Regulatory T-cells inhibit microglia-induced pain hypersensitivity in female mice
Figures
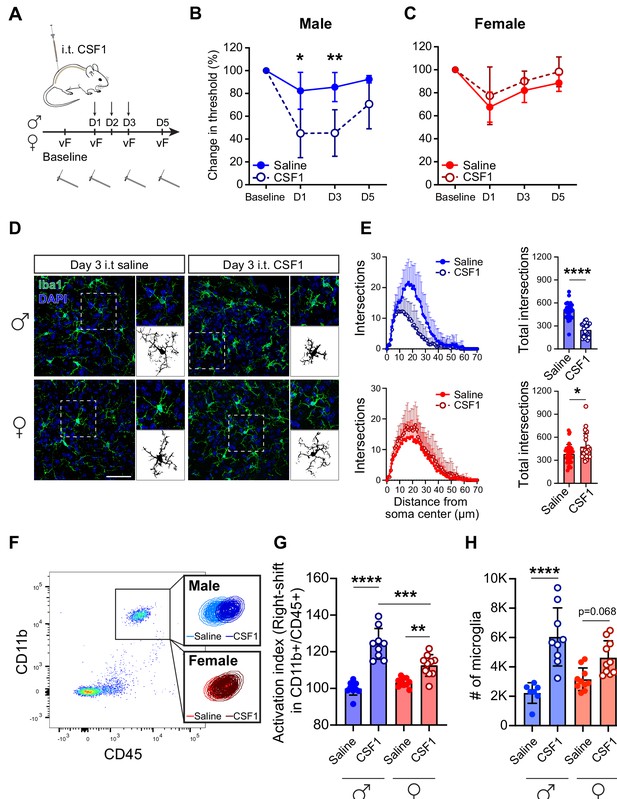
CSF1 induces pain hypersensitivity and microglial activation in male but not female mice.
(A) Schematic depicting 3 days of CSF1 intrathecal injection (i.t.) paradigm with von Frey assay. (B, C) Change in mechanical pain threshold in males and females after saline or CSF1 injection. N=5–7 mice per condition, repeated measures ANOVA. (D) Representative immunohistochemistry of lumbar spinal cord sections after 3 days of CSF1 i.t. injection. Insets indicate single microglia and binary images used for subsequent quantifications. Scale bar=50 µm. (E) Ramification calculated by Scholl analysis in males (blue, top) and females (red, bottom). N=3 mice/condition, 25 cells/group; dots represent individual microglia, Student’s t-test. (F) Representative flow cytometry plot demonstrating right-shift of the CD11b+/CD45+ population in lumbar spinal cord. Insets indicate microglia population gated on CD11b+CD45+Ly-6C−. (G) Microglial activation index calculated from flow-cytometry data as a sum of mean fluorescence intensity of CD11b and CD45 fluorescence intensity. Dots represent individual mice. One-way ANOVA with Tukey’s multiple comparisons. (H) Microglial numbers calculated by flow cytometry data. Dots represent individual mice. One-way ANOVA with Tukey’s multiple comparisons. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. CSF1, colony-stimulating factor 1.
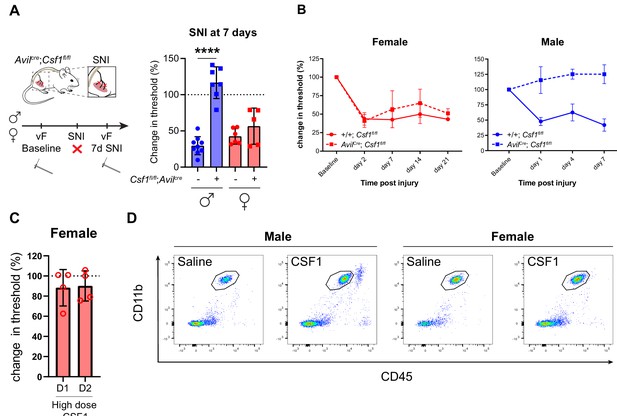
CSF1 deletion in sensory neurons rescues pain in male but not female mice.
