N = 3–4. **p < 0.01; ***p < 0.001; ****p < 0.0001, one-way ANOVA with post hoc multiple comparisons test. (D) UMAP of the human proximal tubular cells from AKI kidneys. (E) Dot plots showing the expression of indicated genes. Note that PT cells in state 3 (DA-PT-like) show increased gene expressions of markers for mouse damage-associated PT cells (SOX9, VCAM1, CDH6) and reduced expression of homeostatic genes (ALDOB, MIOX, and GPX4). (F) Pseudotime trajectory analysis of PT clusters (PT and DA-PT-like cells). A region occupied with ALDOBhigh cells were set as a starting state. Arrow, predicted trajectory from PT cells to DA-PT-like cells.
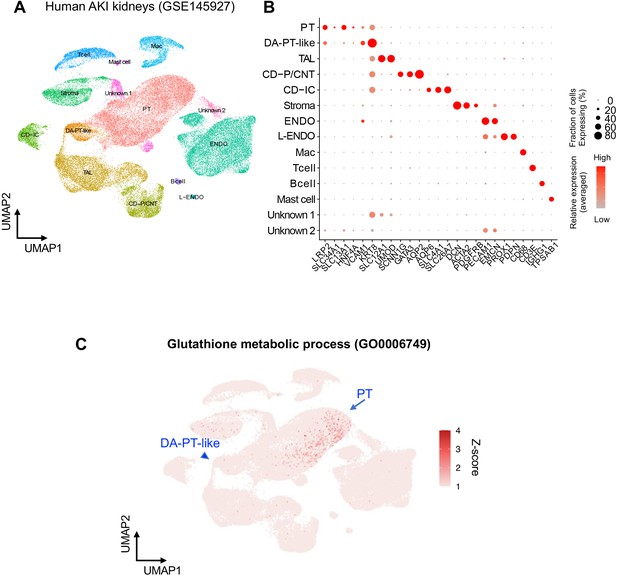
Characterization of human AKI kidney single-cell RNA-seq data.
(A) UMAP plots showing human AKI kidney cells from GSE145927 (43,998 cells). Data from the two non-rejecting AKI tissues were analyzed and integrated. We manually combined two clusters of differentiated proximal tubular cells (PT, state one and PT, state 2) into one cluster (PT) to generate a more coarse-grained cell-type annotation and data visualization. (B) Dot plot showing the gene expression patterns of cluster-enriched canonical markers. Note that DA-PT-like cells (PT, state 3) exhibit reduced expression of homeostatic genes (ex. LRP2, SLC34A1) but are enriched for damage-induced genes (ex. VCAM1, KRT8) as in the case of mouse damage-associated PT (DA-PT) cells. (C) UMAP rendering of signaling pathways. Note that genes for the glutathione-metabolic process are highly expressed in PT and gradually reduced towards DA-PT-like cells as in the predicted trajectory in Figure 6F.

Genetic induction of ferroptotic stress to Sox9-lineage cells augments kidney injury.
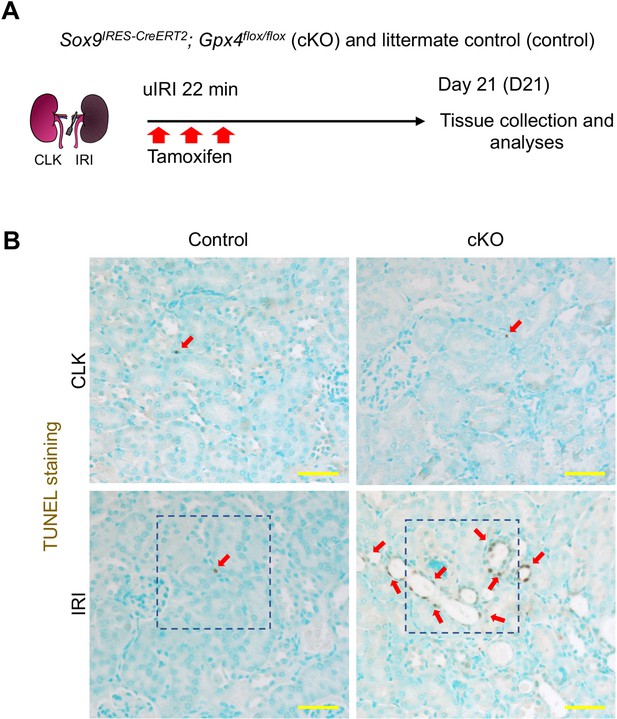
(A) Experimental workflow for Gpx4 deletion in Sox9-lineage cells. uIRI, unilateral IRI (ischemic time 22 min). Kidneys were harvested on day 21 post-IRI. cKO mice and their littermate controls were subjected to the same ischemic stress and tamoxifen treatment. Gpx4 is deleted in Sox9-lineage cells after IRI with tamoxifen administration. (B) The deletion of Gpx4 results in renal atrophy. Relative size of post-IRI kidneys compared to contralateral kidneys (CLK) was quantified. Control, littermate control. N = 7. (C) and (D) Immunostaining for tubular injury markers (KIM1 and KRT8). IRI kidneys from cKO and control littermates are shown. CLK did not show KIM1 or KRT8 staining. Quantification of KIM1 or KRT8-positive area over the DAPI+ area is shown in (D). N = 5–7. (E) and (F) Immunostaining for F4/80 and αSMA. IRI kidneys from cKO and control littermates are shown. Quantification