CA1 pyramidal cell diversity is rooted in the time of neurogenesis
Figures
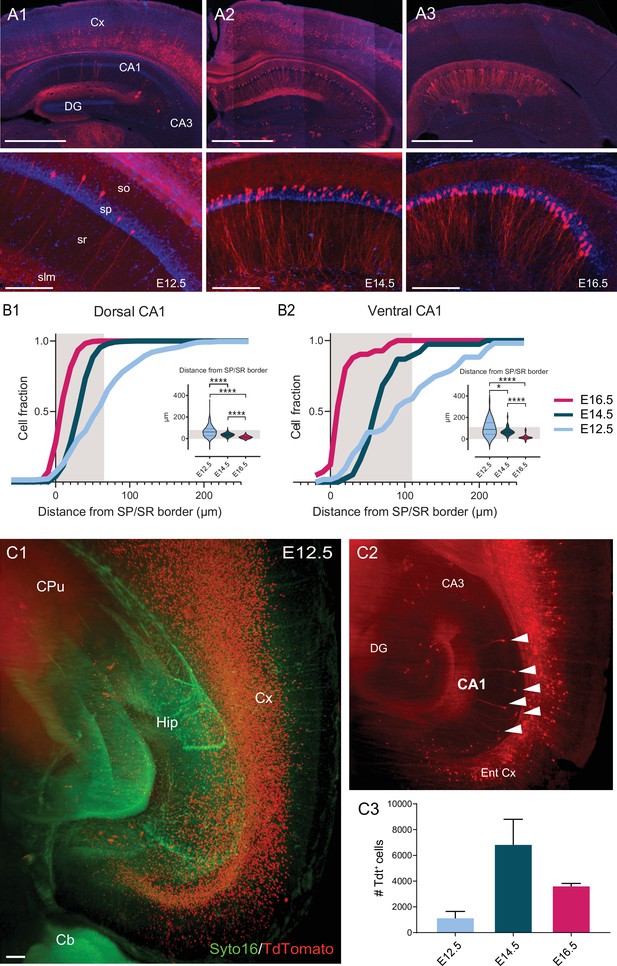
Soma location distribution of CA1PNs changes according to birthdate.
(A1-3) Top, representative sections of the dorsal hippocampus and cortex, illustrating Tdtomato (Tdt) labeling in CA1 pyramidal neurons (PNs) in Neurog2CreER-Tdt mice after tamoxifen induction at embryonic day 12.5 (E12.5), E14.5, and E16.5, from left to right. Bottom, higher magnification on CA1. E12.5 PNs are rare and dispersed, while E14.5 and E16.5 are predominantly found in the deep (upper) and superficial (lower) portion of the pyramidal layer, respectively. Note that in the above cortical areas, E12.5 PNs are restricted to the deepest layers, E14.5 PNs the middle ones (IV-III) and E16.5 PNs are most superficial (layers I-II). Scalebars: top, 1000 µm; bottom, 200 µm. Cx: Cortex; DG: dentate gyrus; so: stratum oriens; sp: stratum pyramidale; sr: stratum radiatum; slm: stratum lacunosum-moleculare. (B1-2) Quantification of the soma location distribution of fate-mapped CA1 PNs. Gray shaded areas represent the thickness of the stratum pyramidale. (B1) Cumulative fraction of E12.5, E14.5 and E16.5 PNs in dorsal CA1, calculated as distance in µm from the superficial (lower) border of the stratum pyramidale. Inset, E12.5 PNs are located further away from the border than E14.5PNs (p<0.0001, CI95% [18.18; 33]) and E16.5 PNs (p<0.0001, CI95% [41; 55.73]). In turn, E14.5PNs also occupy deeper positions than E16.5 PNs (p<0.0001, CI95% [21; 25]). (B2) Cumulative fraction of E12.5, E14.5, and E16.5 PNs in ventral CA1. Inset, similarly to dorsal CA1, in ventral CA1 E12.5 PNs are located deeper than E14.5 PNs (p: 0.0175, CI95% [3.13; 57.72]) and E16.5 PNs (p<0.0001, CI95% [53.93; 109.57]), and E14.5 PNs than E16.5 PNs (p<0.0001, CI95% [44.47; 59.32]). (C1-2) Quantification of cell abundance of fate-mapped CA1PNs. (C1) Two-dimensional view of the 3D reconstruction of a clarified hemisphere obtained by light-sheet microscopy from a Neurog2CreER-Tdt mouse induced at E12.5. In red, fate-mapped glutamatergic neurons within hippocampus (Hip) and deep layers of cortex (Cx). Green fluorescent protein Syto16 is used to visualize brain structures. Cb, cerebellum; CPu, caudate putamen; Hip, hippocampus; Cx, cortex; scale bar: 100 µm. (C2) Optical slice extracted from the same brain as in C1, with focus of hippocampal CA1 area. TdTomato-expressing PNs in CA1 are indicated by white arrows. Ent Cx, entorhinal cortex; DG, dentate gyrus. C3 Histogram representing the abundance of fate-mapped glutamatergic pyramidal neurons in the whole CA1 area (mean ± SD).