(A) Schematic and von Frey assay for AvilCre:Csf1fl/fl at day 7 after peripheral nerve injury. Dots represent individual mice, unpaired Student’s t-test. (B) Full-time course of data summarized in (A). (C) Mechanical hypersensitivity after high dose (30 ng) CSF1 (N=4 mice/group). (D) Flow cytometry plot for CD11b and CD45 highlighting male and female microglia in the naïve and CSF1 group. One representative mouse per condition is shown. N=5 mice per group. CSF1, colony-stimulating factor 1; SNI, spared nerve injury.
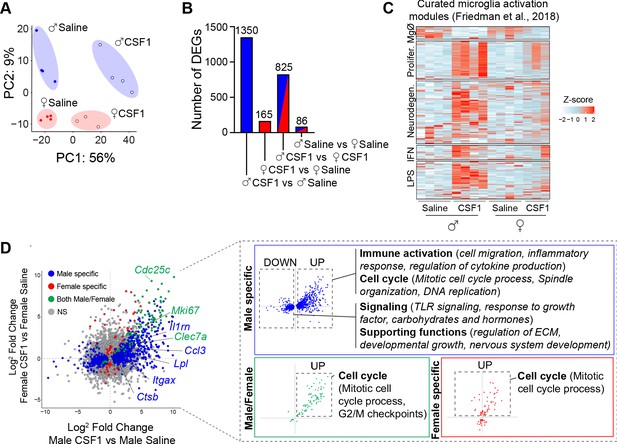
CSF1 promotes immune activation in male but not female microglia.
(A) Principal component analysis of genes expressed by microglia isolated by flow cytometry from male and female mice after 3 days of saline or CSF1 i.t. Dots represent individual mice. (B) Number of differentially expressed genes (DEGs) per comparison (adjusted p-value<0.01). (C) Heatmap of DEGs in male and female microglia after CSF1 overlaid with microglia activation modules curated by Friedman et al., 2018. (D) Four-way plot depicting DEGs (adjusted p-values<0.01) that are male-specific (blue), female-specific (red), or male-female shared (green). Inset highlights gene ontology terms identified in the respective categories. CSF1, colony-stimulating factor 1; i.t., intrathecal injection.
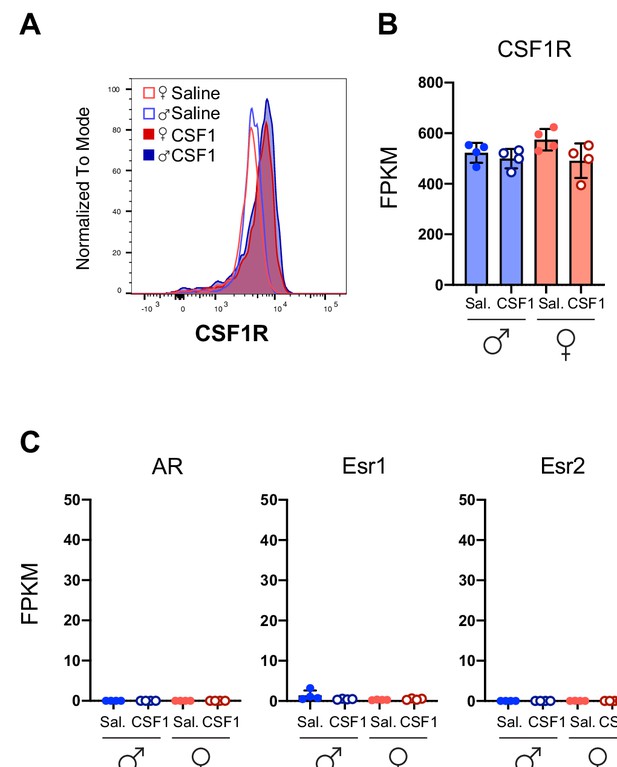
Male and female microglia express equal levels of the CSF1 receptor.
(A) Histogram of CSF1R expression on microglia for each condition by flow cytometry. One representative trace per condition. Original data from n=3 mice per group. (B) CSF1R mRNA expression in microglia as determined by RNA sequencing. Each dot represents one mouse. N=4 mice/group. CSF1, colony-stimulating factor 1; FPKM, fragments per kilobase of transcript per million.