of F4/80 or αSMA-positive area over the DAPI+ area is shown in (F). N = 5–7. Insets: individual fluorescence channels of the dotted box area. (G) and (H) TUNEL staining for evaluating cell death. Quantification of TUNEL-positive nuclei is shown in (H). N = 4. Arrowheads, TUNEL+ nuclei. Abbrev: hpf, high power field. Unpaired t-test for (D) and (F). One-way ANOVA with post hoc multiple comparison test for (H). Scale bars, 100 μm in (C) and (E); 20 μm in (G).

Genetic deletion of Gpx4 leads to augmented ferroptotic stress after mild IRI.
(A) Experimental workflow for genetic deletion of Gpx4 in Sox9-lineage cells. cKO mice and their littermate controls were subjected to the same ischemic stress (ischemic time, 22 min) and tamoxifen treatment. Kidneys were harvested on day 21 post-IRI. Note that Sox9 is only induced in proximal tubular cells after IRI (Sox9 gene is silenced in this tubular segment after the completion of renal development); therefore, Gpx4 deletion occurs after injury in IRI kidneys with tamoxifen injection. The initial dose of tamoxifen was administered immediately before the surgical intervention. Left kidneys were subjected to mild (22 min) ischemia. Only littermate controls were used for phenotypic comparisons. (B) and (C) Immunostaining for GPX4 and quantification. GPX4 immunostaining confirms the deletion of GPX4 in IRI-kidneys from cKO mice (see IRI, cKO). Inlets show the higher magnification and single-color images of dotted areas. N = 5–7. (D) and (E) Immunostaining for ACSL4 and quantification. Inlets show the higher magnification and single-color images of dotted areas. N = 3–5. **p < 0.01, ***p < 0.001, ****p < 0.0001, one-way ANOVA with post hoc multiple comparisons test. Scale bars: 50 μm in (B) and (D).

Genetic deletion of Gpx4 leads to severe kidney injury after mild IRI.
(A) Experimental workflow for genetic deletion of Gpx4 in Sox9-lineage cells. cKO mice and their littermate controls were subjected to the same ischemic stress (ischemic time, 22 min) and tamoxifen treatment. Kidneys were harvested on day 21 post-IRI. (B) Hematoxylin-eosin staining of IRI kidneys. Control; littermate controls. Representative images from IRI kidneys are shown. (C) Higher magnification images. Note that Gpx4 deletion after mild IRI resulted in severe tubular dilatation, flattening of tubular epithelial cells, cast formation, and inflammatory cell infiltration. (D) Quantification of kidney injury score. N = 4–5. ****p < 0.0001, one-way ANOVA with post hoc multiple comparisons test. Scale bars: 50 μm in (B); 20 μm in (C).

Genetic deletion of Gpx4 leads to the accumulation of damage-associated PT cells.
