Epigenome-wide analysis of DNA methylation and coronary heart disease: a nested case-control study
Figures
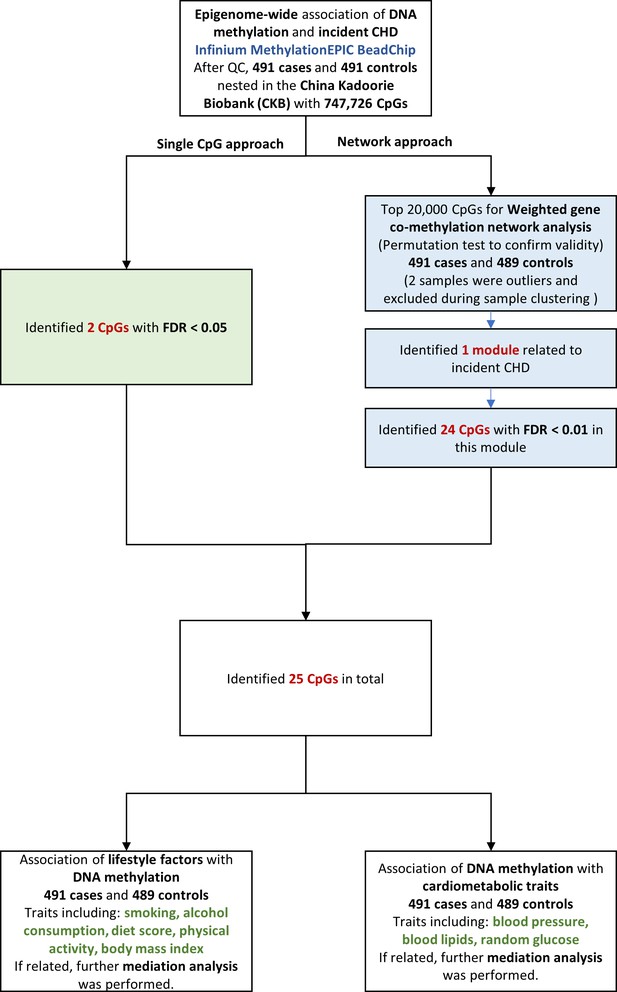
Flowchart of the present study. CHD = coronary heart disease; QC = quality control; CpG = cytosine-phosphate-guanine; FDR = false discovery rate.
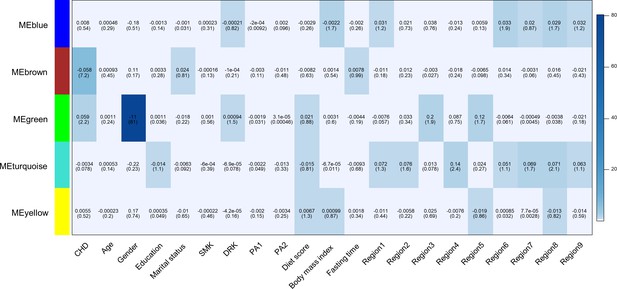
Heatmap of association with methylation network modules.
Correlation coefficient and -log10(P) (inside the bracket) were reported; the degree of -log10(p) is illustrated with the color legend. Linear regressions were fitted with inverse normal transformed module eigengene (ME) as dependent variables; coronary heart disease (CHD1) as indicator; and age, sex, education, marital status, smoking (SMK), drinking (DRK), physical activity (PA1 and PA2 as the second and third tertile respectively), diet score, body mass index, fasting time, study area and all surrogate variables as covariates.

Permutation test to confirm the validity of weighted gene co-methylation network analysis.
Correlation coefficient and -log10(P) (inside the bracket) were reported; the degree of -log10(P) is illustrated with the color legend. Linear regressions were fitted with inverse normal transformed module eigengene (ME) as dependent variables; shuffled case and control status (Permutation_case) as indicator; and age, sex, education, marital status, smoking (SMK), drinking (DRK), physical activity (PA1 and PA2 as the second and third tertile, respectively), diet score, body mass index (BMI), fasting time, study area and all surrogate variables as covariates.
