The genomic consequences of hybridization
Figures
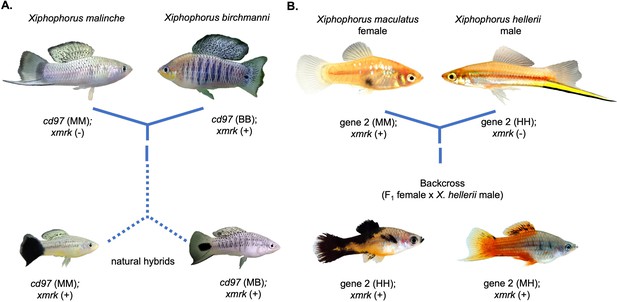
Repeated hybrid incompatibilities in Xiphophorus.
Classic models in evolutionary biology predict that incompatibilities can arise between any pair of interacting genes. Recent empirical work has suggested that certain genes or pathways may be especially prone to becoming involved in hybrid incompatibilities. The gene xmrk independently causes melanoma in hybrids between different swordtail fish species. (A) In crosses between Xiphophorus birchmanni and Xiphophorus malinche, xmrk interacts with the gene cd97 to generate melanoma in a subset of hybrids (Powell et al., 2020). (B) In crosses between distantly related species Xiphophorus maculatus and Xiphophorus hellerii, xmrk interacts with a different region, rab3d, to cause melanoma (Schartl et al., 2010; Kazianis et al., 1998; Lu et al., 2020). Phylogenetic analyses suggest that these incompatibilities with xmrk have arisen independently (Powell et al., 2020). Photos of hybrids in (B) were provided by Manfred Schartl.
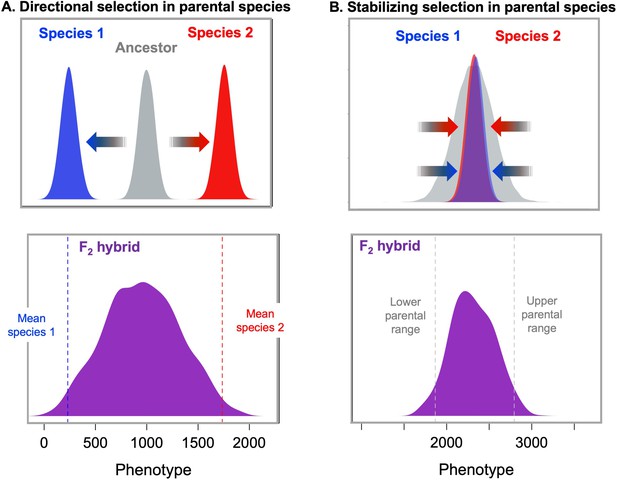
Models of selection on polygenic traits in parental species and their implications for hybrids.
(A, top) If two species have adapted from the ancestral state (gray) towards two different phenotypic optima (blue and red respectively), hybrids between those two species (purple, bottom) are predicted to fall far from the phenotypic optimum of either parental species (Barton, 2001; Thompson et al., 2019b; Thompson, 2020). The distribution shown for F2 hybrids was generated by simulating a phenotype controlled by 10 loci in each of the parental species with an exponential distribution of effect sizes, a mean trait value of 250 for parent species 1 (dashed blue line), a mean trait value of 1750 for parent species 2 (dashed red line), and additive effects at each locus on the phenotype. Simulations were performed in admix’em (Cui et al., 2016). (B, top) Similar principles apply in the case of a polygenic trait that does not differ between the parental species because it has been under stabilizing selection (Slatkin and Lande, 1994; Barton, 2001). In this case, different combinations of trait increasing and trait decreasing alleles are expected to have fixed over time in the two parental species without changing the average trait value across species. As a result, this will generate increased phenotypic variance in F2 and later generation hybrids compared to the parental species (Slatkin and Lande, 1994; Barton, 2001). These higher variance phenotypes in hybrids should be selected against via stabilizing selection. Simulations shown here illustrate this principle; F2 hybrids (purple bottom) have increased trait variance relative to the parental species. Simulations were performed as above but the average trait value was the same in the two parental species (2200). The underlying alleles and their effect sizes for this simulation were drawn from a random exponential distribution. Simulation code can be accessed on GitHub (https://github.com/Schumerlab/hybridization_review, copy archived at swh:1:rev:69a398b89365cc069c6856d990c2b74293b52486 .Schumerlab, 2021).
