Combining hypothesis- and data-driven neuroscience modeling in FAIR workflows
Figures
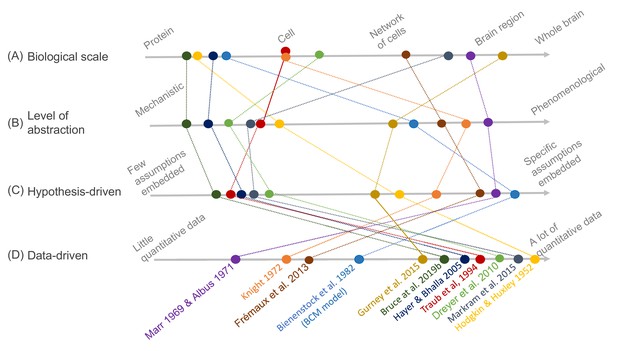
Schematic illustration of different types of models and their relative relationships.
A subset of neuroscience models are visualized based on four criteria: (A) biological scale, (B) level of abstraction, (C) the degree to which a hypothesis-driven, or (D) data-driven approach have been used for the model construction. Illustration includes examples from the text as well as several classical models: Albus, 1971 (hypothesis-driven phenomenological model of the cerebellar circuitry as a pattern recognition system); Bienenstock et al., 1982 (algebraic bidirectional synaptic plasticity rule); Bruce et al., 2019b (mechanistic molecular model of the regulation of adenylyl cyclase 5 by G-proteins in corticostriatal synaptic plasticity induction); Dreyer et al., 2010 (data-driven mechanistic model of tonic and phasic dopamine release and activation of receptors in the dorsal striatum), Frémaux et al., 2013 (hypothesis-driven phenomenological model of a reward-modulated spike-timing-dependent learning rule for an actor-critic network); Gurney et al., 2015 (basal ganglia model with corticostriatal reinforcement learning using data-driven synaptic plasticity rules); Hayer and Bhalla, 2005 (data-driven biochemical bidirectional synaptic plasticity model involving calcium/calmodulin-dependent protein kinase II [CaMKII]); Hodgkin and Huxley, 1952 (classic biophysical model of action potentials); Knight, 1972 (hypothesis-driven phenomenological model of stimulus encoding into a neuronal population); Markram et al., 2015 (large-scale digital reconstruction of somatosensory cortex microcircuitry); Marr, 1969 (cerebellar algorithm model); Traub et al., 1994 (biophysical hippocampal CA3 neuron model showing the importance of dendritic ion channels).
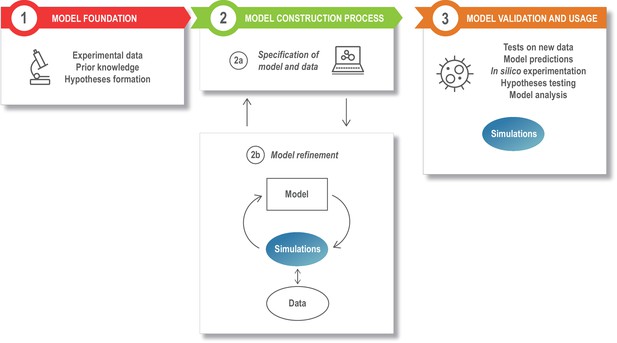
The modeling process.
Model development starts with assembling the information that is the foundation of the modeling study, such as the relevant experimental literature, published models, and additional experimental results (box 1). This is specified in a structured model and data format (box 2a), and the model is next refined and updated (box 2b). The model refinement step is an iterative process, where the model is simulated a large number of times to update the model parameters so that the model captures specific experimental data (quantitatively) or phenomena (qualitatively). Once developed, the model can be exploited for a variety of applications (box 3).