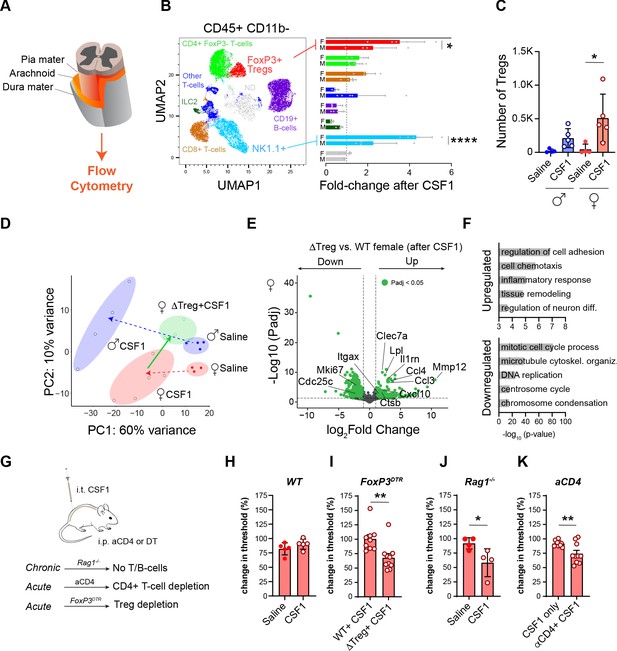
Regulatory T-cells restrict microglial activation and pain behavior in female mice.
(A) Schematic of spinal cord meninges. (B) UMAP plot of lymphoid, non-myeloid cells (CD45+CD11b−) isolated from spinal cord meninges. Image is a pool of all samples colored by cell type specific markers as indicated. Bar graph shows fold-change in indicated populations in males and females after CSF1. Dots in bar graph: individual samples. N=5 mice per group. (C) Quantification of regulatory T-cells (Tregs; CD4+FoxP3+) from (B). (D) Principal component analysis (PCA) of microglial gene expression profiles in select conditions. Red=female, blue=male, green=Treg deficient female (FoxP3DTR). Dots: individual mice. PCA consists of two experiments. The first experiment is depicted in Figure 2A and complemented with a second experiment consisting of WT females with CSF1 and Treg deficient females treated with CSF1. (E) Volcano plot depicting DEGs (adjusted p-values<0.05; green) between female Treg KO mice after CSF1 versus female mice after CSF1. N=4 mice per group. (F) Gene ontology terms for upregulated and downregulated genes from volcano plot in (E). (G) Schematic depicting the approach of using Rag1 KO mice (no T/B cells), antibody against CD4 (aCD4) to deplete T-cells and FoxP3DTR mice, in which Tregs are depleted using diphtheria toxin. (H, I) Change in mechanical hypersensitivity at day 3 after i.t. CSF1 in WT female mice (data from day 3, Figure 1B) or in females lacking regulatory T-cells (FoxP3DTR). Dots: individual mice. (J) Change in mechanical hypersensitivity at day three after CSF1 i.t. in Rag1−/−. Dots: individual mice. (K) Change in mechanical hypersensitivity at day 3 after CSF1 in female mice injected with a CD4 blocking antibody 1 day prior to CSF1 injections. Dots: individual mice. In (I–K) unpaired two-tailed t-test and (C) one-way ANOVA with Tukey’s multiple testing correction. *p<0.05, **p<0.01, ****p<0.0001. DEG, differentially expressed gene; WT, wild-type.