Tables
Age-, sex- and study area-adjusted baseline characteristics of 982 participants according to the case or control status.
| Baseline characteristics | Cases(n = 491) | Controls(n = 491) | P value |
|---|---|---|---|
| Age, year | 50.6 | 49.5 | - |
| Female, % | 43.6 | 43.6 | - |
| Urban area, % | 20.6 | 20.6 | - |
| Middle school and above, % | 43.4 | 45.6 | 0.730 |
| Married, % | 90.4 | 94.7 | 0.028 |
| Family history of heart attack, % | 6.9 | 4.7 | 0.127 |
| Fasting time, h | 4.0 | 4.0 | - |
| Lifestyle factors | |||
| Daily tobacco smoker, % | 46.6 | 40.3 | 0.004 |
| Daily alcohol drinker, % | 9.0 | 10.0 | 0.455 |
| Physical activity, MET-h/day | 22.0 | 23.9 | 0.097 |
| Diet score | 2.3 | 2.5 | 0.001 |
| Vegetables 7 days/week, % | 92.7 | 91.0 | 0.278 |
| Fruit 7 days/week, % | 9.4 | 13.8 | 0.030 |
| Read meat <7 days/week, % | 79.2 | 80.0 | 0.600 |
| Soybean product ≥4 days/week, % | 5.9 | 9.6 | 0.026 |
| Fish ≥1 days/week, % | 24.6 | 28.9 | 0.022 |
| Coarse grains ≥ 4 days/week, % | 22.8 | 24.6 | 0.047 |
| Body mass index, kg/m2 | 23.9 | 23.3 | 0.002 |
| Metabolic risk factors | |||
| Prevalent hypertension, % | 52.5 | 29.9 | < 0.001 |
| Prevalent diabetes, % | 10.0 | 4.5 | 0.004 |
| Blood lipids | |||
| Total cholesterol, mmol/L | 4.69 | 4.52 | 0.005 |
| LDL-C, mmol/L | 2.35 | 2.21 | 0.003 |
| HDL-C, mmol/L | 1.22 | 1.18 | 0.025 |
| Triglyceride, mmol/L | 2.20 | 2.01 | 0.064 |
-
The results are presented as means or percentages. P values were not showed for matched factors. MET = metabolic equivalent of task; LDL-C = low-density lipoprotein cholesterol; HDL-C = high-density lipoprotein cholesterol.
Associations of 25 significant CpGs with the risk of coronary heart disease.
| Chr | Position(hg19) | CpG | SD | Gene | Relation to gene | EWAS | WGCNA* | Odds Ratio† (95% CI) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| β‡ | P | FDR | Module-specific FDR | |||||||
| 9 | 115513036 | cg23398826 | 0.008 | SNX30 | TSS200 | –0.003 | 1.57E-08 | 0.012 | 1.05E-04 | 0.56 (0.45, 0.70) |
| 3 | 49068057 | cg02386575 | 0.016 | IMPDH2 | TSS1500 | 0.006 | 9.61E-08 | 0.036 | 2.02E-04 | 2.00 (1.57, 2.56) |
| QRICH1 | Body | |||||||||
| 19 | 37329330 | cg10400937 | 0.007 | ZNF790 | TSS200 | 0.002 | 1.09E-05 | 0.288 | 0.009 | 1.53 (1.24, 1.89) |
| 12 | 131758671 | cg20562821 | 0.022 | (RPS6P20 §) | 0.005 | 2.42E-05 | 0.288 | 0.009 | 1.72 (1.28, 2.31) | |
| 6 | 34855635 | cg08106661 | 0.016 | TAF11 | 1stExon | 0.003 | 3.16E-05 | 0.305 | 0.009 | 1.87 (1.35, 2.59) |
| ANKS1A | TSS1500 | |||||||||
| 1 | 153203211 | cg11630610 | 0.019 | (MIR584§) | 0.005 | 3.83E-05 | 0.329 | 0.009 | 1.77 (1.36, 2.32) | |
| 1 | 8426319 | cg20302171 | 0.018 | RERE | 5'UTR | –0.004 | 4.29E-05 | 0.340 | 0.009 | 0.55 (0.42, 0.73) |
| 11 | 63909324 | cg26334131 | 0.025 | MACROD1 | Body | –0.005 | 4.44E-05 | 0.340 | 0.009 | 0.53 (0.38, 0.73) |
| 20 | 2444631 | cg07560408 | 0.018 | SNORD119 | TSS1500 | –0.005 | 4.46E-05 | 0.340 | 0.009 | 0.60 (0.47, 0.77) |
| SNRPB | Body | |||||||||
| 19 | 46522185 | cg21210537 | 0.027 | MIR769 | TSS200 | 0.004 | 4.85E-05 | 0.356 | 0.009 | 2.18 (1.47, 3.23) |
| 20 | 60546782 | cg15833447 | 0.021 | (TAF4§) | 0.006 | 5.55E-05 | 0.375 | 0.009 | 1.50 (1.20, 1.88) | |
| 11 | 94963255 | cg02591826 | 0.005 | LOC100129203 | TSS200 | 0.002 | 5.70E-05 | 0.375 | 0.009 | 1.52 (1.23, 1.87) |
| 7 | 100861083 | cg16639138 | 0.006 | ZNHIT1 | 5'UTR/1stExon | 0.002 | 6.46E-05 | 0.375 | 0.009 | 1.52 (1.24, 1.86) |