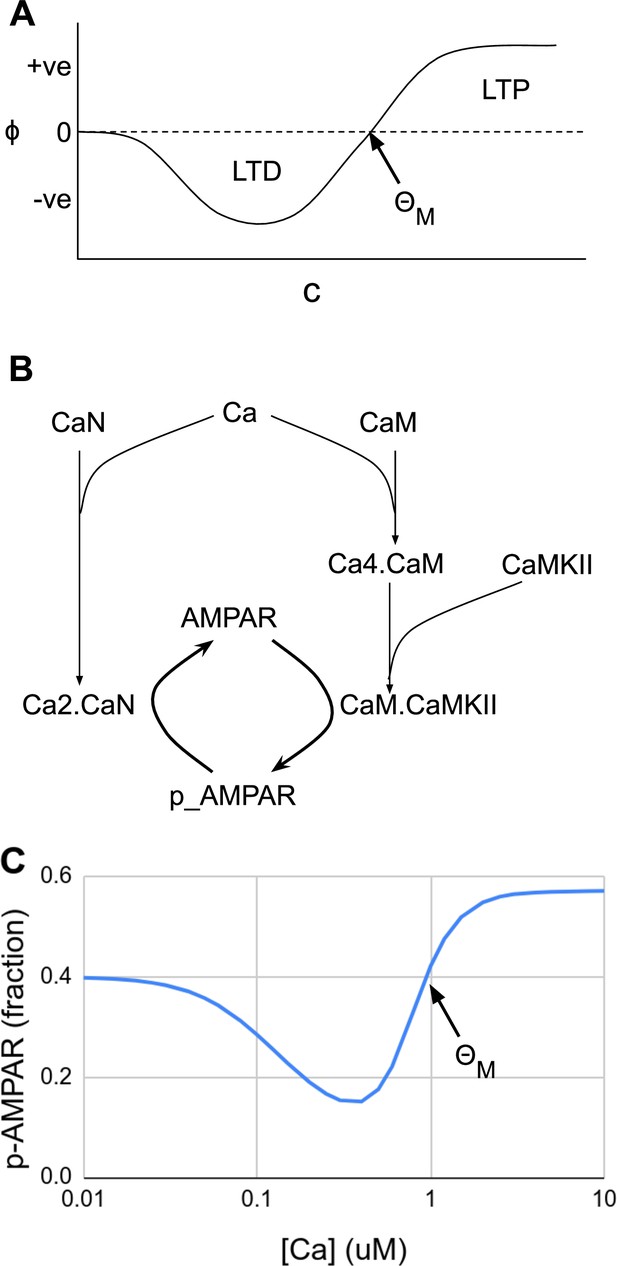
Abstract and mechanistic versions of the Bienenstock–Cooper–Munro (BCM) rule model.
(A) Original version where the rate of plasticity change, , is a function of the stimulus strength . At the threshold the sign of synaptic change flips from negative to positive. (B) Simplified mechanistic (chemical) model based on known pathways that could implement the BCM rule. The calcium stimulus activates both a kinase, CaMKII, and a phosphatase, CaN. These act in an antagonistic manner on the AMPA receptor, leading to its dephosphorylation and removal from the synapse when the calcium concentration, [Ca2+], is moderate, but insertion when [Ca2+] is high. The output from the model is p_AMPAR, the phospho-form of AMPAR, which is inserted into the membrane. (C) Simulated response of the model in Panel B, measured as phosphorylated receptor p_AMPAR. This curve has the same shape as the abstract BCM curve model in Panel A, including a threshold level of [Ca2+] = at which the synaptic change as measured by AMPA phosphorylation changes sign from negative to positive. The basal fraction of p_AMPAR is 0.4. Model from accession 96 on DOQCS (see Table 1, Row 15).

Workflow for uncertainty quantification.
An important aspect of data-driven, mechanistic modeling is to describe the uncertainty in the parameter estimates (inverse uncertainty quantification), that is to find and describe the parameter space that provides a good fit with the selected data. This is often done through Bayesian methodology starting from a model structure, some quantitative data that can be mapped to the output from the model and prior information on the parameters (like assumed ranges or distribution). The parameter space retrieved from this process is referred to as the posterior parameter distribution. This uncertainty is propagated (forward uncertainty propagation) to the predictions that we make from the model, by performing simulations from a sample of parameters representing the possible parameter distribution (corresponding to an ‘ensemble model’). Finally a global sensitivity analysis can be performed based on the posterior distribution (Eriksson et al., 2019). Global sensitivity analysis are also often performed in other settings (not shown here) directly on a preassumed parameter distribution. Figure modified from Eriksson et al., 2019.
