Planarian stem cells sense the identity of the missing pharynx to launch its targeted regeneration
Figures
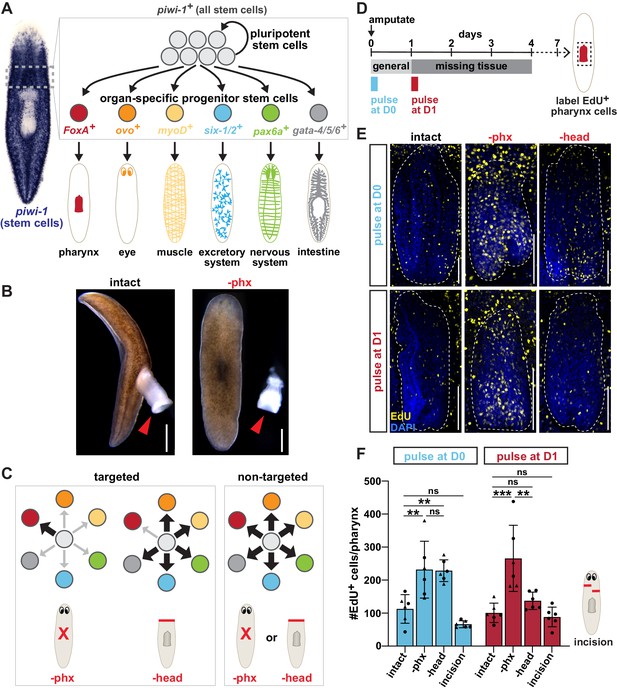
Both targeted and non-targeted mechanisms contribute to pharynx regeneration.
(A) Schematic of planarian stem cell lineage. Left, whole-mount in situ hybridization (WISH) for the stem cell marker piwi-1. Right, cartoon depiction of the dashed boxed region showing planarian stem cells consisting of pluripotent stem cells and organ-specific progenitors that produce planarian organs. Markers of organ-specific progenitors are indicated. (B) Live images of planarians before and after pharynx amputation. Arrows = pharynx; scale bars = 500 μm. (C) Models for targeted and non-targeted regeneration after different amputations (indicated by red lines). Progenitors are color coded as in A. (D) Schematic of F-ara-EdU delivery relative to amputation for E and F. (E) Confocal images of F-ara-EdU (yellow) in the pharynx (dashed outline) of intact animals, or 7 days after pharynx or head amputation. Dashed box in D = region imaged; DAPI = DNA (blue); scale bar = 100 μm. (F) Number of F-ara-EdU+ cells in the entire pharynx (dashed outline in E). Cartoon represents incision injuries. Graphs represent mean ± SD; symbols = individual animals; shapes distinguish biological replicates and; *, p≤0.05; **, p≤0.01; ***, p≤0.001; one-way ANOVA with Tukey test. Raw data can be found in Figure 1—source data 1.
-
Figure 1—source data 1
Quantification of F-ara-EdU+ cells in Figure 1F.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig1-data1-v1.xlsx
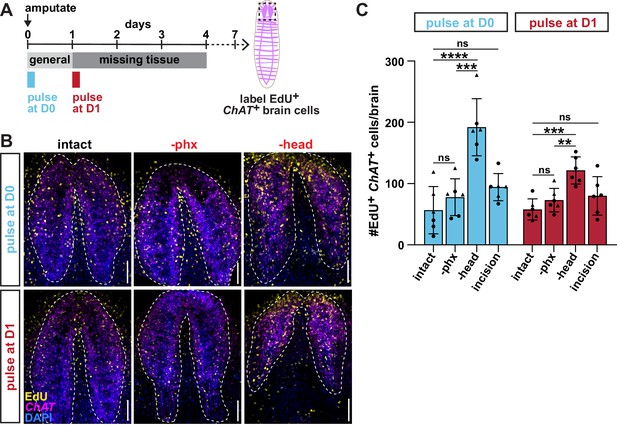
Selective pharynx removal does not increase incorporation of new brain cells.
(A) Schematic of F-ara-EdU delivery relative to amputation for B and C. (B) Confocal images of F-ara-EdU (yellow) and fluorescent in situ hybridization (FISH) for ChAT (magenta) in the brain (dashed outline) of intact and injured animals, 7 days after amputation. Dashed box in A = region shown; DAPI = DNA (blue); scale bar = 50 μm. (C) Number of F-ara-EdU+ ChAT+ cells in the entire brain (dashed outline in B). Graph represents mean ± SD. Symbols = individual animals; shapes distinguish biological replicates; **, p≤0.01; ***, p≤0.001; ****, p≤0.0001, one-way ANOVA with Tukey test. Raw data can be found in Figure 1—figure supplement 1—source data 1.
-
Figure 1—figure supplement 1—source data 1
Quantification of F-ara-EdU+ cells in Figure 1—figure supplement 1C.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig1-figsupp1-data1-v1.xlsx
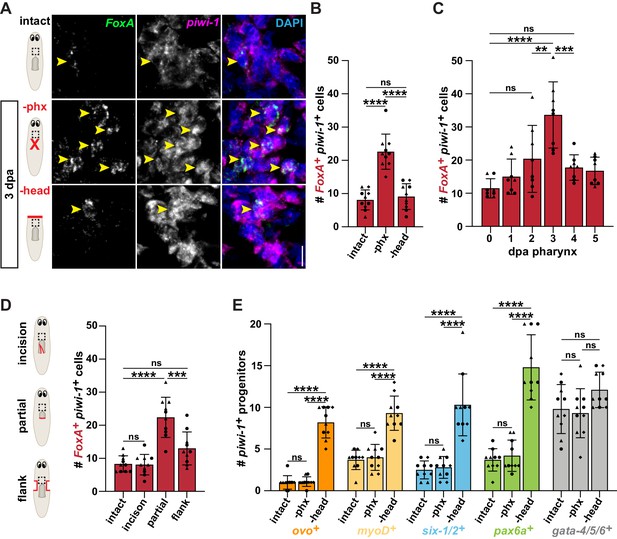
Pharynx tissue loss selectively increases pharynx progenitors.
(A) Confocal images of double fluorescent in situ hybridization (FISH) for FoxA (green) and piwi-1 (magenta) in intact and injured animals, 3 days post-amputation (dpa). Images are partial projections of a portion of the area outlined by dashed boxes. DAPI = DNA (blue); arrows = double-positive cells; scale bar = 10 μm. (B) Number of FoxA+ piwi-1+ cells in the area outlined by dashed boxes in A. (C) Number of FoxA+ piwi-1+ cells at indicated times post-pharynx amputation in the area outlined by dashed boxes in A. (D) Number of FoxA+ piwi-1+ cells in intact and injured animals, 3 days after injury (red lines) in the area outlined by dashed boxes in cartoons. (E) Number of cells double-positive for piwi-1 and the indicated progenitor marker in intact and injured animals, 3 days after amputation in the area outlined by dashed boxes in A. For graphs, a 6000 μm2 region in the same location of the pre-pharyngeal region was analyzed over 20 z-sections, as represented by dashed boxes in A and D. Graphs represent mean ± SD; symbols = individual animals; shapes distinguish biological replicates and; **, p≤0.01; ***, p≤0.001; ****, p≤0.0001, one-way ANOVA with Tukey test. Raw data can be found in Figure 2—source data 1.