| PLOD3 | TSS200 | |||||||||
| 6 | 27863042 | cg01545454 | 0.007 | (HIST1H2BO§) | 0.002 | 7.29E-05 | 0.378 | 0.009 | 1.64 (1.26, 2.13) | |
| 1 | 203242409 | cg07219103 | 0.008 | (CHIT1§) | 0.002 | 7.35E-05 | 0.378 | 0.009 | 1.78 (1.28, 2.47) | |
| 22 | 23994996 | cg05681643 | 0.018 | GUSBP11 | Body | 0.004 | 7.42E-05 | 0.378 | 0.009 | 1.60 (1.24, 2.08) |
| 2 | 88991375 | cg06358566 | 0.009 | RPIA | 1stExon | –0.002 | 7.74E-05 | 0.385 | 0.009 | 0.62 (0.48, 0.80) |
| 2 | 162273185 | cg19583211 | 0.016 | TBR1 | 1stExon | –0.003 | 7.97E-05 | 0.385 | 0.009 | 0.56 (0.41, 0.77) |
| 20 | 3613189 | cg10643850 | 0.025 | ATRN | Body | 0.004 | 8.04E-05 | 0.385 | 0.009 | 1.97 (1.37, 2.82) |
| 17 | 17460905 | cg13311494 | 0.016 | PEMT | Body | –0.005 | 8.50E-05 | 0.397 | 0.009 | 0.64 (0.52, 0.79) |
| 1 | 179852195 | cg11754670 | 0.009 | TOR1AIP1 | Body | 0.001 | 8.84E-05 | 0.398 | 0.009 | 2.04 (1.40, 2.97) |
| 15 | 74928935 | cg05740632 | 0.014 | EDC3 | Body | –0.004 | 9.07E-05 | 0.398 | 0.009 | 0.62 (0.49, 0.78) |
| 11 | 1972510 | cg08484100 | 0.023 | MRPL23 | Body | –0.004 | 9.19E-05 | 0.398 | 0.009 | 0.54 (0.40, 0.74) |
| 1 | 52822428 | cg24792179 | 0.019 | CC2D1B | Body | 0.004 | 9.87E-05 | 0.410 | 0.009 | 1.79 (1.36, 2.35) |
| 7 | 68973036 | cg22794712 | 0.021 | (LOC100507468§) | –0.006 | 1.10E-04 | 0.413 | 0.010 | 0.63 (0.50, 0.80) | |
-
*
cg23398826 in the Turquoise module, all other CpGs in the Brown module.
-
†
Odds ratios were for per standard deviation increase in DNA methylation level.
-
‡
Effect sizes were calculated based on normalized methylation values, denoting the methylation difference between cases and controls.
-
§
For inter-genic CpG sites, R package FDb.InfiniumMethylation.hg19 was used to locate the nearest annotated gene.
-
CpG = cytosine-phosphoguanine site; Chr = chromosome; EWAS = epigenome wide association; WGCNA = weighted gene co-methylation network analysis; FDR = false discovery rate; CI = confidence interval; TSS200 = within 200 bp from transcription start site; TSS1500 = within 1500 bp from transcription start site; Body = the CpG is in gene body; 1stExon = the first exon; and UTR = untranslated region.
Associations between lifestyle factors and methylation level of identified CpGs, and the risk of coronary heart disease mediated through methylation level of CpG sites.
| Effect size(SE) | P | Mediation effect | ||
|---|---|---|---|---|
| Proportion mediated, % | P | |||
| Smoking, no. of cigarettes/day | ||||
| cg08106661 | 1.50E-04 (4.67E-05) | 0.001 | 28.50 | 0.036 |
| Diet score (ranging 0-6) | ||||
| cg21210537 | 3.60E-03 (1.27E-03) | 0.005 | 4.66 | 0.206 |
| cg10643850 | 2.57E-03 (1.26E-03) | 0.042 | -6.91 | 0.088 |
| cg05740632 | 1.37E-03 (6.88E-04) | 0.047 | 11.30 | 0.068 |
| Body mass index, kg/m2 | ||||
| cg20302171 | 3.90E-04 (1.67E-04) | 0.020 | -2.87 | 0.267 |
| cg08484100 | 4.17E-04 (2.10E-04) | 0.048 | -1.91 | 0.373 |
-
Linear regression was fitted by including all five lifestyle factors (smoking, alcohol consumption, physical activity, diet score, and body mass index) simultaneously in the same model, with methylation values as dependent variables, and age, sex, study area, fasting time, education level, marital status and batch as covariates. CpG = cytosine-phosphoguanine site; SE = standard error. Alcohol consumption and physical activity were not associated with any of the coronary heart disease-associated CpGs. Details were reported in the Table 3—source data 1.