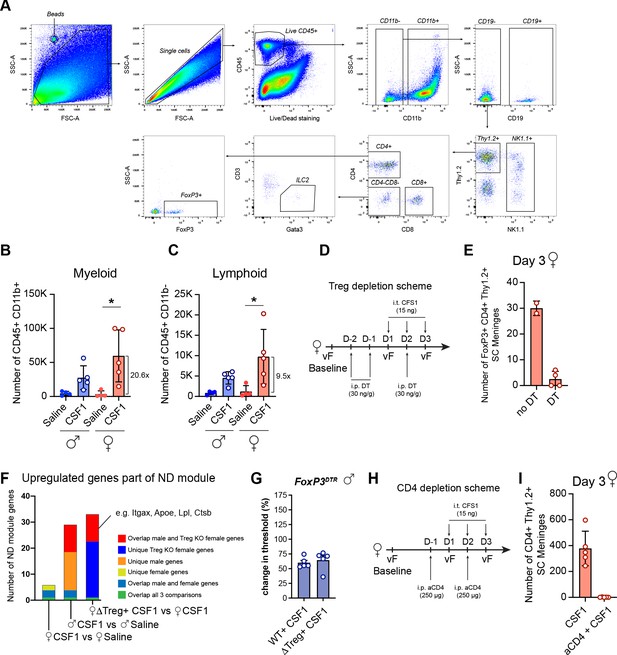
Isolation and depletion of meningeal immune cells.
(A) Flow cytometry gating strategy for spinal cord meningeal immune cells. (B, C) Quantification of myeloid and non-myeloid cells after three daily CSF1 i.t or saline injections. N=4–5 mice/group. Numbers: fold expansion in females after CSF1 over females with saline. (D) Schematic depicting the approach to deplete Tregs in combination with CSF1 injections. (E) Tregs in the SC meninges with and without depletion at day 3. Each dot represents one mouse. (F) Results of microglial sequencing, showing the upregulated ‘neurodegeneration’ related genes from per Friedman et al., 2018. Common genes are upregulated in male and female microglia after Treg depletion (red), as well as genes unique to Treg depletion in females (dark blue). (G) Change in mechanical hypersensitivity at day 3 after i.t. CSF1 in WT males and in males lacking regulatory T-cells (FoxP3DTR). Dots: individual mice. (H) Schematic depicting depletion of CD4+ T-cells in combination with CSF1 injections. (I) CD4+ T-cells in the SC meninges after CSF1, with and without CD4+ depletion at day 3. Each dot represents one mouse. CSF1, colony-stimulating factor 1; i.t., intrathecal injection; WT, wild-type.
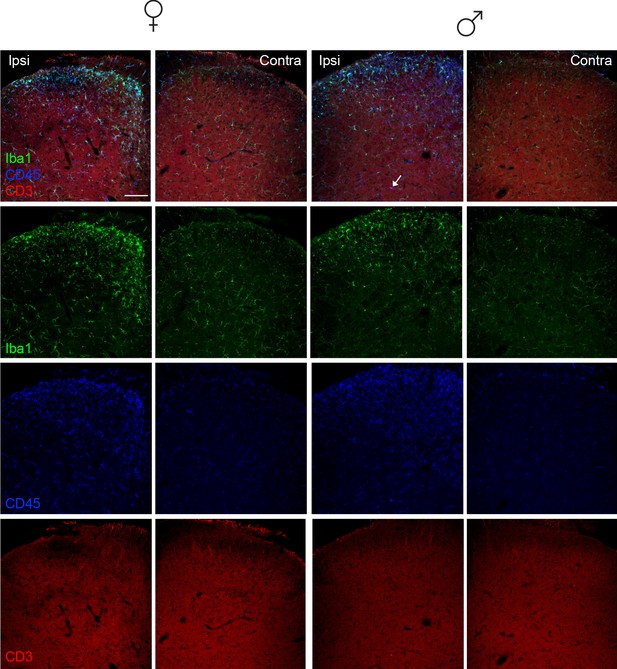
T-cells are rarely detected 7 days post SNI.
Representative immunohistochemistry of lumbar spinal cord sections for Iba1, CD45, and CD3 showing minimal to no T-cell infiltration 7 days after SNI. Scale bar=100 µm. SNI, spared nerve injury.
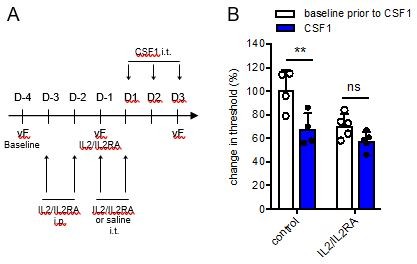
Inducing Treg proliferation with IL2/IL2RA alters mechanical withdrawal thresholds in male mice.