-
Figure 2—source data 1
Quantification of piwi-1+ cells in Figure 2B–E.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig2-data1-v1.xlsx
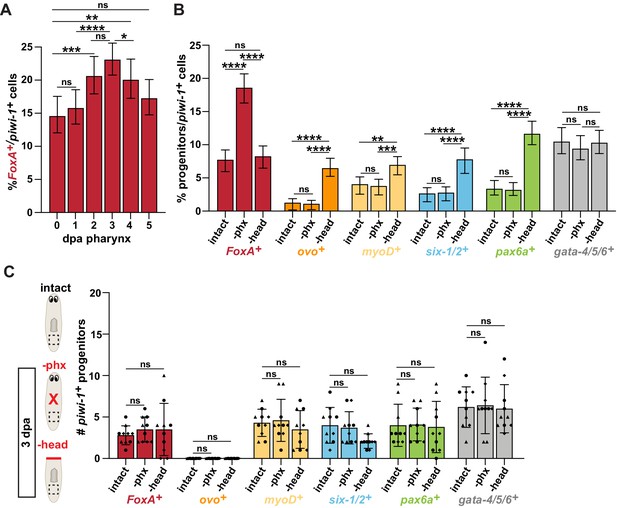
Amputation increases organ-specific progenitors relative to stem cells and is localized to wounds.
(A) Proportion of FoxA+ piwi-1+ cells relative to all piwi-1+ stem cells at the indicated days post-pharynx amputation (dpa), as represented by dashed boxes in cartoons in Figure 2A. n ≥ 631 cells per sample from two independent experiments. (B) Proportion of cells double-positive for piwi-1 and the indicated progenitor marker relative to all piwi-1+ stem cells in intact and injured animals, 3 days after amputation, as indicated by dashed boxes in Figure 2A. n ≥ 790 cells per sample from three independent experiments. (C) Number of cells in the tail (dashed boxes in cartoons) double-positive for piwi-1 and the indicated progenitor marker in intact and injured animals 3 days after amputation. For graphs, a 6000 μm2 region was analyzed over 20 z-sections. Graphs represent a proportion ± 95% confidence intervals (A,B); or mean ± SD (C) where symbols = individual animals; shapes distinguish biological replicates. *, p≤0.05; **, p≤0.01; ***, p≤0.001; ****, p≤0.0001; Fisher’s Exact Test (A,B); one-way ANOVA with Tukey Test (C). Raw data can be found in Figure 2—figure supplement 1—source data 1.
-
Figure 2—figure supplement 1—source data 1
Quantification of piwi-1+ cells in Figure 2—figure supplement 1A–C.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig2-figsupp1-data1-v1.xlsx
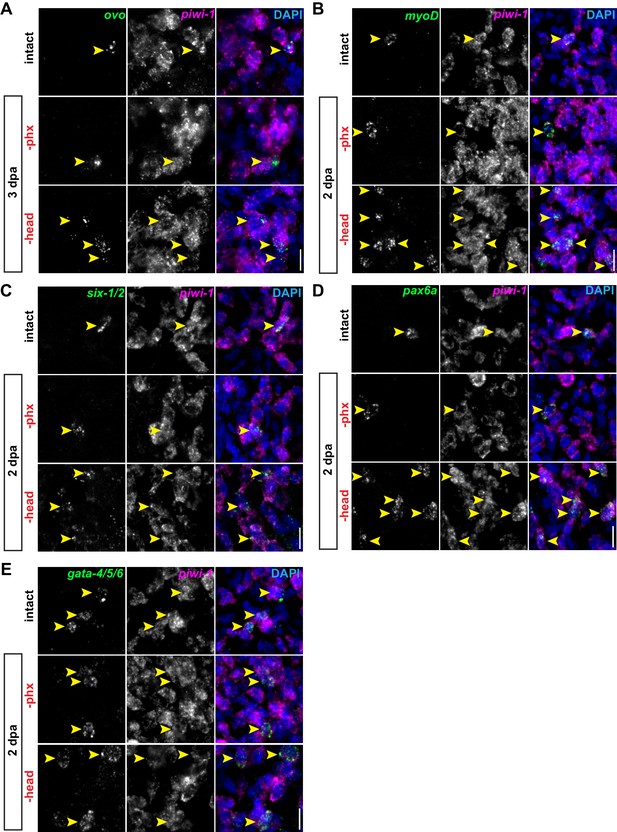
Pharynx loss does not affect non-pharyngeal progenitors.
(A–E) Confocal images of double FISH for ovo (A), myoD (B), six-1/2 (C), pax6a (D) and gata-4/5/6 (E) (green) and piwi-1 (magenta) in intact or injured animals, 3 days post-amputation (dpa). DAPI = DNA (blue); arrows = double-positive cells; scale bar = 10 µm. Images are partial projections of a portion of the area outlined by dashed boxes in Figure 2A.
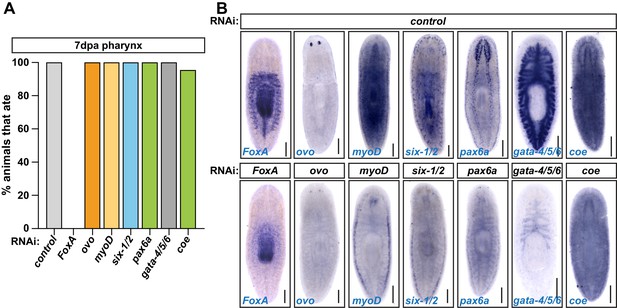
Non-pharyngeal organ-specific transcription factors are not required for pharynx regeneration.
(A) Proportion of animals capable of feeding 7 days after pharynx amputation following knockdown of the indicated genes. n ≥ 20 animals from two independent experiments. (B) WISH of the indicated progenitor marker in control and knockdown animals. Scale bars = 250 µm. Raw data can be found in Figure 2—figure supplement 3—source data 1.