-
Table 3—source data 1
Association between lifestyle factors and identified CpGs.
- https://cdn.elifesciences.org/articles/68671/elife-68671-table3-data1-v2.docx
Associations between quartile methylation level of identified CpGs and cardiometabolic traits, and the risk of coronary heart disease mediated through different cardiometabolic traits.
| Quartile 1 vs. 4 | P for trend | Mediation effect | |||
|---|---|---|---|---|---|
| Effect size(SE)* | P | Proportion mediated, % | P | ||
| Systolic blood pressure†, mmHg | |||||
| cg23398826 | -6.410 (2.118) | 0.003 | <0.001 | 7.65 | 0.003 |
| cg13311494 | -6.580 (2.122) | 0.002 | 0.020 | 15.61 | 0.031 |
| Diastolic blood pressure†, mmHg | |||||
| cg23398826 | -3.574 (1.218) | 0.003 | <0.001 | 6.39 | 0.006 |
| cg13311494 | -3.650 (1.221) | 0.003 | 0.029 | 12.38 | 0.045 |
| Total cholesterol, mmol/L | |||||
| cg26334131 | 0.197 (0.089) | 0.026 | 0.003 | -31.62 | 0.168 |
| cg05740632 | 0.163 (0.089) | 0.066 | 0.013 | -3.21 | 0.126 |
| cg21210537 | 0.175 (0.094) | 0.064 | 0.027 | -8.19 | 0.197 |
| cg19583211 | -0.064 (0.088) | 0.466 | 0.047 | 2.73 | 0.270 |
| Cholesterol in LDL, mmol/L | |||||
| cg26334131 | 0.110 (0.063) | 0.079 | 0.007 | -32.14 | 0.135 |
| cg20302171 | 0.107 (0.063) | 0.09 | 0.029 | -10.48 | 0.161 |
| cg05740632 | 0.109 (0.063) | 0.083 | 0.019 | -3.38 | 0.110 |
| cg19583211 | -0.078 (0.062) | 0.208 | 0.020 | 3.60 | 0.210 |
| cg13311494 | -0.117 (0.062) | 0.06 | 0.027 | 3.70 | 0.208 |
| cg21210537 | 0.126 (0.067) | 0.059 | 0.044 | -8.55 | 0.177 |
| Cholesterol in HDL, mmol/L | |||||
| cg15833447 | 0.037 (0.026) | 0.154 | 0.013 | -10.77 | 0.180 |
| cg21210537 | 0.040 (0.027) | 0.146 | 0.019 | 7.30 | 0.235 |
| Random blood glucose‡, mmol/L | |||||
| cg10400937 | 0.551 (0.231) | 0.017 | 0.003 | 6.72 | 0.107 |
| cg01545454 | 0.203 (0.234) | 0.385 | 0.006 | 9.58 | 0.097 |
| cg11754670 | 0.466 (0.236) | 0.049 | 0.005 | 36.39 | 0.086 |
| cg26334131 | -0.517 (0.234) | 0.028 | 0.018 | 32.70 | 0.109 |
| cg07219103 | 0.578 (0.244) | 0.018 | 0.032 | 6.17 | 0.135 |
| cg20302171 | -0.556 (0.234) | 0.018 | 0.027 | 15.77 | 0.123 |
-
*
Effect sizes denoted the differences of metabolic traits between the top and bottom quartile methylation level. Details of other quartiles were reported in the Table 4—source data 1.
-
†
We added 15 and 10 mmHg to the measured systolic blood pressure and diastolic blood pressure respectively among participants who reported usage of blood pressure-lowering medications.
-
‡
Additionally adjusted for treatment of diabetes (yes or no) at baseline. Multivariable model was adjusted for: age, sex, education level, marital status, smoking, drinking, physical activity, dietary score, body mass index, fasting time, study area, and batch. The CpGs which were significantly associated with any metabolic risk factors were reported. Details of other CpGs were reported in the Supplementary tables. CpG = cytosine-phosphoguanine site; SE = standard error; LDL = low-density lipoprotein; HDL = high-density lipoprotein.