(A:) Schematic showing the timeline to increase Treg proliferation in mice prior to intrathecal CSF1 injections. (B:) Change in mechanical withdrawal threshold in male control and IL2/IL2RA injected mice before and after 3 days of CSF1 i.t. All thresholds are normalized to baseline thresholds prior to IL2/IL2RA treatment.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Mus musculus) | Csf1 | MGI | MGI:1339753NCBI Gene: 12,977 | |
| Gene (M. musculus) | Foxp3 | MGI | MGI:1891436NCBI Gene: 20,371 | |
| Gene (M. musculus) | Avil | MGI | MGI:1333798NCBI Gene: 11,567 | |
| Strain, strain background (M. musculus, male and female) | C57BL/6 J | The Jackson Laboratory | RRID:IMSR_JAX:000664 | |
| Strain, strain background (M. musculus, male and female) | B6.129S7-Rag1tm1Mom/J | The Jackson Laboratory | RRID:IMSR_JAX:002216 | |
| Strain, strain background (M. musculus, male and female) | B6.129(Cg)-Foxp3tm3(DTR/GFP)Ayr/J | The Jackson Laboratory | RRID:IMSR_JAX:016958 | |
| Strain, strain background (M. musculus, male and female) | AvilCre | Zurborg et al., 2011 | ||
| Strain, strain background (M. musculus, male and female) | Csf1fl/fl | Harris et al., 2012 | ||
| Peptide, recombinant protein | CSF1(M. musculus) | Thermo Fisher Scientific | Cat: #PMC2044 | 15 ng or 30 ng in 5 µl (i.t.) |
| Peptide, recombinant protein | Diphtheria Toxin(Corynebacterium diphtheriae) | Sigma-Aldrich | Cat: #D0564 | 30 ng/g in 200 µl (i.p.) |
| Antibody | Monoclonal rat anti-mouse CD4Clone: GK1.5 | Bio X Cell | Cat: #BE0003-1 | 250 µg in 200 µl (i.p.) |
| Antibody | Polyclonal Rabbit anti-mouse Iba1 | WAKO | Cat: #019-19741 | IF: (1:2000) |
| Antibody | Monoclonal Alexa 647-coupled rat anti-mouse CD45(clone 30-F11) | BioLegend | Cat: #103123 | IF: (1:200) |
| Antibody | Monoclonal hamster anti-mouse CD3 (clone 145-2C11) | BD Bioscience | Cat: #553058 | IF: (1:200) |
| Antibody | Monoclonal PE anti-mouse CD11b (clone M01/70) | eBioscience | Cat: #12-0112-81 | FACS (1:200) |
| Antibody | Monoclonal PE/Cy7 anti-mouse CD11b (clone M01/70) | eBioscience | Cat: #25-0112-81 | FACS (1:200) |
| Antibody | Monoclonal Brilliant Violet 605-conjugated anti-CD11b (M1/70) | Thermo Fisher Scientific | Cat: #BDB563015 | FACS (1:400) |
| Antibody | Monoclonal FITC anti-mouse CD45 (clone 30-F11) | eBioscience | Cat: #11-0451-81 | FACS (1:200) |
| Antibody | Monoclonal BUV395 anti-mouse CD45 (clone 30-F11) | BD Biosciences | Cat: #564279 | FACS (1:400) |
| Antibody | Monoclonal PE/Cy7 anti-mouse CD45 (clone 30-F11) | eBioscience | Cat: #25-0451-82 | FACS (1:200) |
| Antibody | Monoclonal APC anti-mouse Ly-6C (clone HK1.4) | BioLegend | Cat: #128016 | FACS (1:150) |