-
Figure 2—figure supplement 3—source data 1
Quantification of feeding behavior in Figure 2—figure supplement 3A.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig2-figsupp3-data1-v1.xlsx
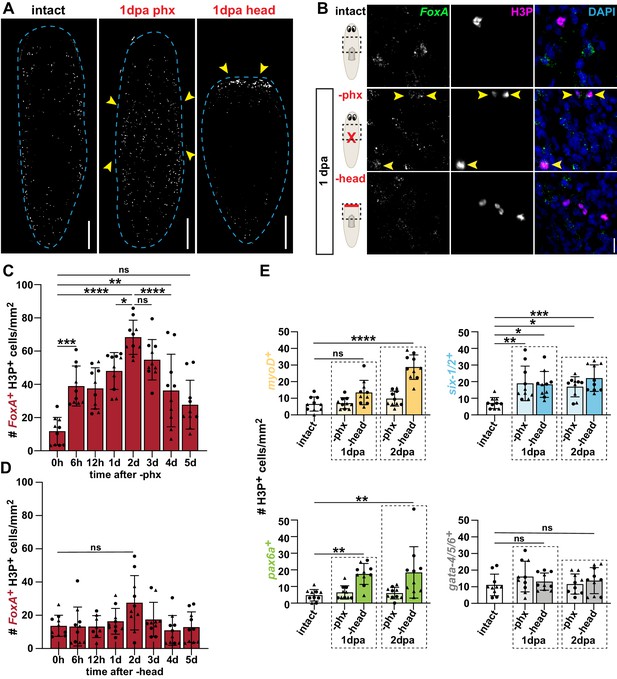
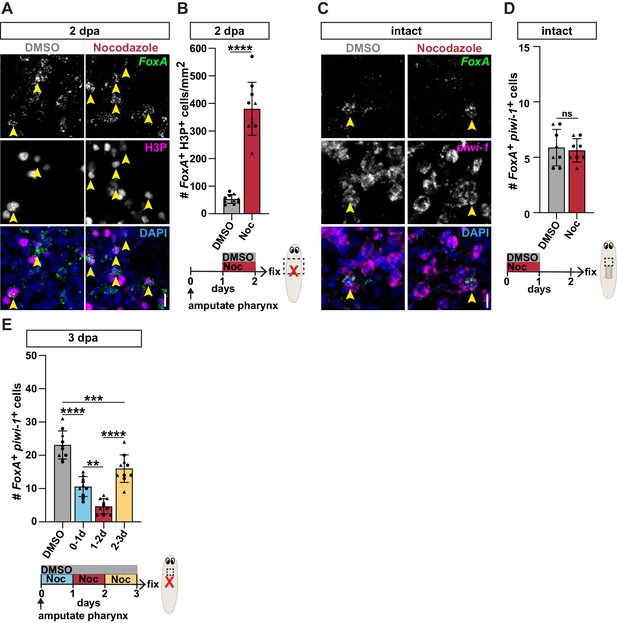
Pharynx loss selectively induces mitosis of pharynx progenitors.
(A) Whole-mount images of animals stained with anti-phosphohistone H3 (H3P) in intact and injured animals, 1 day post-amputation (dpa). Dashed line outlines animal; arrows = areas of increased H3P; scale bars = 250 μm. (B) Confocal images of FISH for FoxA (green) and H3P antibody (magenta) in intact and injured animals, 1 day post-amputation. Images are partial projections of a portion of the area outlined by dashed boxes. DAPI = DNA (blue); arrows = double-positive cells; scale bar = 10 μm. (C) Number of FoxA+ H3P+ cells at indicated times after pharynx amputation in the area outlined by dashed boxes in B. (D) Number of FoxA+ H3P+ cells at indicated times after head amputation in the area outlined by dashed boxes in B. (E) Number of cells double-positive for H3P and the indicated progenitor marker in the area outlined by dashed boxes in B. For H3P quantification, the entire pre-pharyngeal region was analyzed over 30 z-sections, as represented by dashed boxes in B, and normalized to area. Graphs represent mean ± SD; symbols = individual animals; shapes distinguish biological replicates and; *, p≤0.05; **, p≤0.01; ***, p≤0.001; ****, p≤0.0001, one-way ANOVA with Tukey test. Raw data can be found in Figure 3—source data 1.
-
Figure 3—source data 1
Quantification of H3P+ cells in Figure 3C–E.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig3-data1-v1.xlsx
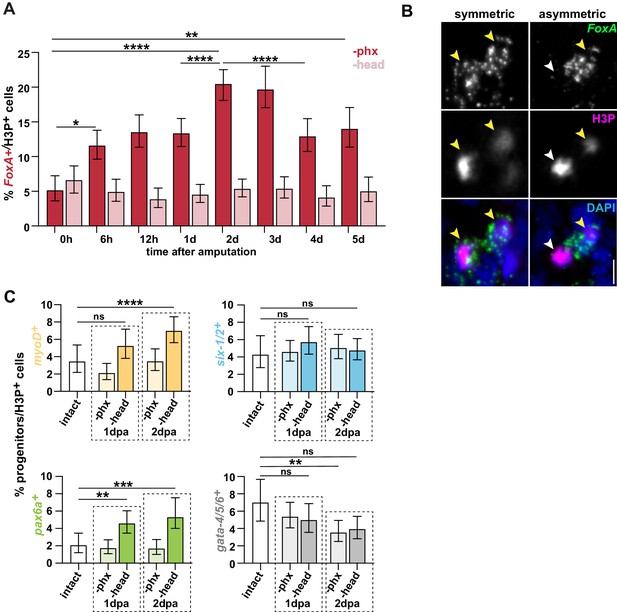
Pharynx loss selectively increases the number of mitotic pharynx progenitors in proportion to all stem cells.
(A) Proportion of FoxA+ H3P+ cells relative to all H3P+ stem cells at the indicated times post-pharynx or head amputation (dpa). n ≥ 515 cells per sample from two independent experiments. (B) Confocal images of FISH for FoxA (green) and H3P antibody (magenta) in anaphase cell, 2 days post-pharynx amputation (dpa). Images are partial projections of a portion of the area outlined by dashed boxes in Figure 3B. DAPI = DNA (blue); yellow arrows = double-positive daughters; white arrows = FoxA- H3P+ daughters; scale bar = 5 μm. (C) Proportion of cells double-positive for H3P and the indicated progenitor marker relative to all H3P+ stem cells in intact and injured animals, 1 and 2 days after pharynx or head amputation. n ≥ 472 cells per sample from two independent experiments. Graphs represent a proportion ± 95% confidence intervals. The entire pre-pharyngeal region was analyzed over 30 z-sections, as represented by dashed boxes in cartoons in Figure 3B. *, p≤0.05; **, p≤0.01; ***, p≤0.001; ****, p≤0.0001; Fisher’s Exact Test. Raw data can be found in Figure 3—figure supplement 1—source data 1.
-
Figure 3—figure supplement 1—source data 1
Quantification of H3P+ cells in Figure 3—figure supplement 1A and C.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig3-figsupp1-data1-v1.xlsx
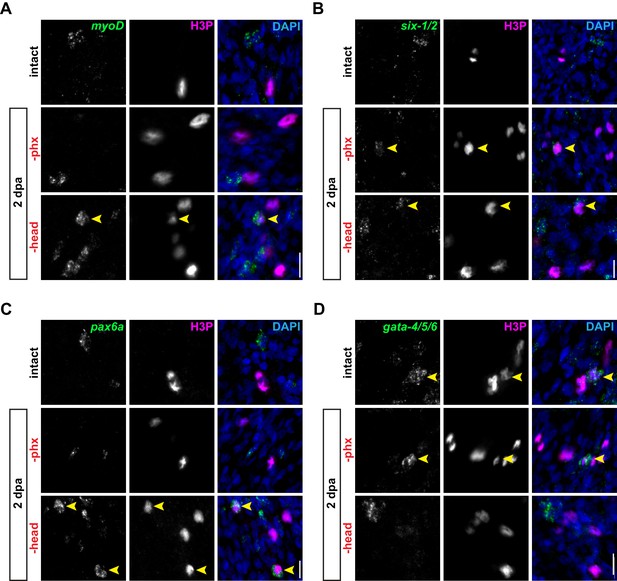
Division of non-pharyngeal progenitors is not triggered by pharynx loss.