-
Table 4—source data 1
Association between quartile methylation level of identified CpGs and systolic blood pressure (mmHg).
- https://cdn.elifesciences.org/articles/68671/elife-68671-table4-data1-v2.docx
-
Table 4—source data 2
Association between quartile methylation level of identified CpGs and diastolic blood pressure (mmHg).
- https://cdn.elifesciences.org/articles/68671/elife-68671-table4-data2-v2.docx
-
Table 4—source data 3
Association between quartile methylation level of identified CpGs and total cholesterol (mmol/L).
- https://cdn.elifesciences.org/articles/68671/elife-68671-table4-data3-v2.docx
-
Table 4—source data 4
Association between quartile methylation level of identified CpGs and cholesterol in low-density lipoprotein (mmol/L).
- https://cdn.elifesciences.org/articles/68671/elife-68671-table4-data4-v2.docx
-
Table 4—source data 5
Association between quartile methylation level of identified CpGs and cholesterol in high-density lipoprotein (mmol/L).
- https://cdn.elifesciences.org/articles/68671/elife-68671-table4-data5-v2.docx
-
Table 4—source data 6
Association between quartile methylation level of identified CpGs and random glucose (mmol/L).
- https://cdn.elifesciences.org/articles/68671/elife-68671-table4-data6-v2.docx
-
Table 4—source data 7
Association between quartile methylation level of identified CpGs and triglyceride (mmol/L).
- https://cdn.elifesciences.org/articles/68671/elife-68671-table4-data7-v2.docx
| Outcome | CpG name | beta coefficient in ourstudy | p-value in our study | beta coefficient in the previous study | p-valuein theprevious study | Gene |
|---|---|---|---|---|---|---|
| CHD | cg22617878 | -5.89E-04 | 3.75E-01 | −0.3719 | 1.99E-08 | ATP2B2 |
| cg13827209 | 8.39E-04 | 4.47E-01 | 0.268 | 3.76E-08 | TGFBR1 | |
| cg14185717 | / | / | −0.2878 | 1.38E-07 | BNC2 | |
| cg10307345 | -3.62E-03 | 1.88E-01 | −0.1480 | 1.86E-07 | PTPN5 | |
| cg13822123 | -6.12E-04 | 1.45E-01 | 0.4138 | 2.03E-07 | PSME4 | |
| cg23245316 | 1.04E-03 | 1.18E-01 | −0.4674 | 2.17E-07 | TSSC1 | |
| cg24977276 | 2.83E-03 | 4.60E-02 | −0.3256 | 2.54E-07 | GTF2I | |
| cg24447788 | 1.17E-03 | 3.20E-01 | −0.2679 | 4.33E-07 | (PTBP1**) | |
| cg08422803 | 1.35E-03 | 3.47E-01 | 0.1994 | 7.52E-07 | ITGB2 | |
| cg01751802 | -1.43E-03 | 5.85E-01 | 0.1473 | 9.35E-07 | KANK2 | |
| cg02449373 | 2.25E-05 | 9.77E-01 | 0.3715 | 9.98E-07 | FUT1 | |
| cg02683350 | -1.19E-03 | 3.62E-01 | −0.5062 | 1.55E-06 | ADAMTS2 | |
| cg05820312 | 2.89E-04 | 7.87E-01 | 0.5031 | 1.60E-06 | TRAPPC9 | |
| cg06639874 | -8.25E-04 | 6.41E-01 | −0.2506 | 1.83E-06 | MLPH | |
| cg06582394 | -2.56E-03 | 1.49E-01 | 0.1657 | 1.90E-06 | CASR | |
| cg02155262 | -4.62E-04 | 3.07E-01 | 0.477 | 1.97E-06 | AGA | |
| cg12766383 | 2.26E-03 | 3.11E-02 | −0.6194 | 1.98E-06 | UBR4 | |
| cg05892484 | 5.55E-05 | 9.60E-01 | −0.5020 | 2.01E-06 | MAD1L1 | |
| cg03031868 | 1.46E-04 | 8.65E-01 | 0.3461 | 2.29E-06 | ESD | |
| cg25497530 | -5.63E-03 | 1.45E-03 | −0.2225 | 2.62E-06 | PTPRN2 | |