(A) and (B) Immunostaining for KIM1, KRT8, F4/80, and αSMA. Post-IRI kidneys on day 21 were analyzed. Note that Gpx4 deletion increased tubular injury markers (KIM1 and KRT8) and led to the accumulation of myeloid cells (F4/80) and myofibroblasts (αSMA) in post-IRI kidneys from cKO mice on day 21 (ischemic time, 22 min). (C) Real-time PCR analyses of indicated genes. N = 6–7. *p < 0.05; **p < 0.01, one-way ANOVA with post-hoc multiple comparisons test. (D) Immunostaining for VCAM1, EMCN (endomucin), and F4/80 using post-IRI kidney on day 21. Note that VCAM1+EMCN–F4/80– cells (damage-associated PT cell state) accumulated in Gpx4-deleted kidneys after mild IRI on day 21. (E) In situ hybridization for Cdh6 expression using post-IRI kidney on day 21. Cdh6 is induced in tubular epithelial cells in Gpx4-deleted kidneys after mild IRI on day 21. Arrowheads: Cdh6+ tubular epithelial cells. These data indicate that damage-associated PT (DA-PT) cells accumulate even after mild IRI when the Gpx4 gene is deleted. Note that littermate control kidneys underwent the same ischemic time (22 min) recovered without accumulating myofibroblasts, macrophages, and damage-associated PT cells, indicative of successful recovery. Gpx4-deleted kidneys underwent mild IRI (22 min ischemia) mimicked the failed renal repair phenotype observed after severe ischemic injury. Scale bars: 50 μm in (A), (B), and (D) and 20 μm in (E).
Genetic deletion of Gpx4 leads to cell death of tubular epithelial cells.
(A) Experimental workflow for genetic deletion of Gpx4 in Sox9-lineage cells. cKO mice and their littermate controls were subjected to the same ischemic stress (ischemic time, 22 min) and tamoxifen treatment. Kidneys were harvested on day 21 post-IRI. (B) TUNEL staining for evaluating cell death. The dotted areas are shown in Figure 7G. See quantification for Figure 7H. Scale bar, 50 μm. N=4. Arrows, TUNEL+ nuclei.

Genetic induction of ferroptotic stress induces the accumulation of damage-associated PT cells after mild injury.
(A) Schematic representation of experimental workflow. tdTomato-lineage tracing was employed to detect Sox9-lineage cells. cKO mice and their littermate controls were subjected to the same ischemic stress (ischemic time, 22 min) and tamoxifen treatment. Kidneys were harvested on day 21 post-IRI. (B) and (C) Immunostaining for VCAM1, EMCN (endomucin), and F4/80. IRI kidneys from cKO and control littermates (control) are shown. Quantification of VCAM1+EMCN–F4/80– area over the DAPI+ area is shown in (C). N = 6. (D) Immunostaining for VCAM1, ACSL4, and native tdTomato (TdT) fluorescence. Insets: individual fluorescence channels. (E) Real-time PCR analyses of indicated gene expression. Whole kidney lysates were used. N = 6–7. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, one-way ANOVA with post hoc multiple comparisons test for (C) and (E). (F) Immunostaining for SOX9 and VCAM1. Arrowheads indicate double-positive cells (damage-associated PT cells). (G) ISH for Cdh6 expression. Red arrowheads indicate Cdh6-positive renal tubular cells. Scale bars, 100 μm in (B); 20 μm in (D); 50 μm in (F); and 10 μm in (G). (H) Schematic illustration of PT cell state dynamics. Differentiated/mature PT cells are activated, transit into a damage-associated inflammatory PT cell state (DA-PT), and redifferentiate to their original state after mild injury. Ferroptotic stress prevents the redifferentiation of damage-associated PT cells into normal PT cell state, leading to the accumulation of the pathologic PT cells that actively produce inflammatory signals.