(A–D) Confocal images of FISH for myoD (A), six-1/2 (B), pax6a (C) and gata-4/5/6 (D) (green) and H3P antibody (magenta) in intact or injured animals, 2 days post-amputation. DAPI = DNA (blue); arrows = double-positive cells; scale bar = 10 µm. Images are partial projections of a portion of the area outlined by dashed boxes in Figure 3B.
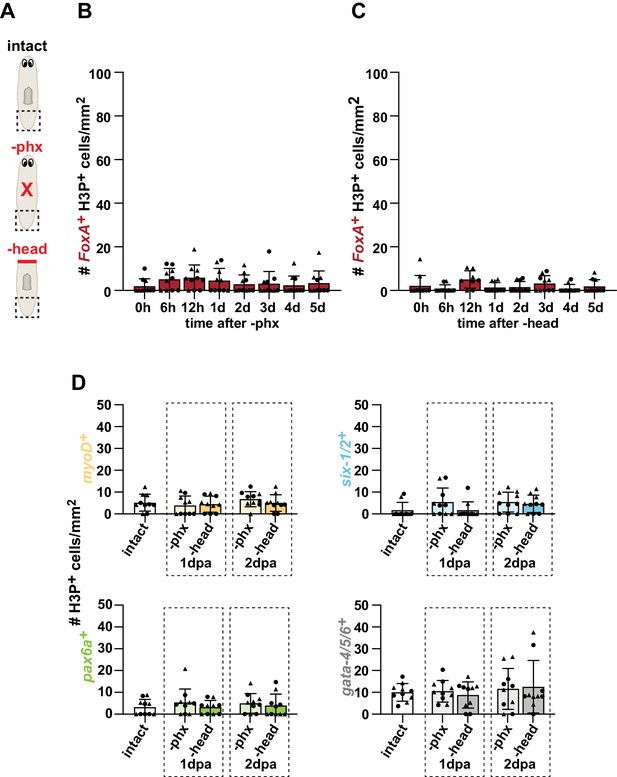
Amputation-induced division of organ-specific progenitors is localized to wounds.
(A) Cartoons depicting different amputation conditions. For B-D, the entire tail region was analyzed over 30 z-sections, as represented by dashed boxes, and normalized to area. (B) Number of FoxA+ H3P+ cells at indicated times after pharynx amputation. (C) Number of FoxA+ H3P+ cells at indicated times after head amputation. (D) Number of cells double-positive for H3P and the indicated progenitor marker, 1 and 2 days post-amputation (dpa). Graphs represent mean ± SD; symbols = individual animals; shapes distinguish biological replicates. No significant differences were detected by one-way ANOVA. Raw data can be found in Figure 3—figure supplement 3—source data 1.
-
Figure 3—figure supplement 3—source data 1
Quantification of H3P+ cells in Figure 3—figure supplement 3B–D.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig3-figsupp3-data1-v1.xlsx
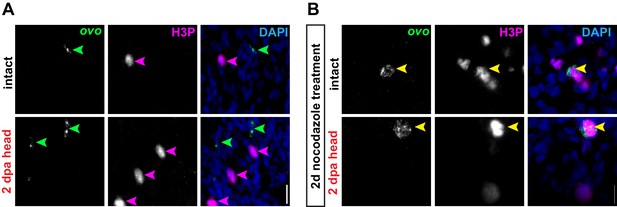
Eye progenitors do not divide in response to amputation.
(A) Confocal images of FISH for ovo (green) and H3P antibody (magenta) in intact or decapitated animals, 2 days post-amputation (dpa). (B) Confocal images of FISH for ovo (green) and H3P antibody (magenta) in intact or decapitated animals, 2 days post-amputation treated with nocodazole beginning immediately after amputation for 2 days (schematic). DAPI = DNA (blue); green arrows = ovo+; magenta arrows = H3P+; yellow arrows = double-positive; scale bar = 10 μm. Images are partial projections of a portion of the area outlined by dashed boxes in Figure 3B.
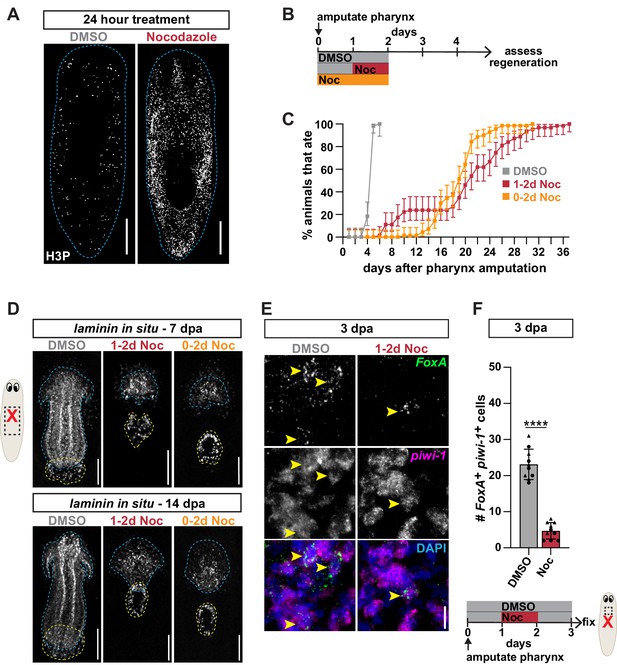
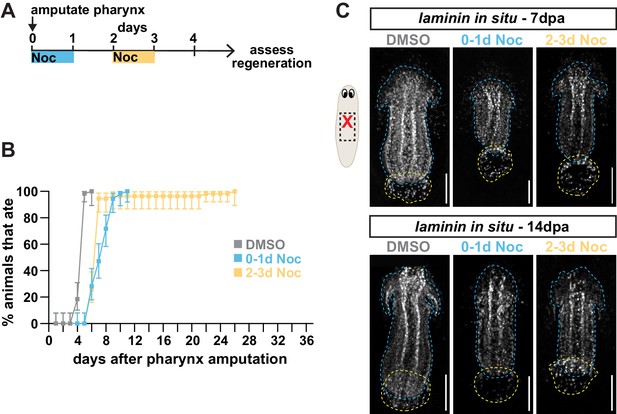
Stem cell division within a critical window is required for pharynx regeneration.