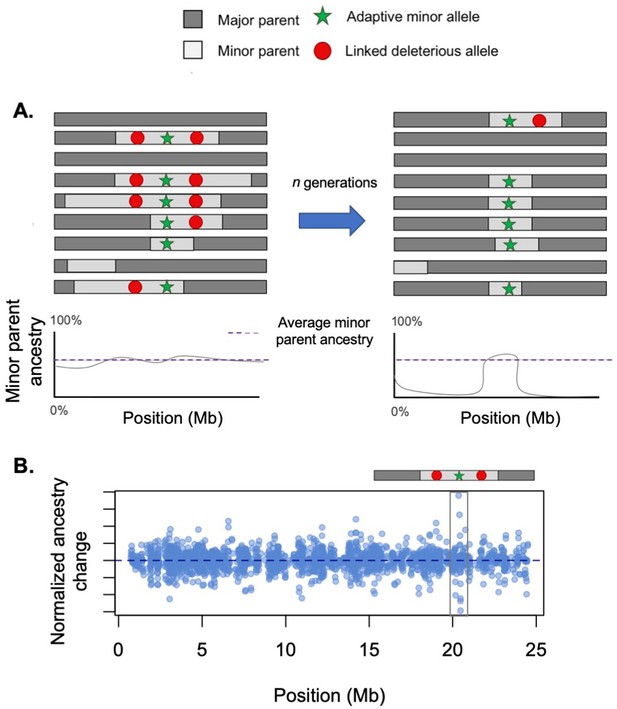
Conflicting selection between linked alleles.
Hybridization-derived haplotypes can be deleterious, neutral, or adaptive. In some cases, selection on deleterious and adaptive sites may interfere with each other. (A) Here, we illustrate a case in which there is tight physical linkage between sites that are deleterious in hybrids (such as Dobzhansky–Muller hybrid incompatibilities [DMIs]) and a site that is beneficial. Left – When positively and negatively selected sites are linked on the same haplotype, selection will act on the average of their selection coefficients. In this case, due to interference between positive and negative selection, ancestry is relatively stable in this region when selected sites are linked on the same haplotype. (Right) After a recombination event occurs and breaks apart this linkage, the positively selected haplotype will begin to rapidly increase in frequency. (B) Although not easily detectable using existing methods, such interference effects are potentially detectable using sharp transitions in ancestry over a short distance. Here, we illustrate the results of a simulation using the hybrid population simulator admix’em (Cui et al., 2016) where an adaptive locus (s = 0.05) is flanked on either side with loci deleterious in hybrids (each s = −0.05, 50 kb away). The admixture proportions simulated were 75% parent 1 and 25% parent 2, and the simulation was conducted for 200 generations. In this simulation, a haplotype arises where recombination events have unlinked the adaptive and deleterious sites, allowing the haplotype harboring the adaptive allele to begin to sweep to fixation. Long before fixation has occurred, however, the adaptive haplotype (gray box) is detectable due to the sharp ancestry change surrounding it.
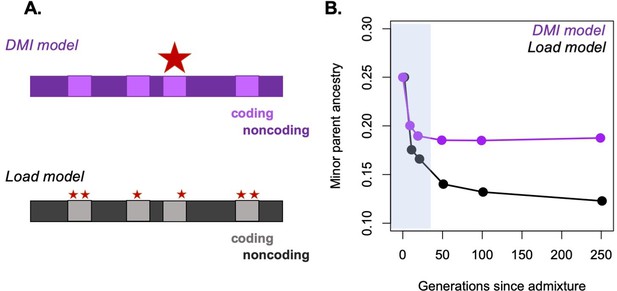
Possible approaches to differentiating between selective forces in simulations.
A major challenge in the field is distinguishing between possible sources of selection driving particular patterns of ancestry in hybrids. One promising approach is to use simulations to begin to distinguish between these possibilities. (A) As an example, we simulate ancestry change under two models, the Dobzhansky–Muller hybrid incompatibility (DMI) model and the hybridization load model after a pulse of admixture. Selected sites are shown as red stars, with the size of the star in the schematic corresponding to the strength of selection on individual sites. (B) We performed simulations using SLiM under these two models of selection on hybrids (Haller and Messer, 2019). Admixture proportions for both simulations were set at 75% parent 1 and 25% parent 2, and F1 fitness was 0.85. Ancestry was tracked on a 25 Mb chromosome in a diploid hybrid population (N = 2000). In the simulation shown in purple, selection on hybrids is driven by selection on three hybrid incompatibilities with dominance of 0.5, randomly positioned along the chromosome. In the simulation shown in black, selection on hybrids mimics a load model, with a total of 160 sites under selection along the chromosome. In this case, the selected sites are deleterious in all genetic backgrounds. The shaded area indicates the period of ‘fast’ initial purging (Principle 1), which is followed by a slower period of long-term purging in the hybridization load simulation. Although differences in the dynamics of purging between the two models are partly driven by the number of loci under selection in hybrids, the DMI model differs from other models of selection because not all minor parent alleles are disfavored (see Figure 4—figure supplement 1). Simulation code can be accessed on GitHub (https://github.com/Schumerlab/hybridization_review).