| cg06596307 | -2.06E-03 | 1.24E-01 | −0.4198 | 2.99E-06 | IGF1R | |
| cg10702366 | -2.35E-03 | 1.47E-01 | −0.1093 | 3.09E-06 | FGGY | |
| cg26470101 | 1.24E-03 | 4.46E-01 | 0.3052 | 3.09E-06 | (DLX2**) | |
| cg26042024 | 1.66E-03 | 4.16E-01 | −0.3109 | 3.13E-06 | ZFAT | |
| cg00466121 | 7.27E-04 | 4.26E-01 | 0.4646 | 3.16E-06 | ZNHIT6 | |
| cg04987302 | -1.96E-03 | 1.85E-01 | −0.3378 | 3.71E-06 | (OTX2-AS1**) | |
| cg08853494 | 1.82E-04 | 7.27E-01 | 0.221 | 4.03E-06 | RCHY1;THAP6 | |
| cg26467725 | 1.34E-03 | 2.02E-01 | −0.4225 | 4.22E-06 | SLCO3A1 | |
| cg06442192 | 9.07E-04 | 5.78E-01 | −0.5241 | 4.89E-06 | ZNF541 | |
| cg00393373 | 1.11E-03 | 2.92E-01 | −0.3156 | 4.91E-06 | ZNF518B | |
| MI | cg22871797 | 4.77E-04 | 6.85E-01 | −0.599 | 5.29E-08 | CYFIP1 |
| cg24977276 | 2.83E-03 | 4.60E-02 | −0.366 | 9.97E-08 | GTF2I | |
| cg18598861 | 1.43E-03 | 1.98E-01 | −0.671 | 1.61E-07 | IRF9 | |
| cg09777776 | 7.95E-04 | 7.38E-01 | 0.287 | 2.25E-07 | ZNF254 | |
| cg20545941 | -3.22E-04 | 6.69E-01 | −0.885 | 2.47E-07 | MPPED1 | |
| cg19935845 | 9.61E-04 | 5.30E-01 | −0.336 | 4.65E-07 | TNXB | |
| cg24423782 | -2.37E-03 | 1.33E-01 | −0.398 | 5.37E-07 | MIR182 | |
| cg00393373 | 1.11E-03 | 2.92E-01 | −0.401 | 7.68E-07 | ZNF518B | |
| cg00466121 | 7.27E-04 | 4.26E-01 | 0.487 | 7.79E-07 | ZNHIT6 | |
| cg19227382 | / | / | −0.504 | 8.12E-07 | CDH23 | |
| cg03467256 | / | / | −0.408 | 8.33E-07 | HPCAL1 | |
| cg25196881 | 3.45E-04 | 7.46E-01 | −0.269 | 1.05E-06 | (THBS1**) | |
| cg02321112 | 9.68E-05 | 8.99E-01 | 0.39 | 1.08E-06 | (MNX1-AS1**) | |
| cg00355799 | 2.79E-04 | 8.27E-01 | −0.216 | 1.40E-06 | (LOC339529**) | |
| cg17556588 | -1.20E-03 | 3.90E-01 | −0.154 | 1.45E-06 | PRRG4 | |
| cg04987302 | -1.96E-03 | 1.85E-01 | −0.428 | 1.50E-06 | (OTX2-AS1**) | |
| cg07289306 | -1.45E-03 | 3.24E-02 | 0.616 | 1.71E-06 | (MIR138-1**) | |
| cg05892484 | 5.55E-05 | 9.60E-01 | −0.551 | 1.84E-06 | MAD1L1 | |
| cg10702366 | -2.35E-03 | 1.47E-01 | −0.150 | 2.11E-06 | FGGY | |
| cg22618720 | / | / | −0.424 | 2.37E-06 | (MIR5095**) | |
| cg14010194 | 3.46E-04 | 7.84E-01 | −0.484 | 2.71E-06 | GUCA1B | |
| cg13827209 | 8.39E-04 | 4.47E-01 | 0.285 | 2.71E-06 | TGFBR1 | |
| cg24318598 | -1.75E-03 | 2.59E-01 | −0.254 | 2.79E-06 | ANO1 | |
| cg07015775 | 1.01E-03 | 1.36E-01 | 0.479 | 3.13E-06 | ZNHIT6 | |
| cg21018156 | 2.00E-03 | 1.39E-01 | −0.135 | 3.17E-06 | (LINC01312**) | |
| cg07475527 | -1.64E-03 | 2.46E-01 | −0.225 | 3.77E-06 | (RCAN3**) | |
| cg20000562 | 2.06E-03 | 8.15E-02 | 0.218 | 3.93E-06 | SFTA3 | |
| cg07436807 | 3.57E-04 | 7.88E-01 | −0.779 | 4.10E-06 | STAMBPL1; ACTA2 | |
| cg14029912 | -1.62E-03 | 1.39E-01 | −0.367 | 4.29E-06 | (BHLHE40**) | |
| cg22871797 | 4.77E-04 | 6.85E-01 | −0.599 | 5.29E-08 | CYFIP1 |
| CpG | Basic model | Full model | ||
|---|---|---|---|---|
| β | P | β | P | |
| cg23398826 | -0.003 | 3.44E-08 | -0.003 | 1.57E-08 |
| cg02386575 | 0.005 | 2.76E-07 | 0.006 | 9.61E-08 |
| cg10400937 | 0.002 | 1.28E-05 | 0.002 | 1.09E-05 |
| cg20562821 | 0.005 | 2.95E-05 | 0.005 | 2.42E-05 |