(A) Whole-mount images of intact animals stained with H3P after treatment with DMSO or nocodazole for 24 hr. Dashed line outlines animal; scale bars = 250 µm. (B) Schematic of nocodazole treatment relative to pharynx amputation for graph in C and images in D. (C) Proportion of animals capable of feeding after pharynx amputation, treated as indicated in B and assayed daily. Error bars represent ± 95% confidence intervals. n ≥ 54 animals from three independent experiments. (D) Whole-mount FISH for the pharynx marker laminin 7 and 14 days post-pharynx amputation (dpa) in animals treated as indicated in B. Dashed blue line outlines pharynx; dashed yellow line outlines mouth; scale bars = 100 µm. n ≥ 23 animals from three independent experiments. (E) Confocal images of FISH for FoxA (green) and piwi-1 (magenta) 3 days post-pharynx amputation in animals treated with DMSO or nocodazole, 1 day after amputation for 24 hr. DAPI = DNA (blue); arrows = double-positive cells; scale bar = 10 µm. (F) Number of FoxA+ piwi-1+ cells quantified in animals after indicated treatments (schematic) in the area outlined by dashed box in cartoon. Graph represents mean ± SD; symbols = individual animals; shapes distinguish biological replicates and; ****, p≤0.0001, unpaired t-test. Raw data can be found in Figure 4—source data 1.
-
Figure 4—source data 1
Raw data for feeding assay (Figure 4C) and quantification of piwi-1+ cells (Figure 4F).
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig4-data1-v1.xlsx
Stem cell division outside a critical window is not required for pharynx regeneration.
(A) Schematic of nocodazole treatment relative to pharynx amputation for graph in B and images in C. (B) Proportion of animals capable of feeding after pharynx amputation, treated as indicated in A and assayed daily. Error bars represent ± 95% confidence intervals. n ≥ 53 animals from three independent experiments. (C) Whole-mount FISH for the pharynx marker laminin 7 and 14 days post-pharynx amputation (dpa) in animals treated as indicated in A. Dashed blue line outlines pharynx; dashed yellow line outlines mouth; scale bars = 100 µm. n ≥ 30 animals from three independent experiments. Raw data can be found in Figure 4—figure supplement 1—source data 1.
-
Figure 4—figure supplement 1—source data 1
Raw data for feeding assay in Figure 4—figure supplement 1B.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig4-figsupp1-data1-v1.xlsx
Inhibiting stem cell division for 24 hr reduces pharynx progenitors during regeneration.
(A) Confocal images of FISH for FoxA (green) and H3P (magenta) 2 days post-pharynx amputation (dpa) in animals treated with DMSO or nocodazole beginning 1 day after amputation (schematic). DAPI = DNA (blue); arrows = double-positive cells; scale bar = 10 µm. (B) Number of FoxA+ H3P+ cells 2 days post-pharynx amputation in animals treated with DMSO or nocodazole beginning 1 day after amputation (schematic) in the area outlined by the dashed box in the cartoon. (C) Confocal images of FISH for FoxA (green) and piwi-1 (magenta) in intact animals treated with DMSO or nocodazole, beginning immediately after amputation for 1 day (schematic). DAPI = DNA (blue); arrows = double-positive cells; scale bar = 10 µm. (D) Number of FoxA+ piwi-1+ cells in intact animals treated with DMSO or nocodazole for 1 day (schematic), in the area outlined by the dashed box in the cartoon. (E) Number of FoxA+ piwi-1+ cells 3 days post-pharynx amputation in animals treated with DMSO or nocodazole in 24 hr increments (schematic) in the area outlined by the dashed box in the cartoon. Graphs represent mean ± SD; symbols = individual animals; shapes distinguish biological replicates and **, p≤0.01; ***, p≤0.001; ****, p≤0.0001; unpaired t-test (B, D), one-way ANOVA with Tukey test (E). Raw data can be found in Figure 4—figure supplement 2—source data 1.
-
Figure 4—figure supplement 2—source data 1
Quantification of H3P+ cells (Figure 4—figure supplement 2B), and piwi-1+ cells (Figure 4—figure supplement 2D and E).
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig4-figsupp2-data1-v1.xlsx
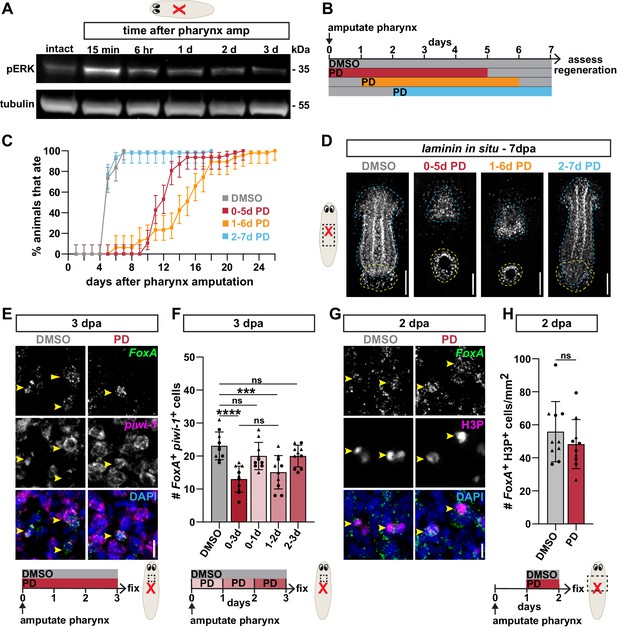
ERK phosphorylation is required to produce pharyngeal progenitors.
(A) Western blot for phosphorylated ERK (pERK) and tubulin (loading control) in intact animals and at the indicated times after pharynx amputation. (B) Schematic of PD0325901 (PD) exposure relative to pharynx amputation for graph in C and images in D. (C) Proportion of animals capable of feeding after pharynx amputation, treated as indicated in B and assayed daily. Error bars represent ± 95% confidence intervals. n ≥ 47 animals from three independent experiments. (D) Whole-mount FISH for the pharynx marker laminin 7 days post-pharynx amputation (dpa) in animals treated as indicated in B. Dashed blue line outlines pharynx; dashed yellow line outlines mouth. Scale bars = 100 µm. n ≥ 18 animals from two independent experiments. (E) Confocal images of FISH for FoxA (green) and piwi-1 (magenta) 3 days post-pharynx amputation in animals treated with DMSO or PD (schematic). DAPI = DNA (blue); dashed box = region imaged; arrows = double-positive cells; scale bar = 10 µm. (F) Number of FoxA+ piwi-1+ cells 3 days post-pharynx amputation after indicated treatments (schematics E and F). (G) Confocal images of FoxA FISH (green) and H3P antibody (magenta) 2 days post-pharynx amputation in animals treated with DMSO or PD, 1 day after amputation for 24 hr. DAPI = DNA (blue); arrows = double-positive cells; scale bar = 10 μm. (H) Number of FoxA+ H3P+ cells 2 days post-pharynx amputation in animals treated with DMSO or PD (schematic). Bar graphs represent mean ± SD; symbols = individual animals; shapes distinguish biological replicates and; ***, p≤0.001; ****, p≤0.0001; one-way ANOVA with Tukey test (F), unpaired t-test (H). Raw data can be found in Figure 5—source data 1.
-
Figure 5—source data 1
Original, uncropped images of western blots in Figure 5A.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig5-data1-v1.zip
-
Figure 5—source data 2
Raw data for feeding assay (Figure 5C), quantification of piwi-1+ cells (Figure 5F) and H3P+ cells (Figure 5H).