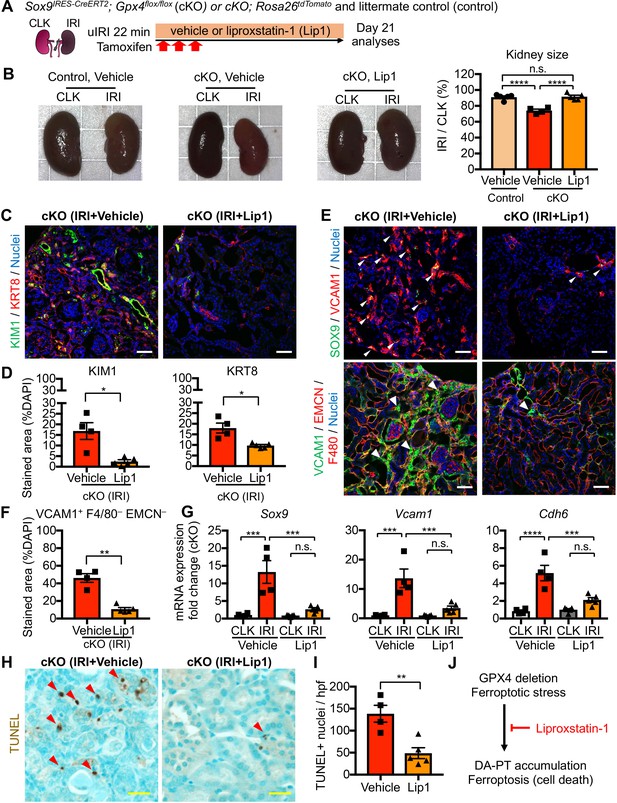
Pharmacological inhibition of ferroptotic stress blunts the accumulation of damage-associated PT cells and cell death.
(A) Schematic representation of experimental workflow. All mice (cKO and control littermates) were subjected to the same ischemic stress (ischemic time, 22 min, unilateral IRI) and tamoxifen treatment. The same volume of vehicle was administered to the control groups (control vehicle and cKO vehicle). Kidneys were harvested on day 21 post-IRI. (B) Liproxstatin-1 prevents renal atrophy. Relative size of post-IRI kidneys compared to contralateral kidneys (CLK) was quantified. Control, littermate control. N = 4–5. (C and D) Immunostaining for KIM1 and KRT8. IRI kidneys from cKO are shown. Quantification of immunostained area over the DAPI+ area is shown in (D). N = 4–5. (E and F) Immunostaining for SOX9 and VCAM1. Quantification of VCAM1+EMCN–F4/80– area over the DAPI+ area is shown in (F). Arrowheads indicate damage-associated PT cells. (G) Real-time PCR analyses of indicated gene expression. Whole kidney lysates were used. N = 4–5. (H) and (I) TUNEL staining for evaluating cell death. Quantification of TUNEL-positive nuclei is shown in (I). N = 4–5. Red arrowheads indicate TUNEL+ tubular epithelial cells. Scale bars, 50 μm in (C) and (E); and 20 μm in (H). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001, unpaired t-test for (D), (F). and (I); One-way ANOVA with post hoc multiple comparisons test for (G). (J) Liproxstatin-1 improves renal repair after IRI.

Liproxstatin-1 potently reduced ferroptotic stress in the absence of GPX4.
(A) Experimental workflow for liproxstatin-1 (Lip-1) treatment to Gpx4 cKO mice. cKO mice and their littermate controls were subjected to the same ischemic stress (ischemic time, 22 min) and tamoxifen treatment. The same volume of vehicle was administered to cKO mice and littermate Cre-negative mice. Kidneys were harvested on day 21 post-IRI. (B) Distribution of tdTomato-expressing cells (Sox9-lineage cells) in contralateral (CLK) and IRI kidneys on day 21. Note that liproxstatin-1-treated kidneys and vehicle-treated kidneys show comparable targeting (n=three for vehicle, and n=two for liproxstatin-1). (C) Immunostaining for GPX4 and quantification. GPX4 immunostaining confirms the deletion of GPX4 in IRI-kidneys from cKO mice both vehicle and liproxstatin-1-treated group. N = 4–5. (D) Immunostaining for ACSL4 and quantification using kidneys from cKO mice (day 21 post-IRI). Liproxstatin-1 treatment reduced the expression of ACSL4 in IRI kidneys in the absence of GPX4. p<0.05, unpaired t-test. Scale bars, 100 μm in (B) and 50 μm in (C) and (D).

Liproxstatin-1 potently mitigated ferroptotic stress-induced pathologic changes in the absence of GPX4.
