Single-molecule imaging reveals the concerted release of myosin from regulated thin filaments
Figures
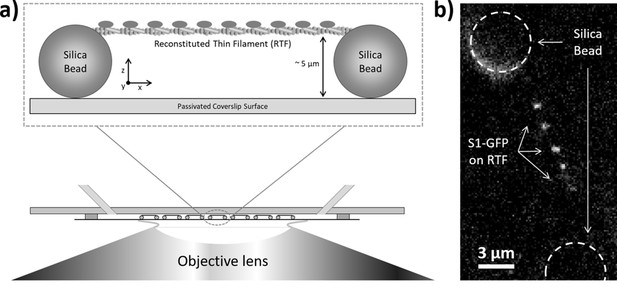
Imaging individual GFP-tagged myosins interacting with regulated thin filaments.
(a) Regulated thin filaments (RTFs) are suspended between surface-immobilised beads (top) using a microfluidic chamber (bottom). The silica beads are functionalised with poly-L-lysine that adheres them to the passivated (methoxypolyethylene glycol (mPEG)-coated) coverslip surface and to the RTFs. Illumination is achieved at an oblique angle to reduce background, and fluorescence detection occurs through the objective lens. (b) Top-down view of GFP-tagged myosin-S1 (S1-GFP) molecules bound to an RTF suspended between silica beads (dashed circles). Each pixel is 126.4 × 126.4 nm.
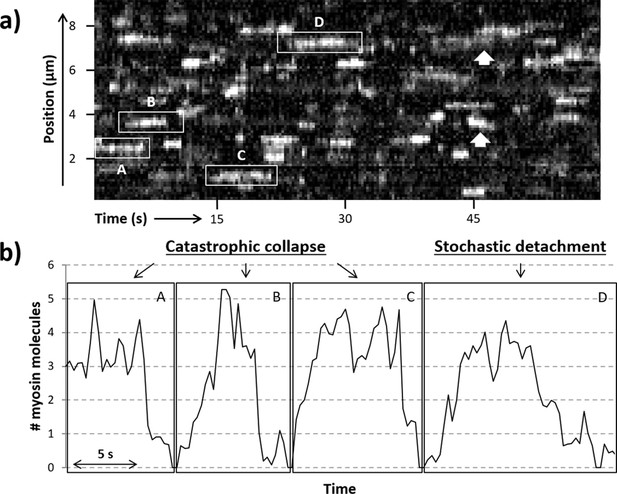
Active regions on an RTF tightrope.
(a) A representative kymograph of a regulated thin filament (RTF) in sub-maximal activating conditions, with some of the locally active regions (highlighted in boxes) labelled A to D. (b) The number of GFP-tagged myosin-S1 (S1-GFP) molecules bound in the highlighted active regions is shown. Collapse of the active region is seen in particular for areas A, B, and C, whereas region D shows both stepwise attachments and detachments. White arrows highlight example regions where the active cluster is seen to move along the thin filament. Data were obtained for 5 nM S1-GFP at pCa 6 and 0.1 µM adenosine triphosphate (ATP).
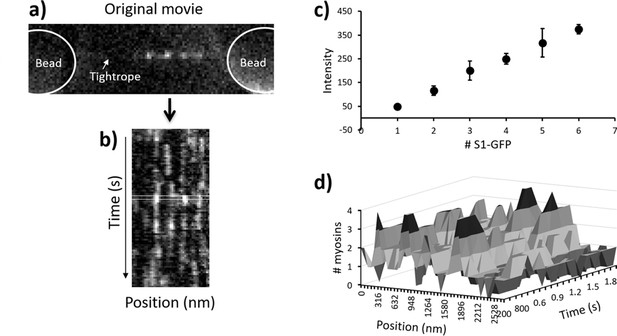
Pipeline for semi-automated data analysis.
(a) A representative snapshot from a metastable regulated thin filament (RTF), the beads being greyed out to highlight the active regions. (b) This region is transformed into a kymograph where vertical streaks indicate active regions. During analysis, a running window of one frame (rectangular box) is moved across the data through time, and each line profile of that box is fitted to a sum of multiple Gaussians using MATLAB. All fitted intensity values are plotted as a histogram and fitted again to a sum of Gaussians (see Desai et al., 2015). (c) The resulting mean intensity values are plotted to reveal a straight line (R2 = 0.999); the error bars represent 2 x SD for the Gaussian fit. The mean ± 2 x SD represents the range of intensity values used to define the presence of that specific number of myosins. These are used to rescale the kymograph into the number of myosin molecules present. (d) A section of the kymograph in (b) rescaled to the number of myosins. Subsequent analysis is used to define the myosins that belong to a specific active region (see main text). The data shown here were obtained with 10 nM GFP-tagged myosin-S1 (S1-GFP) at pCa 6 and 0.1 µM adenosine triphosphate (ATP).