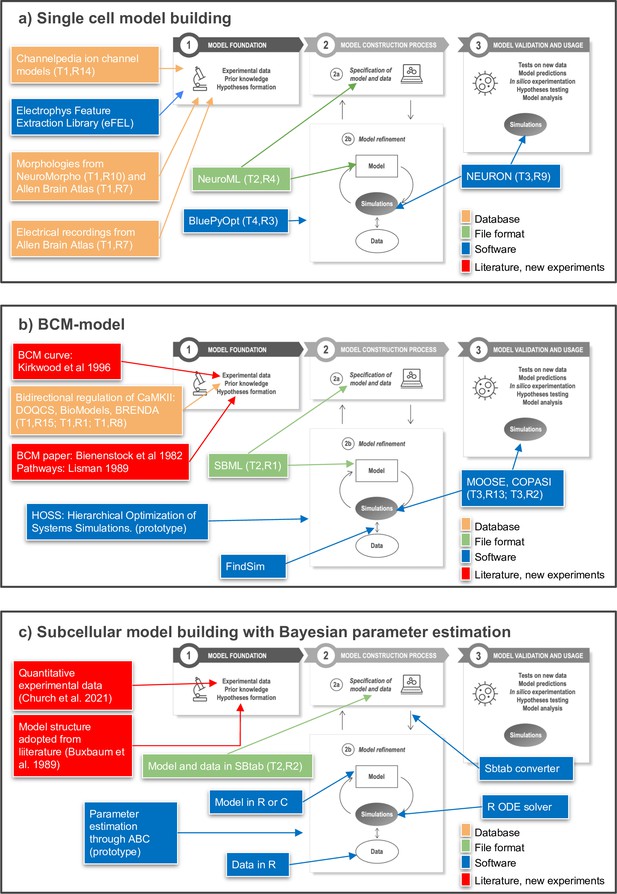
Toward FAIR (Findable, Accessible, Interoperable, Reusable) workflows in neuroscience.
Three examples of workflows at different biological scales. Several of the workflow components can be found in the tables of the article as indicated by T (Table) and R (Row) in the figure, for example T1,R4. (a) To build a single neuron multicompartmental electrical model, morphologies from databases such as NeuroMorpho or Allen Brain Atlas (http://celltypes.brain-map.org/) can be used (Ascoli et al., 2007; Gouwens et al., 2020). Electrical recordings from Allen Brain Atlas are also available. Features from experimental traces can be extracted using eFEL (https://github.com/BlueBrain/eFEL), and kinetic parameters for ion channel models obtained from Channelpedia. During the neuron model reconstruction step, the model is represented in NeuroML and model parameters such as conductance density are optimized with BluePyOpt. In silico experiments can be performed using NEURON. (b) Example workflow for building a chemical kinetic model to implement the Bienenstock–Cooper–Munro (BCM) curve. The expected shape of the curve is obtained from the classic Bienenstock et al., 1982 study, and a first pass of likely chemical pathways from Lisman, 1989. Detailed chemistry and parameters are from databases that cover models (DOQCS, BioModels) and from BRENDA, which hosts enzyme kinetics. Both model databases support SBML, which can be used to define the model and parameters. Several simulators including MOOSE and COPASI can run the SBML model during the optimization step and subsequently for model predictions. For optimization, the FindSim framework (Viswan et al., 2018) compares model outcome to experiments. The score from these comparisons is used by HOSS (Hierarchical Optimization of Systems Simulations https://github.com/upibhalla/HOSS) to carry out parameter fitting. (c) An example of a workflow used in subcellular model building with Bayesian parameter estimation. A prototype of the workflow was used in Church et al., 2021, where the experimental data is described. The model structure was adopted from Buxbaum and Dudai, 1989. A smaller demo version can be found at https://github.com/icpm-kth/uqsa (copy archived at https://doi.org/10.5281/zenodo.6625529). Experimental data and model structure are taken from literature and saved in the SBtab format. Scripts written in R convert the SBtab file to R code and the Bayesian parameter estimation is performed with the UQSA software written in R (https://github.com/icpm-kth/uqsa, copy archived at https://doi.org/10.5281/zenodo.6625529). The SBtab files can subsequently be updated with the refined model with new parameter estimates including uncertainties.
Tables
Databases in cellular neuroscience and systems biology.