| Antibody | Monoclonal APC/Cy7 anti-mouse Ly-6C (clone HK1.4) | BioLegend | Cat: #128025 | FACS (1:150) |
| Antibody | Monoclonal PE anti-mouse CSF1R (clone AFS98) | BioLegend | Cat: #135505 | FACS (1:100) |
| Antibody | Monoclonal Brilliant Violet 421-conjugated anti-Thy1.2 (clone 53-2.1) | BioLegend | Cat: #140327 | FACS (1:400) |
| Antibody | Monoclonal PEDazzle594-conjugated anti-CD19 (6D5) | BioLegend | Cat: #115553 | FACS (1:400) |
| Antibody | Monoclonal Brilliant Violet 711-conjugated anti-CD4 (RM4-5) | BioLegend | Cat: #100549 | FACS (1:200) |
| Antibody | Monoclonal Brilliant Violet 785-conjugated anti-CD8a (53-6.7) | BioLegend | Cat: #100749 | FACS (1:200) |
| Antibody | Monoclonal Brilliant Violet 650-conjugated anti-NK1.1 (PK136) | BioLegend | Cat: #108735 | FACS (1:400) |
| Antibody | Monoclonal Alexa Fluor 700-conjugated anti-CD3 (17A2) | BioLegend | Cat: #100215 | FACS (1:200) |
| Antibody | Monoclonal AF488-conjugated anti-FoxP3 (FJK-16s) | eBioscience | Cat: #53-5773-82 | FACS (1:200) |
| Antibody | Monoclonal PE-conjugated anti-Gata3 (TWAJ) | eBioscience | Cat: #12-9966-42 | FACS (1:100) |
| Antibody | Monoclonal anti-mouse CD16/32 antibody | eBioscience | Cat: #14-0161-82 | FACS (1:200) |
| Commercial assay or kit | Foxp3/Transcription Factor Staining Buffer Set | eBioscience (Thermo Fisher Scientific) | Cat. no.: 00-5523-00 | |
| Commercial assay or kit | RNeasy Plus Micro Kit | Qiagen | Cat. no./ID: 74034 | |
| Commercial assay or kit | Agilent RNA 6000 Pico Kit | Agilent | Part no.: 5067-1513 | |
| Commercial assay or kit | Ovation RNA-Seq System V2 Kit | NuGen | Part no.: 7102 | |
| Commercial assay or kit | Trio RNA-Seq Kit | NuGen | Part no.: 0506 | |
| Commercial assay or kit | Qubit dsDNA HS Assay Kit | Thermo Fisher Scientific | Cat no.: Q32851 | |
| Software, algorithm | Fiji (ImageJ) | Schindelin et al., 2012 | RRID:SCR_002285 | |
| Software, algorithm | FastQC | Babraham Institute | RRID:SCR_011106 | |
| Software, algorithm | STAR(version 2.5.4b) | Dobin et al., 2013 | ||
| Software, algorithm | HTSeq(version 0.9.0) | Anders et al., 2015 | RRID:SCR_005514 | |
| Software, algorithm | DESeq2(version 1.24.0) | Love et al., 2014 | RRID:SCR_015687 | |
| Software, algorithm | Limma | Ritchie et al., 2015 | RRID:SCR_010943 | |
| Software, algorithm | Metascape | Zhou et al., 2019 | RRID:SCR_016620 | |
| Other | Zombie NIR(fixable viability dye) | BioLegend | Cat: #423105 | FACS1:1000 |
| Other | DAPI | Sigma-Aldrich | Cat: #9542 | 1:1000 |
| Other | RLT+ | Qiagen | Cat: # 1053393 |
Statistical reporting.