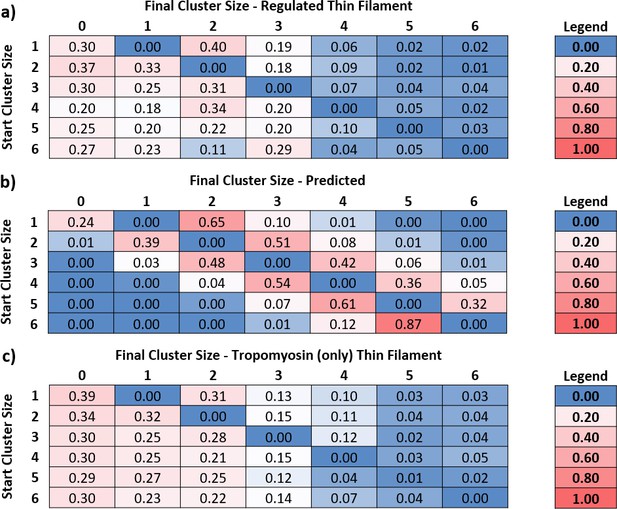
Predicted and measured transitions between cluster sizes.
By measuring the number of myosins in each cluster, the transition rates between them can be determined. (a) Using the transition rates measured for regulated thin filaments (RTFs), a transition probability table was constructed, where the rows are the starting cluster size and the columns, the final. The central diagonal is zero because this table measures the probability of leaving that cluster; all numbers to the right of the diagonal are binding events, and to the left are detachments. Data were obtained at 5 and 10 nM GFP-tagged myosin-S1 (S1-GFP), pCa 6, and 0.1 µM adenosine triphosphate (ATP) and pooled for analysis. A total of 23 kymographs were used to yield 8140 transitions for this analysis. (b) Using the probability of association empirically determined from (a) and known kinetic parameters that govern myosin release, it is possible to construct an expected table of transition probabilities (see ‘Discussion’ for more details). The overall pattern of behaviour between measured (a) and predicted (b) transition probabilities is similar to the right of the central diagonal; however, to the left, there is increased probability of myosin release. (c) The transition probabilities for tropomyosin alone follow a similar pattern to that of the regulated thin filament, indicating that the accelerated release of myosin is mediated mostly by tropomyosin. Data were obtained at 5, 15, and 20 nM S1-GFP in 0.1 µM ATP and pooled for analysis. A total of 19 kymographs were used to yield 2332 transitions for this analysis. Data are available in Figure 4—source data 1.
-
Figure 4—source data 1
Excel spreadsheet of compiled data used for Figure 4a–c.
- https://cdn.elifesciences.org/articles/69184/elife-69184-fig4-data1-v1.xlsx
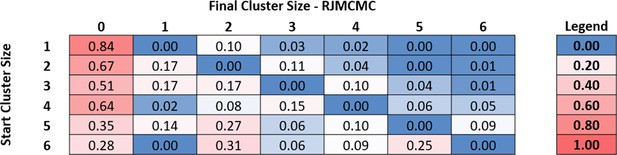
RTF transition table measured using RJMCMC.
This transition table is obtained from an independent analysis using Reversible Jump Markov Chain Monte Carlo analysis. The data depicts a similar qualitative view of the concerted release of myosins shown in Figure 4a. For implementation of this analysis see ‘Materials and Methods’.
-
Figure 4—figure supplement 1—source data 1
Excel spreadsheet containing source data for Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/69184/elife-69184-fig4-figsupp1-data1-v1.xlsx
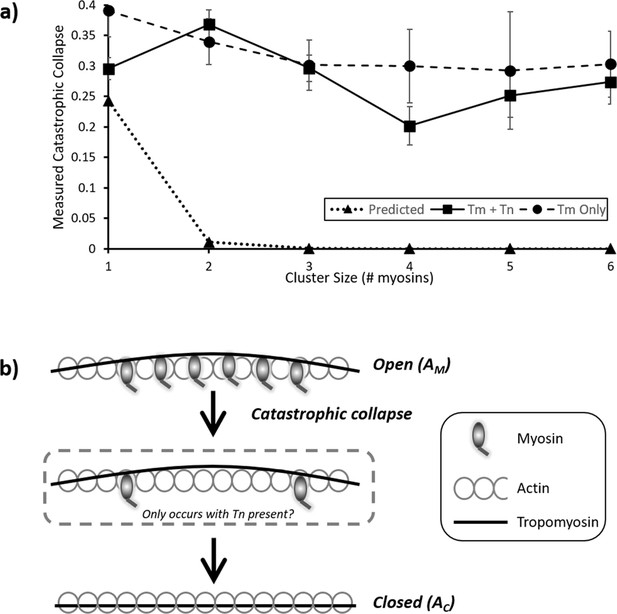
Probability and mechanism of catastrophic collapse.