These are some of the commonly used databases for creating and constraining models at the intracellular and cellular scale.
| Database, alphabetically | Purpose/focus | Reference | Homepage | |
|---|---|---|---|---|
| Computational models | ||||
| 1 | BioModels Database | Physiologically and pharmaceutically relevant mechanistic models in standard formats | Li et al., 2010 | http://www.ebi.ac.uk/biomodels/ |
| 2 | MoDEL Central Nervous System | Atomistic-MD trajectories for relevant signal transduction proteins | http://mmb.irbbarcelona.org/MoDEL-CNS/ | |
| 3 | NeuroML-DB | Models of channels, cells, circuits, and their properties and behavior | Birgiolas et al., 2015 | https://neuroml-db.org/ |
| 4 | NeuroElectro | Extract and compile from literature electrophysiological properties of diverse neuron types | Tripathy et al., 2014 | https://neuroelectro.org |
| 5 | ModelDB | Computational neuroscience model | McDougal et al., 2017 | https://senselab.med.yale.edu/modeldb/ |
| 6 | Ion Channel Genealogy | Ion channel models | Podlaski et al., 2017 | https://icg.neurotheory.ox.ac.uk/ |
| Experimental data | ||||
| 7 | Allen Brain Atlas | Human and mouse brain data | Lein et al., 2007 | http://www.brain-map.org/ |
| 8 | BRENDA | Enzyme kinetic data | Chang et al., 2021 | https://www.brenda-enzymes.org/ |
| 9 | CRCNS - Collaborative Research in Computational Neuroscience | Forum for sharing tools and data for testing computational models and new analysis methods | Teeters et al., 2008 | https://CRCNS.org |
| 10 | NeuroMorpho | Neuronal cell 3D reconstructions | Ascoli et al., 2007 | http://neuromorpho.org/ |
| 11 | Protein Data Bank (PDB) | 3D structures of proteins, nucleic acids, and complex assemblies | wwPDB consortium, 2019 | http://www.wwpdb.org/ |
| 12 | Sabio-RK | Curated database on biochemical reactions, kinetic rate equations with parameters and experimental conditions | Wittig et al., 2012 | http://sabio.h-its.org/ |
| 13 | Yale Protein Expression Database (YPED) | Proteomic and small molecules | Colangelo et al., 2019 | https://medicine.yale.edu/keck/nida/yped/ |
| Experimental data and models | ||||
| 14 | Channelpedia | Ion channel data and channel models | Ranjan et al., 2011 | https://channelpedia.epfl.ch/ |
| 15 | DOQCS The Database of Quantitative Cellular Signaling | Kinetic data for signaling molecules and interactions | Sivakumaran et al., 2003 | http://doqcs.ncbs.res.in/ |
| 16 | EBRAINS (including EBRAINS Knowledge Graph) | Digital research infrastructure that gathers data, models and tools for brain-related research | https://ebrains.eu (https://search.kg.ebrains.eu) | |
| 17 | FAIRDOMHub | The FAIRDOMHub is a repository for publishing FAIR Data, Operating procedures and Models for the Systems Biology community | Wolstencroft et al., 2017 | https://fairdomhub.org/ |
| 18 | Open Source Brain | A resource for sharing and collaboratively developing computational models of neural systems | Gleeson et al., 2019 | https://www.opensourcebrain.org/ |
Model standards and file formats in cellular neuroscience and systems biology.
The formats described in this table allow standardized representation of models and their porting across simulation platforms.