| cg08106661 | 0.003 | 6.75E-05 | 0.003 | 3.16E-05 |
| cg11630610 | 0.005 | 4.22E-05 | 0.005 | 3.83E-05 |
| cg20302171 | -0.004 | 3.86E-05 | -0.004 | 4.29E-05 |
| cg26334131 | -0.005 | 3.51E-05 | -0.005 | 4.44E-05 |
| cg07560408 | -0.004 | 5.73E-05 | -0.005 | 4.46E-05 |
| cg21210537 | 0.0043 | 4.81E-05 | 0.004 | 4.85E-05 |
| cg15833447 | 0.005 | 0.000107 | 0.006 | 5.55E-05 |
| cg02591826 | 0.002 | 4.97E-05 | 0.002 | 5.70E-05 |
| cg16639138 | 0.002 | 0.000122 | 0.002 | 6.46E-05 |
| cg01545454 | 0.002 | 6.38E-05 | 0.002 | 7.29E-05 |
| cg07219103 | 0.002 | 6.77E-05 | 0.002 | 7.35E-05 |
| cg05681643 | 0.004 | 7.33E-05 | 0.004 | 7.42E-05 |
| cg06358566 | -0.002 | 8.02E-05 | -0.002 | 7.74E-05 |
| cg19583211 | -0.003 | 6.46E-05 | -0.003 | 7.97E-05 |
| cg10643850 | 0.004 | 0.000104 | 0.004 | 8.04E-05 |
| cg13311494 | -0.004 | 8.04E-05 | -0.005 | 8.50E-05 |
| cg11754670 | 0.001 | 0.000101 | 0.001 | 8.84E-05 |
| cg05740632 | -0.004 | 4.51E-05 | -0.004 | 9.07E-05 |
| cg08484100 | -0.005 | 3.69E-05 | -0.004 | 9.19E-05 |
| cg24792179 | 0.004 | 0.000115 | 0.004 | 9.87E-05 |
| cg22794712 | -0.005 | 0.000173 | -0.006 | 1.10E-04 |
| Basic model | Full model | |||
|---|---|---|---|---|
| Proportion mediated, % | Proportion mediated, % | Proportion mediated, % | Proportion mediated, % | |
| Smoking, no. of cigarettes/daycg08106661 | 26.80 | 0.038 | 28.50 | 0.036 |
| Diet score (ranging 0-6)cg21210537 | 3.89 | 0.256 | 4.66 | 0.206 |
| cg10643850 | -6.73 | 0.083 | -6.91 | 0.088 |
| cg05740632 | 11.03 | 0.062 | 11.30 | 0.068 |
| Body mass index, kg/m2cg20302171 | -2.90 | 0.269 | -2.87 | 0.267 |
| cg08484100 | -2.32 | 0.304 | -1.91 | 0.373 |
| Systolic blood pressure*, mmHg cg23398826 | 13.21 | 0.002 | 7.65 | 0.003 |
| cg13311494 | 3.14 | 0.082 | 15.61 | 0.031 |
| Diastolic blood pressure*, mmHg cg23398826 | 16.32 | 0.002 | 6.39 | 0.006 |
| cg13311494 | 3.80 | 0.079 | 12.38 | 0.045 |
| Total cholesterol, mmol/L cg26334131 | -5.67 | 0.449 | -31.62 | 0.168 |
| cg05740632 | -33.04 | 0.020 | -3.21 | 0.126 |
| cg21210537 | -6.70 | 0.169 | -8.19 | 0.197 |
| cg19583211 | 8.67 | 0.114 | 2.73 | 0.270 |
| Cholesterol in LDL, mmol/L cg26334131 | -3.25 | 0.406 | -32.14 | 0.135 |
| cg20302171 | -4.47 | 0.263 | -10.48 | 0.161 |
| cg05740632 | -17.77 | 0.032 | -3.38 | 0.110 |
| cg19583211 | 9.03 | 0.175 | 3.60 | 0.210 |
| cg13311494 | 9.02 | 0.177 | 3.70 | 0.208 |
| cg21210537 | -5.50 | 0.175 | -8.55 | 0.177 |
| Cholesterol in HDL, mmol/L cg15833447 | -3.69 | 0.293 | -10.77 | 0.180 |
| cg21210537 | 3.79 | 0.260 | 7.30 | 0.235 |
| Random blood glucose‡, mmol/L cg10400937 | 8.92 | 0.056 | 6.72 | 0.107 |
| cg01545454 | 6.08 | 0.122 | 9.58 | 0.097 |
| cg11754670 | 1.26 | 0.613 | 36.39 | 0.086 |
| cg26334131 | 1.50 | 0.612 | 32.70 | 0.109 |
| cg07219103 | 6.76 | 0.102 | 6.17 | 0.135 |
| cg20302171 | 2.49 | 0.397 | 15.77 | 0.123 |
-
* We added 15 and 10 mmHg to the measured systolic blood pressure and diastolic blood pressure respectively among participants who reported usage of blood pressure-lowering medications.