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig5-data2-v1.xlsx
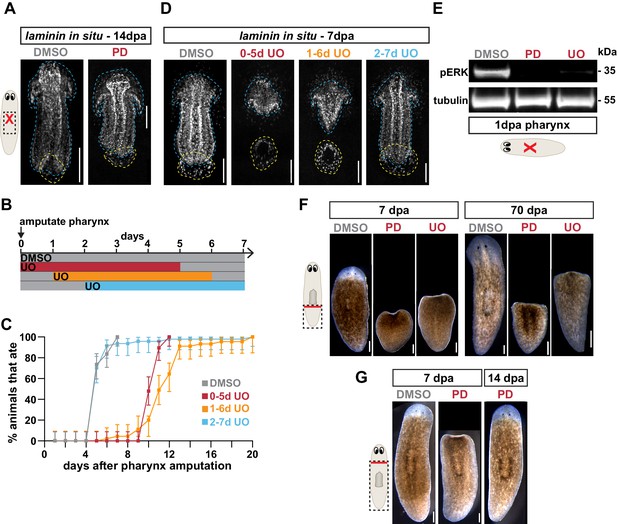
MEK inhibitors U0126 and PD0325901 prevent ERK phosphorylation and tissue regeneration.
(A) Whole-mount FISH for the pharynx marker laminin 14 days post-pharynx amputation (dpa) in animals, treated with DMSO or PD0325901 (PD) for 5 days beginning immediately after amputation. Dashed blue line outlines pharynx; dashed yellow line outlines mouth; scale bars = 100 µm. n ≥ 33 animals from three independent experiments. (B) Schematic of UO126 (UO) exposure relative to pharynx amputation for graph in C and images in D. (C) Proportion of animals capable of feeding after pharynx amputation, treated as indicated in B and assayed daily. Error bars represent ± 95% confidence intervals. n ≥ 45 animals from three independent experiments. (D) Whole-mount FISH for the pharynx marker laminin 7 days post-pharynx amputation in animals, treated as indicated in B. Dashed blue line outlines pharynx; dashed yellow line outlines mouth; scale bars = 100 µm. n ≥ 14 animals from two independent experiments. (E) Western blot for phosphorylated ERK (pERK) and tubulin (loading control) 1 day post-pharynx amputation in animals treated with DMSO, PD, or UO beginning immediately after amputation. (F) Tail fragments of planarians treated with DMSO, PD, or UO for 5 days beginning immediately after amputation, and imaged 7 or 70 days after amputation. Scale bars = 250 µm. n ≥ 28 animals from three independent experiments. (G) Planarians treated with DMSO or PD for 5 days beginning immediately after head amputation, and imaged 7 or 14 days post-amputation. Scale bars = 250 µm. n ≥ 22 animals from two independent experiments. Raw data can be found in Figure 5—figure supplement 1—source data 1.
-
Figure 5—figure supplement 1—source data 1
Raw data for feeding assay in Figure 5—figure supplement 1C.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig5-figsupp1-data1-v1.xlsx
-
Figure 5—figure supplement 1—source data 2
Original, uncropped images of western blots in Figure 5—figure supplement 1E.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig5-figsupp1-data2-v1.zip
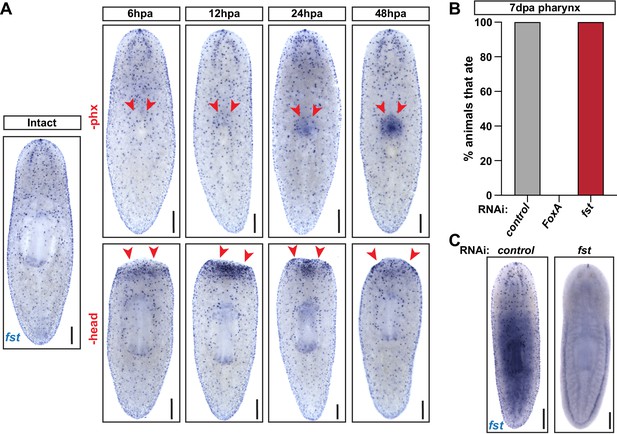
ERK-dependent pharynx regeneration is independent of follistatin.
(A) Follistatin (fst) WISH in intact and injured animals, at the indicated hours post- pharynx or head amputation (hpa). Arrows = amputation site; scale bars = 250 µm. (B) Proportion of animals capable of feeding 7 days post-pharynx amputation (dpa), in control (unc-22), FoxA(RNAi) and fst(RNAi) animals. n ≥ 21 animals from two independent experiments. (C) fst WISH showing expression after knockdown. Scale bars = 250 µm. Raw data can be found in Figure 5—figure supplement 2—source data 1.
-
Figure 5—figure supplement 2—source data 1
Quantification of feeding behavior in Figure 5—figure supplement 2B.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig5-figsupp2-data1-v1.xlsx
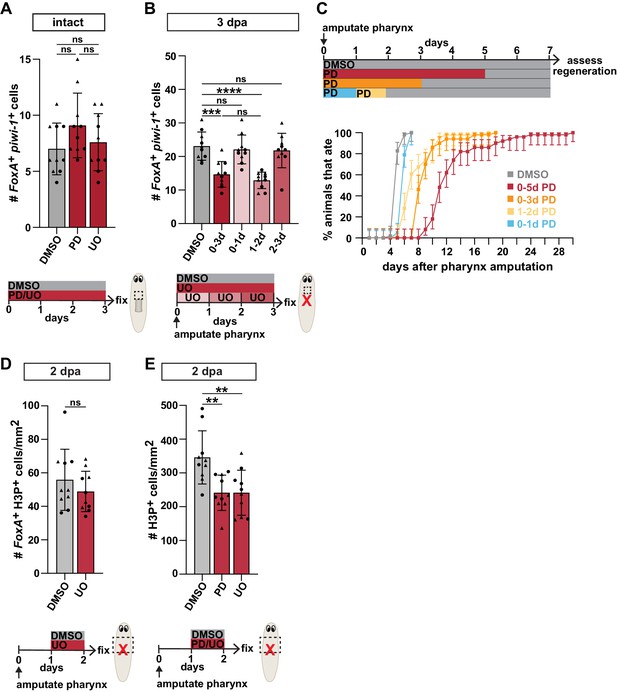
Inhibiting ERK phosphorylation reduces pharynx progenitors during regeneration.
(A) Number of FoxA+ piwi-1+ cells in intact animals, treated with DMSO, PD0325901 (PD) or U0126 (UO) for 3 days (schematic). (B) Number of FoxA+ piwi-1+ cells 3 days post-pharynx amputation (dpa) after treatments with DMSO or UO (schematic). (C) Proportion of animals capable of feeding after pharynx amputation, treated with DMSO or PD as indicated (schematic) and assayed daily. Error bars represent ± 95% confidence intervals. n ≥ 46 animals from three independent experiments. (D) Number of FoxA+ H3P+ cells 2 days post-pharynx amputation in animals treated with DMSO or UO, 1 day after amputation for 24 hr (schematic). (E) Number of H3P+ cells 2 days post-pharynx amputation in animals treated with DMSO, PD, or UO, 1 day post-pharynx amputation for 24 hr (schematic). Bar graphs represent mean ± SD; symbols = individual animals; shapes distinguish biological replicates and; **, p≤0.01; ***, p≤0.001; ****, p≤0.0001; one-way ANOVA with Tukey test (A,B,E), unpaired t-test (D). Raw data can be found in Figure 5—figure supplement 3—source data 1.