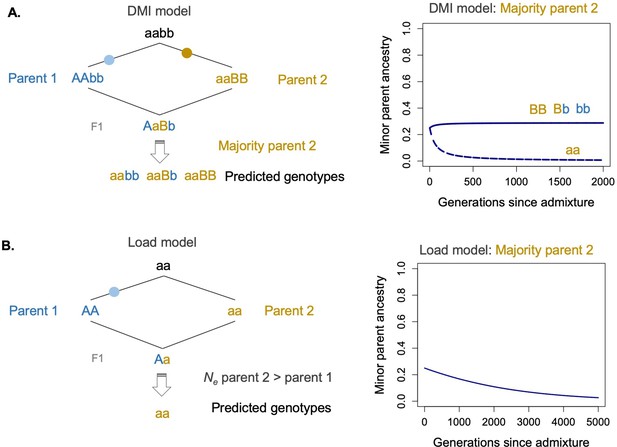
The Dobzhansky–Muller hybrid incompatibility (DMI) model and model of selection against hybridization load differ not just in assumptions about the number of sites under selection and strength of selection coefficients but also in the types of genotypes under selection.
(A) Specifically, under a two-locus DMI model, minor parent ancestry at one of the loci can be neutral or even favored by selection. This is because recombination between the parental types can regenerate ancestral (aabb) and transitional genotypes (AAbb,aaBB), which are globally compatible (left panel). Models predict that this will lead to retention of minor parent ancestry at one of the two loci involved in the DMI (Bank et al., 2012; Lewontin and Kojima, 1960, right panel). Solid line shows ancestry at locus 1 and dashed line show ancestry at locus 2 under a deterministic model of selection on hybrid incompatibilities (Lewontin and Kojima, 1960) with a starting admixture proportion of 0.25, selection coefficient of 0.1, and dominance of 0.5. (B) In contrast, when minor parent ancestry is globally deleterious at sites under selection, purging is always expected. An example is shown in the left panel of fixation of a deleterious site in parent species 1 as a result of less efficient selection due to a lower historical effective population size. After admixture (right panel), parent 1 ancestry is predicted to be purged at this locus, assuming that hybrid population sizes are sufficiently large. See the main text for a detailed discussion of this issue. Solid line shows ancestry at the selected locus with a starting admixture proportion of 0.25, selection coefficient of 0.001, and dominance of 0.5. R scripts for implementation of these models are available on GitHub (https://github.com/Schumerlab/hybridization_review).
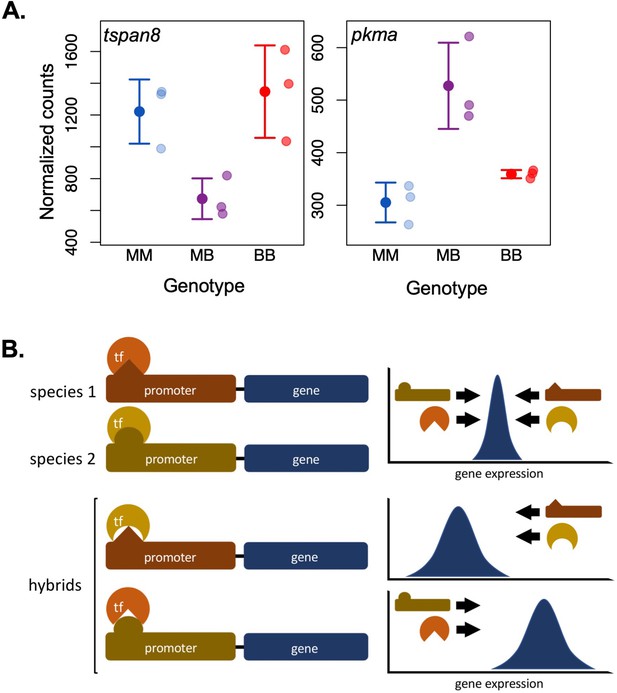
Selection on gene expression in hybrids.
Hybridization can generate mismatches between cis- and trans-acting regulatory factors that have co-evolved within the parental lineages to regulate expression of target genes around an expression optimum (i.e., through stabilizing selection). This can result in an incompatibility generated by misregulation and transgressive expression of such genes in hybrids. (A) tspan8 (left) and pkma (right) are examples of genes for which swordtail hybrids exhibit low and high misexpression, respectively (MM: X. malinche; BB: X. birchmanni; MB: F1 hybrids; data from Powell et al., 2021). (B) This simplified diagram illustrates how mismatches in co-evolved regulatory elements can cause misexpression. Promoters and transcription factors (TFs) are a classic example of cis and trans regulatory elements that interact to promote or suppress expression of target genes. Promoters and TFs can evolve to have opposing regulatory effects on target genes to achieve optimal expression (top), leading to differences in structure, interacting residues, or binding affinity between diverged populations. In hybrids, divergent binding sites within the promoter and changes in binding affinity of the TF may result in over- or underexpression of target genes, leading to misexpression (bottom).