-
‡ Additionally adjusted for treatment of diabetes (yes or no) at baseline.
| Present study | Guarrera S, et al.13 | Agha Golareh, et al.1 | |
|---|---|---|---|
| No. of cases | 491 | 292 | 1,895 |
| Sample size | 982 | 584 | 11,461 |
| Mean age (years) | 50.1 | 52.4 | 64 |
| Mean Follow-up time (years) | 7.6 | 12.9 | 11.2 |
| Model 1 | +adjusted for CC | |||
|---|---|---|---|---|
| Effect size(SE) | P | Effect size(SE) | P | |
| Smoking, no. of cigarettes/day cg08106661 | 1.50E-04 (4.67E-05) | 0.001 | 9.00E-05 (4.39E-05) | 0.041 |
| Systolic blood pressure, mmHg cg23398826 | -313.75 (91.72) | <0.001 | -255.60 (94.88) | 0.007 |
| cg13311494 | -108.86 (48.16) | 0.020 | -101.84 (47.02) | 0.031 |
| Diastolic blood pressure, mmHg cg23398826 | -184.53 (52.74) | <0.001 | -151.73 (55.07) | 0.006 |
| cg13311494 | -59.54 (27.30) | 0.029 | -54.51 (27.30) | 0.046 |
Additional files
-
Supplementary file 1
Members of the China Kadoorie Biobank collaborative group.
- https://cdn.elifesciences.org/articles/68671/elife-68671-supp1-v2.docx
-
Supplementary file 2
Detailed results and interpretation of the present study.
(A) Number of methylation markers associated with incident coronary heart disease across the range of P thresholds in the epigenome-wide association study. (B) Manhattan plot (A) and QQ plot (B) of the P values of the associations between each cytosine-phosphoguanine (CpG) site and incident coronary heart disease. In the Manhattan plot, the red line represents -log10(P) at false discovery rate (FDR) = 0.05. (C) Gene enrichment analysis of 2106 probes from the brown module which was significantly associated with coronary heart disease. (D) Associations of 25 significant CpGs with risk of coronary heart disease among 880 participants without usage of blood pressure lowering drug. (E) Association between quartile methylation level of identified CpGs and systolic blood pressure* (mmHg). (F) Association between quartile methylation level of identified CpGs and diastolic blood pressure* (mmHg). (G) The annotated or nearest annotated gene of the identified CHD-associated CpGs in our study and the previous GWAS finding.
- https://cdn.elifesciences.org/articles/68671/elife-68671-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68671/elife-68671-transrepform1-v2.docx
-
Source code 1
Single DNA methylation marker and incident CHD.
- https://cdn.elifesciences.org/articles/68671/elife-68671-supp3-v2.zip
-
Source code 2
Weighted gene co-methylation network and incident CHD.
- https://cdn.elifesciences.org/articles/68671/elife-68671-supp4-v2.zip
-
Source code 3
Mediation analysis.
- https://cdn.elifesciences.org/articles/68671/elife-68671-supp5-v2.zip
-
Source code 4
Baseline characteristics according to the case or controlstatus.
- https://cdn.elifesciences.org/articles/68671/elife-68671-supp6-v2.zip
-
Reporting standard 1
STROBE Statement—Checklist of items that should be included in reports of case-control studies.
- https://cdn.elifesciences.org/articles/68671/elife-68671-repstand1-v2.doc