-
Figure 5—figure supplement 3—source data 1
Raw data for quantification of piwi-1+ cells (Figure 5—figure supplement 3A and B), feeding assay (Figure 5—figure supplement 3C), and H3P+ cells (Figure 5—figure supplement 3D and E).
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig5-figsupp3-data1-v1.xlsx
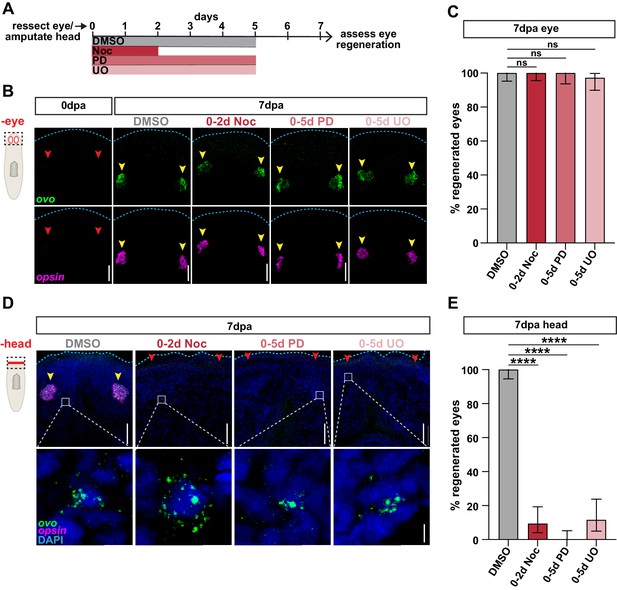
Stem cell division and ERK activation are not required for eye regeneration following selective removal.
(A) Schematic of drug treatment relative to injuries for B-E. (B) Confocal images of FISH for ovo (green) and opsin (magenta) immediately (0dpa), or 7 days post-eye resection in animals treated as in A. Dashed box in cartoon represents area imaged. Blue dashed line outlines anterior edge of worm; red arrows = missing eyes; yellow arrows = regenerated eyes; scale bars = 50 µm. (C) Proportion of ovo+ opsin+ eyes that regenerated 7 days post-eye resection. n ≥ 34 animals from three independent experiments. (D) Confocal images of FISH for ovo (green) and opsin (magenta) 7 days post-head amputation in animals treated as in A. Dashed box in cartoon represents area in top images. DAPI = DNA (blue); blue dashed line outlines anterior edge of worm; red arrows = missing eyes; yellow arrows = regenerated eyes; scale bars = 50 µm. Bottom = zoom of ovo+ cells from the regions outlined by gray boxes in top images. Scale bars = 5 µm. (E) Proportion of ovo+ opsin+ eyes that regenerated 7 days post-head amputation. n ≥ 32 animals from three independent experiments. In graphs, error bars = ± 95% confidence intervals and; ****, p≤0.0001; Fisher's Exact Test. Raw data can be found in Figure 6—source data 1.
-
Figure 6—source data 1
Quantification of eye regeneration in Figure 6C and E.
- https://cdn.elifesciences.org/articles/68830/elife-68830-fig6-data1-v1.xlsx
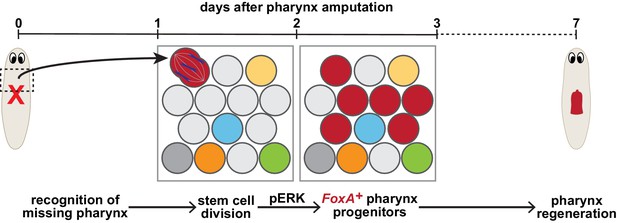
Model for targeted pharynx regeneration.
Soon after pharynx loss, stem cells recognize the pharynx is missing and target regeneration toward the pharynx by selectively inducing division of existing FoxA-expressing stem cells (red), or expression of FoxA in division-competent stem cells, within 1–2 days. This division drives an ERK-dependent increase in pharynx progenitors 3 days after pharynx loss, which is required for pharynx regeneration.
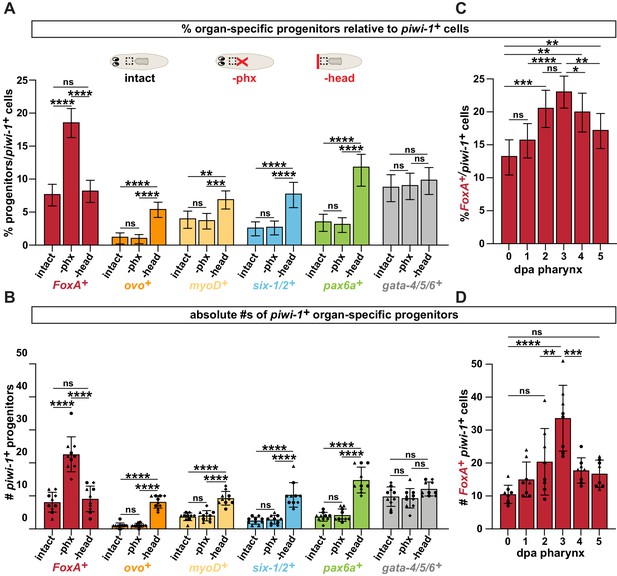
Pharynx loss selectively increases pharynx progenitors in proportion to stem cells.
(A) Proportion of cells double positive for the indicated progenitor marker and piwi-1+ relative to all piwi-1+ stem cells in the area outlined by dashed boxes in cartoons. Cartoons depict different amputation conditions. n ≥ 790 cells per experimental group from 3 independent experiments. (B) Average number of FoxA+ piwi-1+ cells in the same animals and regions as A. Same data as is in Figure 2B, E of manuscript. (C) Proportion of FoxA+ piwi-1+ cells at indicated times post-pharynx amputation relative to all piwi-1+ stem cells in the area outlined by dashed boxes in A. n ≥ 631 cells per experimental group from 3 independent experiments. (D) Average number of FoxA+ piwi-1+ cells in the same animals and regions analyzed as C. Same data as in Figure 2C of manuscript. For all graphs a 6000μm2 region in the same location of the pre-pharyngeal region was analyzed over 20 z-sections, as represented by dashed boxes in A. Graphs represent a proportion ± 95% confidence intervals (A, C) or the mean ± SD with symbols = individual animals; shapes distinguish biological replicates (B, D). *, p ≤ 0.05 **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001, Fisher’s Exact Test (A, C) or one-way ANOVA with Tukey test (B, D).