(A) Experimental workflow for liproxstatin-1 (Lip-1) treatment to Gpx4 cKO mice. cKO mice and their littermate controls were subjected to the same ischemic stress (ischemic time, 22 min) and tamoxifen treatment. The same volume of vehicle was administered to cKO mice and littermate Cre-negative mice. Kidneys were harvested on day 21 post-IRI. IRI, ischemia-reperfusion-injured kidneys. CLK, contralateral kidneys. (B) and (C) Immunostaining for KIM1 and LTL in Sox9-lineage-tagged kidneys. Sox9-lineage cells express native tdTomato red fluorescence (TdT). Note that liproxstatin-1 treatment potently reduced the expression of KIM1 and restored the expression of LTL in tdTomato-positive cells in ischemia-reperfusion-injured cKO kidneys. Insets: individual fluorescence channels. Note that Inlets show the higher magnification and single-color images of dotted areas. (D) TUNEL staining for evaluating cell death. The dotted areas are shown in Figure 9H. See quantification for Figure 9I. N=4–5. Arrows, TUNEL+ nuclei. Scale bars, 50 μm in (B) – (D).
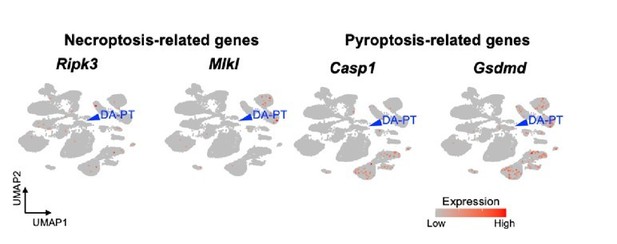
UMAP plots showing genes associated with necroptosis and pyroptosis.
Blue arrowheads indicate DA-PT cells. These genes are not highly expressed in the DA-PT cell population.
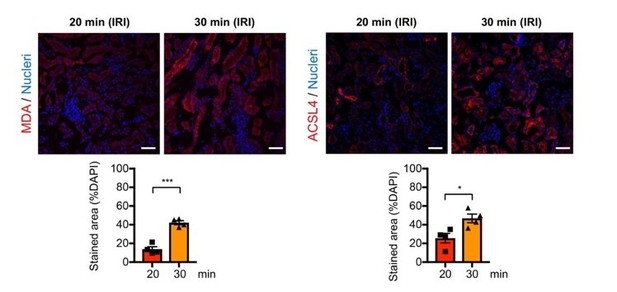
Immunostaining and quantification for malondialdehyde (MDA) and ACSL4.
The post-IRI kidneys harvested on 6-hrs and 1-day after ischemia were used to detect MDA and ACSL4, respectively. *P<0.05 and ***P<0.001 unpaired t-test. Scale bars, 50µm. Note tha 30-min ischemia induces more ferroptotic stress (MDA and ACSL4) in damaged kidneys compares to 20-min ischemia.
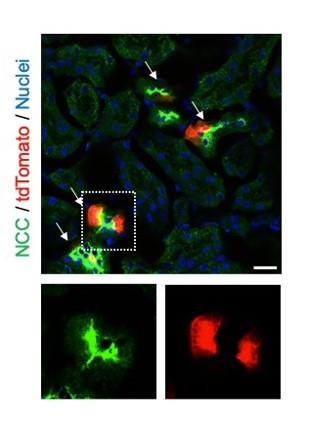
Distal convoluted tubules express SOX9 protein in uninjured kidneys.