| Name | Purpose | Webpage | Reference | |
|---|---|---|---|---|
| Formats for intracellular models in systems biology | ||||
| 1 | SBML | Systems biology markup language, for storing and sharing models | https://sbml.org | Hucka et al., 2003 |
| 2 | SBtab | Systems Biology tables, for storing models (and data for parameter estimation) in spreadsheet form | https://sbtab.net/ | Lubitz et al., 2016 |
| 3 | CellML | Store and exchange mathematical models, primarily in Biology | https://www.cellml.org/ | Kohl et al., 2001 |
| Formats for cellular and network-level models in Neuroscience | ||||
| 4 | NeuroML | A XML-based description language that provides a common data format for defining and exchanging descriptions of neuronal cell and network models | http://www.neuroml.org/ | Gleeson et al., 2010 |
| 5 | NineML | Unambiguous description of neuronal network models | https://github.com/INCF/nineml-spec | Raikov et al., 2011 |
| 6 | NestML | Domain-specific language for the specification of neuron models (python) | https://github.com/nest/nestml | Plotnikov et al., 2016 |
| Custom formats for specific simulators | ||||
| 7 | sbproj | Simbiology Project file | https://se.mathworks.com/products/simbiology.html | Schmidt and Jirstrand, 2006 |
| 8 | COPASI project file | COPASI native format for models and simulations | http://copasi.org/ | Hoops et al., 2006 |
| 9 | SONATA | Efficient descriptions of large-scale neural neworks | https://github.com/AllenInstitute/sonata | Dai et al., 2020 |
| 10 | JSON (HillTau) | JSON files for FindSim and HillTau model reduction method | https://github.com/BhallaLab/HillTau | Bhalla, 2020 |
| 11 | MOD | Expanding NEURON’s repertoire of mechanisms with NMODL | https://www.neuron.yale.edu/neuron/ | Hines and Carnevale, 2000 |
| Formats for specification of parameter estimation problems | ||||
| 12 | SBtab | Systems Biology tables, for storing both models and data for parameter estimation in spreadsheet form | https://sbtab.net/ | Lubitz et al., 2016 |
| 13 | PEtab | Interoperable specification of parameter estimation problems in systems biology | https://github.com/PEtab-dev/PEtab | Schmiester et al., 2021 |
| Formats for specification of experiments and data | ||||
| 14 | SED-ML | Simulation Experiment Description Markup Language | https://sed-ml.org | Waltemath et al., 2011 |
Software for model simulation.
These tools span a wide range of scales and levels of abstraction.
| Name | Purpose | Interchange file formats supported | Homepage | RRID | Reference | |
|---|---|---|---|---|---|---|
| Molecular level | ||||||
| 1 | BioNetGen | Rule-based modeling framework (NFsim) | BNGL, SBML | http://bionetgen.org/ | Harris et al., 2016 | |
| 2 | COPASI | Biochemical system simulator | SBML | http://copasi.org/ | SCR_014260 | Hoops et al., 2006 |
| 3 | IQM Tools | Systems Biology modeling toolbox in MATLAB; successor to SBPOP | SBML | https://iqmtools.intiquan.com/ | ||
| 4 | MCell | Simulation tool for modeling the movements and reactions of molecules within and between cells by using spatially realistic 3D cellular models and specialized Monte Carlo algorithms | SBML | https://mcell.org/ | SCR_007307 | Stiles et al., 1996; Stiles and Bartol, 2001, Kerr et al., 2008 |
| 5 | NeuroRD | Stochastic diffusion simulator to model intracellular signaling pathways | XML | http://krasnow1.gmu.edu/CENlab/software.html | SCR_014769 | Oliveira et al., 2010 |
| 6 | Simbiology | MATLAB’s systems biology toolbox (Mathworks) | sbproj | https://www.mathworks.com/products/simbiology.html | Schmidt and Jirstrand, 2006 | |
| 7 | STEPS | Simulation tool for cellular signaling and biochemical pathways to build systems that describe reaction–diffusion of molecules and membrane potential | SBML | http://steps.sourceforge.net/STEPS/default.php | SCR_008742 | Hepburn et al., 2012 |
| 8 | VCell | Simulation tool for deterministic, stochastic, and hybrid deterministic–stochastic models of molecular reactions, diffusion and electrophysiology | SBML, CellML | https://vcell.org/ | SCR_007421 | Schaff et al., 1997 |
| Cellular level | ||||||