(a) Plots of the values from the first columns of each table in Figure 4; this provides the probability of complete collapse. For the predicted data (triangles), an expected power law dependence correlates with cluster size is seen. However, both for the measured RTF data (squares) and tropomyosin only (circles) a much higher probability of collapse is observed. For RTFs this peaks at two and decreases marginally with larger clusters whereas Tm only shows a steady decline. Error bars represent SEM and were calculated from the values in the underlying kymographs, the number of observations is the same as for Figure 4. Data is available in Figure 5—source data 1. (b) Cartoon representation of the process of catastrophic collapse. Several myosins bind to actin holding tropomyosin in an open (AM) configuration (top). However, all myosins release in one step allowing the tropomyosin to relax into the closed (AC) state (bottom). In the presence of troponin there may be a transitory state with two myosins bound (middle).
-
Figure 5—source data 1
Excel spreadsheet of compiled data used for Figure 5a.
- https://cdn.elifesciences.org/articles/69184/elife-69184-fig5-data1-v1.xlsx
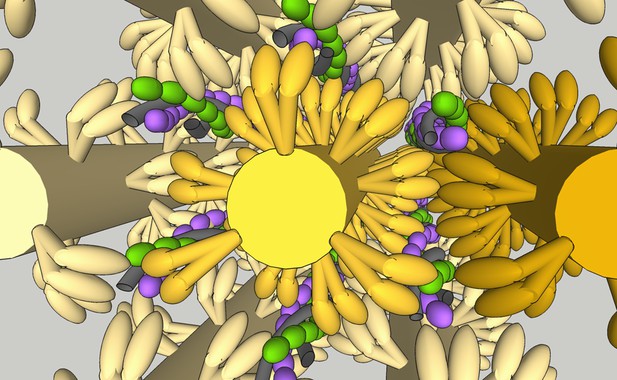
Pictorial representation of a sarcomere.
The very high density of protein within a sarcomere is clear from this image constructed from recent cryo-electron tomography (cryo-ET) measurements of the positions of the sarcomeric components (Burbaum et al., 2020; Wang et al., 2021). All components are to scale; thick filaments are shown in yellow, actin filaments in purple/green, and tropomyosin in grey. The reach of the two heads could span between thin filaments. The challenge presented by the close proximity of myosin and actin to relaxation of contraction is immediately apparent. Image is produced in Google Sketchup Make 2017 (data available in Figure 6—source data 1).
-
Figure 6—source data 1
Google Sketchup file used to create Figure 6.
This file contains an approximate to-scale representation of the sarcomere built from the data in Burbaum et al., 2020 and Wang et al., 2021. It is available to use for free on Sketchup online or with any version of Sketchup version 2017 and above. The file enables a 3D fly around the sarcomere.
- https://cdn.elifesciences.org/articles/69184/elife-69184-fig6-data1-v1.skp
Tables
The mean, variance, and weight of pixel intensity for a maximum of six or eight myosin binders.
| Total of six binders (seven components) | Total of eight binders (nine components) | |||||
|---|---|---|---|---|---|---|
| Mean = μ6 | Variance = σ6 | Weight = w6 | Mean = μ8 | Variance = σ8 | Weight = w8 | |
| Zero binders | 57.17 | 34.73 | 0.12 | 24.52 | 35.58 | 0.10 |
| One binder | 150.75 | 42.07 | 0.21 | 138.57 | 41.54 | 0.15 |
| Two binders | 198.26 | 50.68 | 0.20 | 173.87 | 46.04 | 0.16 |
| Three binders | 254.73 | 60.71 | 0.15 | 210.90 | 51.35 | 0.16 |
| Four binders | 314.23 | 69.58 | 0.14 | 264.22 | 55.51 | 0.14 |
| Five binders | 401.37 | 85.73 | 0.11 | 322.20 | 62.04 | 0.11 |
| Six binders | 551.86 | 111.40 | 0.07 | 382.67 | 80.58 | 0.08 |
| Seven binders | - | - | - | 490.73 | 72.01 | 0.05 |
| Eight binders | - | - | - | 652.38 | 91.13 | 0.04 |