Sox9-lineage tagged uninjured kidneys were used to localize Sox9-lineage cells (tdTomato). Sodium chloride co-transporter (NCC, Slc12a3) is a marker for distal convoluted tubular cells. Insets: Individual fluorescent channels of dotted area are shown. Scale bar: 20 µm. Arrows: double-positive cells.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | C57BL/6J | The Jackson laboratory | RRID:IMSR_JAX:020940 | |
| Genetic reagent (M. musculus) | Sox9IRESCreERT2 | The Jackson laboratory | RRID:MGI:4947114 | |
| Genetic reagent (M. musculus) | Rosa26tdTomato | The Jackson laboratory | RRID:IMSR_JAX:007914 | |
| Genetic reagent (M. musculus) | Gpx4flox | The Jackson laboratory | RRID:IMSR_JAX: 027964 | |
| Antibody | Anti-SOX9 (Rabbit monoclonal) | Abcam (ab196450) | RRID:AB_2665383 | Clone EPR14335 IF: 1:200 |
| Antibody | Anti-SOX9 (Rabbit monoclonal) | Abcam (ab185966) | RRID:AB_2728660 | Clone EPR14335-78 IF: 1:200 |
| Antibody | Anti-KIM1 (goat polyclonal) | R and D systems (AF1817) | RRID:AB_2116446 | IF: 1:400 |
| Antibody | Anti-NGAL (rat monoclonal) | Abcam (ab70287) | RRID:AB_2136473 | IF: 1:400 |
| Antibody | Anti-GPX4 (Rabbit monoclonal) | Abcam (ab125066) | RRID:AB_10973901 | Clone EPNCIR144 IF: 1:200 |
| Antibody | Anti-F4/80 (Rat monoclonal) | Bio-Rad (MCA497) | RRID:AB_2098196 | Clone C1:A3-1 IF: 1:200 |
| Antibody | Anti-Endomucin (Rat monoclonal) | Abcam (ab106100) | RRID:AB_10859306 | Clone V.7C7.1 IF: 1:200 |
| Antibody | Anti-KRT8 (Rat monoclonal) | DSHB (TROMA-I) | RRID:AB_531826 | IF: 1:200 |
| Antibody | Anti-αSMA (mouse monoclonal) | Sigma (C6198) | RRID:AB_476856 | Clone 1A4 IF: 1:200 |
| Reagent, commercial | LTL | Vector laboratories (B-1325 and FL-1321) | RRID:AB_2336558 | IF: 1:200 |
| Antibody | Anti-MDA (rabbit polyclonal) | Abcam (Ab6463) | RRID:AB_305484 | IF: 1:200 |
| Antibody | Anti-ACSL4 (rabbit monocolonal) | Abcam (Ab204380) (Ab155282) | RRID:AB_2714020 | Clone: EPR8640 IF: 1:200 |
| Antibody | Anti-VCAM1 (rabbit monocolonal) | CST 39036S 39301S | RRID:AB_2799146 | Clone: D8U5V IF: 1:100 |
| Commercial assay, kit | RNAScope probe-Mm-Cdh6 | Advance Cell Diagnosis (Cat. 519541) | ||
| Commercial assay, kit | RNAscope Intro Pack 2.5 HD Reagent Kit Brown Mm | Advance Cell Diagnosis (Cat. 322371) | ||
| software, algorithm | ImageJ | NIH, Bethesda, MD (Version 1.52P) | RRID:SCR_003070 | https://imagej.nih.gov/ij/ |
| Software, algorithm | GraphPad Prism | RRID:SCR_002798 | https://www.graphpad.com/scientific-software/prism/ | |
| Software, algorithm | Seurat | RRID:SCR_016341 | Stuart et al., 2019 https://satijalab.org/seurat/get_started.html | |
| Software, algorithm | Monocle 3 | RRID:SCR_018685 | Cao et al., 2019 https://cole-trapnell-lab.github.io/monocle3/ | |
| Software, algorithm | Velocyto.R | La Manno et al., 2018 https://github.com/velocyto-team/velocyto.R | ||
| Software, algorithm | NicheNet | Browaeys et al., 2020 https://github.com/saeyslab/nichenetr/blob/master/vignettes/seurat_wrapper.md | ||
| Software, algorithm | RStudio | RRID:SCR_000432 | http://www.rstudio.com/ | |
| Commercial reagnet | Liberase | Roche (291963) | 0.3 mg/ml | |
| Commercial reagnet | Hyaluronidase | Sigma (H4272) | 10 μg/mL | |
| Commercial reagnet | Trypsin | Corning (45000–664) | 0.25% | |
| Chemical compound, drug | Tamoxifen | Sigma (T5648) | 100 mg/kg | |
| Chemical compound, drug | Liproxstatin-1 | Selleckchem (S7699) | 10 mg/kg | |
| Chemical compound, drug | Aristolochic acid | Sigma (A9451) | 6 mg/kg | |
| Commercial assay, kit | TUNEL staining | Abcam (Ab206386) |
Additional files
-
Supplementary file 1
Cluster-enriched genes in Figure 1.
- https://cdn.elifesciences.org/articles/68603/elife-68603-supp1-v2.xlsx
-
Supplementary file 2
Differentially expressed genes in PT cells.
- https://cdn.elifesciences.org/articles/68603/elife-68603-supp2-v2.xlsx
-
Supplementary file 3
Gene ontology analyses of PT cells.
- https://cdn.elifesciences.org/articles/68603/elife-68603-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68603/elife-68603-transrepform-v2.docx