| 9 | NEURON | Simulation environment to build and use computational models of neurons and networks of neurons; also subcellular simulations with the reaction–diffusion module | SONATA (after conversion) for networks, but can also use NeuroML and SBML | https://neuron.yale.edu/neuron/ | SCR_005393 | Carnevale and Hines, 2009; Hines and Carnevale, 1997 |
| Network level | ||||||
| 10 | BRIAN | Simulation tool for spiking neural networks | SONATA | https://briansimulator.org/ | SCR_002998 | Goodman and Brette, 2008; Stimberg et al., 2019 |
| 11 | NEST | Simulation tools for large-scale biologically realistic neuronal networks | SONATA (after conversion) | https://www.nest-initiative.org/ | SCR_002963 | Diesmann et al., 1999 |
| 12 | PyNN | A Common Interface for Neuronal Network Simulators | SONATA | http://neuralensemble.org/PyNN/ | SCR_005393 | Davison et al., 2008 |
| Multiscale | ||||||
| 13 | MOOSE | Multiscale object-oriented simulation environment to simulate subcellular components, neurons, circuits, and large networks. | SBML, NeuroML | https://moose.ncbs.res.in/ | SCR_002715 | Ray and Bhalla, 2008 |
| 14 | NetPyNe | Multiscale models for subcellular to large network levels | NeuroML/SONATA | netpyne.org | SCR_014758 | Dura-Bernal et al., 2019 |
| 15 | PottersWheel | Comprehensive modeling framework in MATLAB | https://potterswheel.de/ | SCR_021118 | Maiwald and Timmer, 2008 | |
| 16 | SYCAMORE | Building, simulation, and analysis of models of biochemical systems | SBML | http://sycamore.h-its.org/sycamore/ | SCR_021117 | Weidemann et al., 2008 |
| 17 | The Virtual Brain | Create personalized brain models and simulate multiscale networks | hdf5, Nifti, GIFTI | https://thevirtualbrain.org/ | SCR_002249 | Sanz-Leon et al., 2015 |
Tools for model refinement and analysis.
This table exemplifies some common and new tools for parameter estimation and different types of model analyses.
| Name | Purpose | Interchange file formats supported | Homepage | RRID | Reference | |
|---|---|---|---|---|---|---|
| 1 | Ajustador | Data-driven parameter estimation for Moose and NeuroRD models. Provides parameter distributions. | CSV, MOOSE, NeuroRD | https://neurord.github.io/ajustador/ | Jedrzejewski-Szmek and Blackwell, 2016 | |
| 2 | AMICI | High-level language bindings to CVODE and SBML support. | SBML | https://amici.readthedocs.io/en/latest/index.html | Fröhlich et al., 2021 | |
| 3 | BluePyOpt | Data-driven model parameter optimization. | https://github.com/BlueBrain/BluePyOpt | SCR_014753 | Van Geit et al., 2016 | |
| 4 | PottersWheel | Parameter estimation, profile likelihood: determination of identifiabilty and confidence intervals for parameters. | SBML | https://potterswheel.de/ | SCR_021118 | Maiwald and Timmer, 2008; Raue et al., 2009 |
| 5 | pyABC | Parameter estimation through Approximate Bayesian Computation (likelihood free Bayesian approch). | PEtab, SBML via AMICI | https://pyabc.readthedocs.io/en/latest/ | Klinger et al., 2018 | |
| 6 | Simbiology | MATLAB’s systems biology toolbox (Mathworks), performs, for example, parameter estimation, local and global sensitivity analysis, and more. | SBML | https://se.mathworks.com/products/simbiology.html | Schmidt and Jirstrand, 2006 | |
| 7 | Uncertainpy | Global Sensitivity Analysis | NEURON and NEST models | https://github.com/simetenn/uncertainpy | Tennøe et al., 2018 | |
| 8 | XPPAUT/AUTO | Model analysis including phase plane analyses, stability analysis, vector fields, null clines, and more XPPAUT contains a frontend to AUTO for bifurcation analysis. | http://www.math.pitt.edu/~bard/xpp/xpp.html | SCR_001996 | Ermentrout, 2002 (XPPAUT) | |
| 9 | pyPESTO | Toolbox for parameter estimation | SBML, PEtab | https://github.com/ICB-DCM/pyPESTO/ | SCR_016891 | Stapor et al., 2018 |
| 10 | COPASI | Simulation and analysis of biochemical network models | SBML | https://copasi.org | SCR_014260 | Hoops et al., 2006 |
| 11 | PyBioNetFit | Parameterizing biological models | BNGL, SBML, BPSL | https://bionetfit.nau.edu/ | Mitra et al., 2019 | |
| 12 | Data2Dynamics | Establishing ODE models based on experimental data | SBML | https://github.com/Data2Dynamics/d2d | Raue et al., 2015 | |
| 13 | HippoUnit | Testing scientific models | HOC language | https://github.com/KaliLab/hippounit | Sáray et al., 2021 |