| Figure | N | Statistical test | Exact p-value | 95% confidence interval |
|---|---|---|---|---|
| Figure 1b | Male mice saline=3, male mice CSF1=6 | two-way ANOVA, repeated measures, Sidak’s multiple comparison | Treatment=0.0009 | D1=34.46–75.46; D3=34.13–75.13; D5=8.754–49.75 |
| Figure 1c | Female mice saline=5, female mice CSF1=5 | Two-way ANOVA, repeated measures, Sidak’s multiple comparison | Treatment=0.1890 | D1 = −30.41 to 10.58; D3=−28.58 to 12.41; D5=−30.13 to 10.86 |
| Figure 1e | 25 cells/group from 3 mice/condition | Unpaired t-test, two-tailed | Males<0.0001; females=0.0184 | Males=−309 to –195.1; females=16.78–174.6 |
| Figure 1g | Control males=10 mice, CSF1 males=9 mice, control females=10 mice, CSF1 females=10 mice | Ordinary one-way ANOVA, Tukey’s multiple comparisons | Males<0.0001; females=0.0034; male vs. female CSF1=0.0002 | Males=−31.11 to –17.53; females=−15.81 to –2.591; male vs. female CSF1=4.976–18.56 |
| Figure 1h | Male saline=7, male CSF1=9, female saline=10, female CSF1=10 | Ordinary one-way ANOVA, Tukey’s multiple comparisons | Males<0.0001; females=0.0677 | Males=−55.56 to –20.86; females=−30.01 to 77.82 |
| Figure 1—figure supplement 1a. | Female WT=6, female KO=5, male wt=9, male KO=7 | Unpaired students t-test for each sex | Females=0.2424; males< 0.0001 | Females=−11.33–39.37; males=62.83–114.3 |
| Figure 1—figure supplement 1b. | female WT = 6, female KO = 5, male WT = 3, male KO = 2 | N/A | N/A | N/A |
| Figure 1—figure supplement 1c. | 4 mice/group | Unpaired two-tailed t-test (each time point vs. baseline) | D1=0.2238; D2=0.1794 | D1=−0.2804 to 0.0804; D2=−0.2232 to 0.05217 |
| Figure 2—figure supplement 1b. | 4 mice/group | Two-way ANOVA | Interraction=0.2397, sex=0.3858, treatment=0.0501 | |
| Figure 3b | 5 mice/group | Unpaired t-test between males and females for each cell type | ||
| Figure 3c | Control=4 mice/sex, CSF1=5 mice/sex | One-way ANOVA, Tukey’s multiple comparison test | Males=0.5422; females=0.0229 | Males=−595.9 to 215.9; females=−870.4 to –58.58 |
| Figure 3h | 5 mice/group | Unpaired two-tailed t-test | 0.22 | –5.987 to 22.16 |
| Figure 3i | WT=9 mice, FoxP3DTR=10 | Unpaired two-tailed t-test | 0.01 | –54.72 to –10.71 |
| Figure 3j | 4 mice/group | Unpaired two-tailed t-test | 0.04 | –65.47 to –1.634 |
| Figure 3k | 10 mice/group | Unpaired two-tailed t-test | 0.01 | –29.61 to –5.255 |
| Figure 3—figure supplement 1b. | Saline=4 mice/group, CSF1=5 mice/group | One-way ANOVA, Tukey’s multiple comparison test | Males=0.4533; females=0.0111 | Males=−67049 to 21037; females=−100227 to –12141 |
| Figure 3—figure supplement 1c. | Saline=4 mice/group, CSF1=5 mice/group | One-way ANOVA, Tukey’s multiple comparison test | Males=0.4797; females=0.0198 | Males=−10975 to 3602; females=−15820 to –1244 |
| Figure 3—figure supplement 1e. | No DT=2, DT=4 | N/A | N/A | N/A |
| Figure 3—figure supplement 1g. | 5 mice/group | unpaired two tailed t-test | 0.2622 | –14.75 to 23.55 |
| Figure 3—figure supplement 1i. | 5 mice/group | Unpaired two-tailed t-test | 0 | –515 to –238.9 |
Additional files
-
Supplementary file 1
Transcriptomic profiling of CSF1-induced genes in microglia in males and females (Excel file).
- https://cdn.elifesciences.org/articles/69056/elife-69056-supp1-v2.xlsx
-
Supplementary file 2
FPKM values for each sample from transcriptomic profiling of males and females after saline or CSF1 (Excel file).
- https://cdn.elifesciences.org/articles/69056/elife-69056-supp2-v2.xlsx
-
Supplementary file 3
Transcriptomic profiling of CSF1-induced genes in microglia from females with and without Treg depletion (Excel file).
- https://cdn.elifesciences.org/articles/69056/elife-69056-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69056/elife-69056-transrepform1-v2.docx