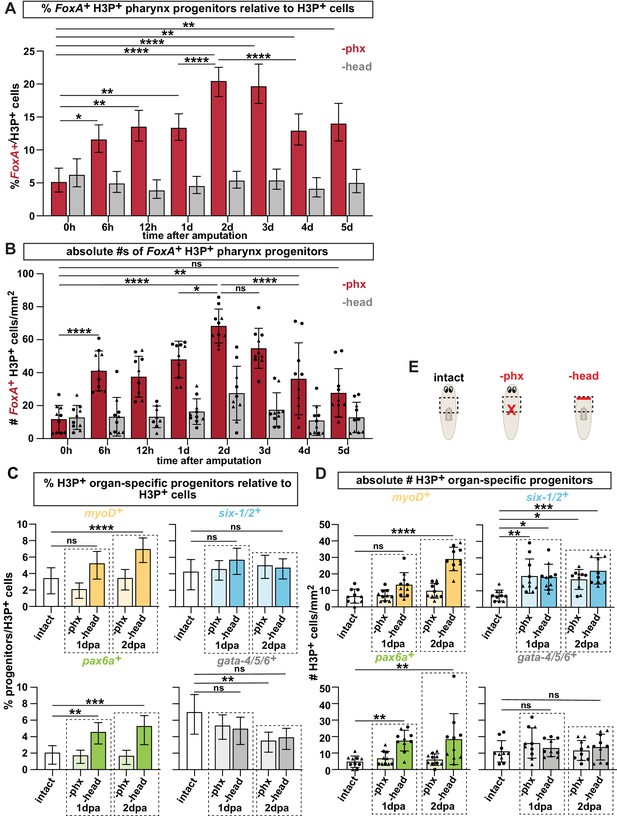
Pharynx tissue loss selectively increases mitotically active pharynx progenitors.
(A) Proportion of FoxA+ H3P+ cells relative to all H3P+ stem cells at indicated times after pharynx or head amputation in the area outlined by dashed boxes in E. n ≥ 515 cells per experimental group from 2 independent experiments. (B) Average number of FoxA+ H3P+ cells quantified in the same animals and regions as A. Same data as is in Figure 3C, D of manuscript. (C) Proportion of cells double-positive for the indicated progenitor marker and H3P+ relative to all H3P+ stem cells in the area outlined by dashed boxes in E. n ≥ 472 cells per experimental group from 2 independent experiments. (D) Average number of cells double-positive for the indicated progenitor marker and H3P+ quantified in the same animals and regions as C. Same data as in Figure 3E of manuscript. (E) Cartoons depicting different amputation conditions. For A-D, the entire pre-pharyngeal region was analyzed over 30 z-sections, as represented by dashed boxes. Graphs represent a proportion ± 95% confidence intervals (A, C) or the mean ± SD with symbols = individual animals; shapes distinguish biological replicates (B, D). *, p ≤ 0.05 **, p ≤ 0.01; ***, p < 0.001; ****, p ≤ 0.0001, Fisher’s Exact Test (A, C) or one-way ANOVA with Tukey test (B, D)
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-DIG-AP (sheep polyclonal) | Roche | Cat#11093274910, RRID:AB_514497 | in situ: 1:3000 |
| Antibody | Anti-DIG-POD (sheep polyclonal) | Roche | Cat# 11207733910, RRID:AB_514500 | in situ: 1:1000 |
| Antibody | Anti-DIG_FITC (sheep polyclonal) | Roche | Cat# 11426346910, RRID:AB_840257 | in situ: 1:1000 |
| Antibody | Anti-phosphohistone H3 (Ser10) (rabbit monoclonal) | Abcam | Cat# Ab32107, RRID:AB_732930 | IF: 1:1000 |
| Antibody | Anti-Oregon Green-HRP (rabbit polyclonal) | Thermo Fisher | Cat# A21253, RRID:AB_2535819 | IF: 1:1000 |
| Antibody | Anti-tubulin (mouse monoclonal) | Sigma/Millipore | Cat# T5168, RRID:AB_477579 | WB: 1:1000 |
| Antibody | Anti-Phospho-p44/42 MAPK (Erk1/2) (rabbit monoclonal) | Cell Signaling Technologies | Cat# 4370S, RRID:AB_2315112 | WB: 1:1000 |
| Antibody | Goat anti-mouse Alexa Flour 488 (polyclonal) | Thermo Fisher | Cat# A11029, RRID:AB_2534088 | WB: 1:4000 |
| Antibody | Goat anti-rabbit IRDye 800CW (polyclonal) | LI-COR | Cat# 926–32211, RRID:AB_621843 | WB: 1:20,000 |
| Chemical compound, drug | F-ara-EdU | Sigma | Cat# T511293 | dilution: 0.5 mg/mL |
| Chemical compound, drug | Oregon Green 488 azide | Thermo Fisher | Cat# O10180 | F-ara-EdU development: 100 µM |
| Chemical compound, drug | Proteinase K | Thermo Fisher | Cat# 25530049 | F-ara-EdU development: 10 µg/mL in situ: 4 µg/mL |
| Chemical compound, drug | Roche Western Blocking Reagent | Roche | Cat# 11921673001 | dilution: 0.5% |
| Chemical compound, drug | Horse serum | Sigma | Cat# H1138-500mL | dilution: 5% |
| Chemical compound, drug | Nocodazole | Sigma | Cat# M1404 | dilution: 50 ng/mL |
| Chemical compound, drug | PD0325901 | EMD Millipore/Calbiochem | Cat# 4449685 MG | dilution: 10 µM |
| Chemical compound, drug | UO126 | Cell Signaling Technologies | Cat# 9903S | dilution: 25 µM |
| Chemical compound, drug | Western blot lysis buffer | Zanin et al., 2011 | PMID:22118282 | |
| Chemical compound, drug | Pierce Protease Inhibitor | Thermo Fisher | Cat# A32965 | |
| Chemical compound, drug | Pierce Phosphatase Inhibitor | Thermo Fisher | Cat# A32957 | |
| Chemical compound, drug | Bolt LDS sample buffer | Life Technologies | Cat# B0007 | |
| Chemical compound, drug | Bolt 4–12% Bis-Tris polyacrylamide gel | Invitrogen | Cat# NW04125BOX | |
| Chemical compound, drug | Odyssey blocking buffer | LI-COR | Cat# 927–40000 | |
| Software, algorithm | GraphPad Prism | GraphPad Prism (https://graphpad.com) | RRID:SCR_002798 | Version 9 |
| Software, algorithm | GraphPad QuickCalcs | GraphPad QuickCalcs (https://graphpad.com/quickcalcs/) | RRID:SCR_000306 | |
| Software, algorithm | ImageJ | Image J https://imagej.net/ | RRID:SCR_003070 | |
| Other | DAPI stain 5 µg/mL | Thermo Fisher | dilution: 1:5000 | |
| Other | Aqua-Polymount | Polysciences Inc | Cat# 18606 | |
| Other | PVDF Immobilon membrane | Merck Millipore | Cat# IPFL00010 |
Additional files
-
Supplementary file 1
Table of primers and plasmids.
- https://cdn.elifesciences.org/articles/68830/elife-68830-supp1-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/68830/elife-68830-transrepform-v1.pdf