-
Figure 1—source data 1
Source data for cell distribution of fate-mapped PNs in dorsal and ventral CA1.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig1-data1-v2.csv
-
Figure 1—source data 2
Source data for cell abundance of fate-mapped CA1 PNs from whole CA1 in clarified brains.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig1-data2-v2.csv
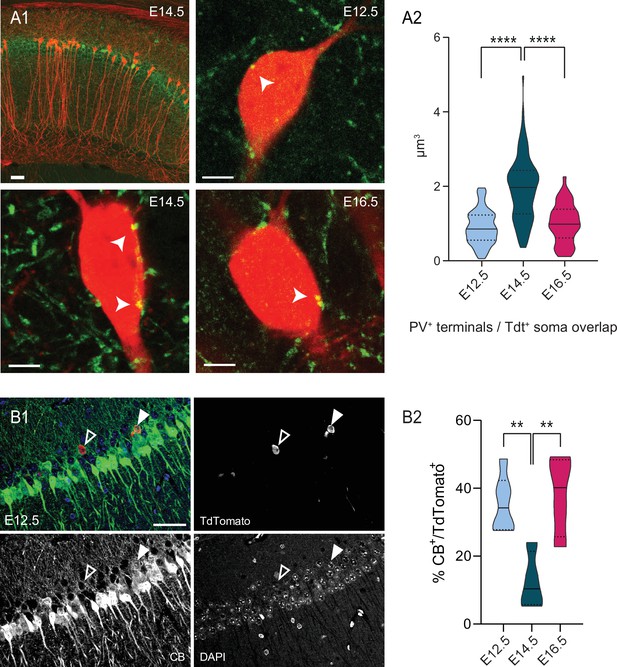
PV innervation and CB expression are biased by birthdate for pioneer cells.
(A1) Top left, representative view of the dorsal CA1 area in a Neurog2CreER-Tdt mouse tamoxifen-induced at E14.5, with Tdt+ cells (red) and PV cells (green). Top right, close-up on the soma of E12.5 PNs (red) displaying putative somatic PV boutons (indicated by a white arrow). Bottom left, close-up on E14.5 PNs. Bottom right, close-up on E16.5 PNs. (A2) Quantification of the volume overlap (µm3) of putative PV terminals with Tdt+ soma depending on the birthdate in dorsal CA1. E14.5 PNs present more putative boutons than E12.5 (p < 0.0001, CI95% [–1.264; –0.81]) and E16.5 (p < 0.0001, CI95% [0.66; 1.22]), while the latter two are not significantly different (p: 0.7038, CI95% [–0.389; 0.19]). Scalebars: top left: 50 µm; top right, bottom left and bottom right: 5 µm. (B1) Representative view of a dorsal CA1 section, after immunohistochemical staining for CB in a Neurog2CreER-Tdt mouse tamoxifen-induced at E12.5 (merge, top left), with Tdt+ cells (red, top right) and CB (green, bottom left) and DAPI (blue, bottom right). The black and white arrow indicate a CB-negative and a CB-positive E12.5 CA1PNs, respectively. Scalebar: 50 µm. (B2) Percentage of CB-expressing Tdt+-PNs depending on the birthdate in CA1. Both E12.5PNs (p:0.0084, CI95% [6.39; 39.5]) and E16.5 PNs (p: 0.0075, CI95% [–43.7; –7.4]) have a higher CB expression rate than E14.5 PNs, while no difference is detected between E12.5 and E16.5 PNs (p:0.90). One-way ANOVA with Tukey’s post hoc test, F (2, 11) = 9.2091; p:0.0045. Violin plots present medians (center), interquartile ranges (bounds), minima and maxima. Color-code: E12.5: light blue, E14.5: dark blue, E16.5: magenta. **p < 0.01, ****p < 0.0001.
-
Figure 2—source data 1
Source data for putative PV bouton volume on fate-mapped PNs in dorsal CA1.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig2-data1-v2.csv
-
Figure 2—source data 2
Source data for CB expression in fate-mapped CA1 PNs in whole CA1.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig2-data2-v2.csv
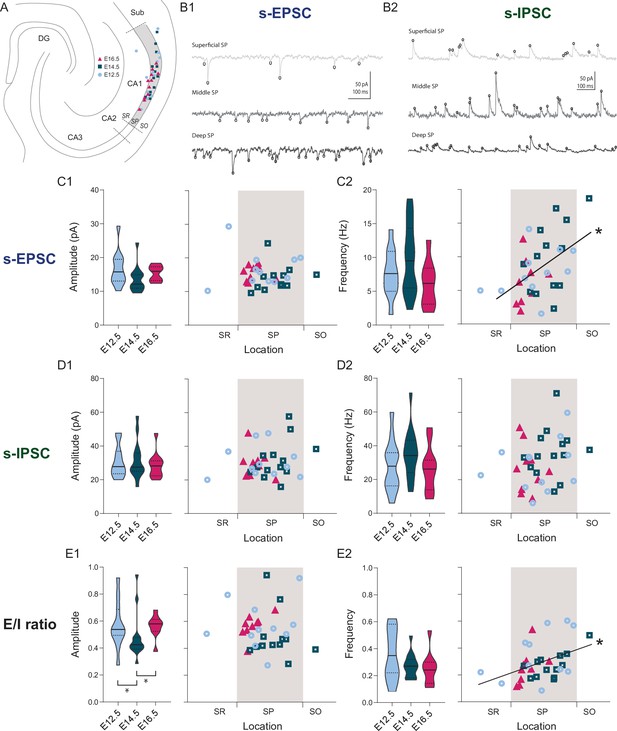
Overall synaptic drive is defined by the time of neurogenesis.
(A) Anatomical location of neurobiotin-filled CA1PNs recorded from acute horizontal slices in voltage-clamp experiments. DG: dentate gyrus, Sub: subiculum, SR: stratum radiatum, SP: stratum pyramidale, SO: stratum oriens. (B1–B2) Representative traces of spontaneous excitatory synaptic currents (s-EPSCs) and inhibitory synaptic currents (s-IPSCs) from CA1PNs located respectively in the deep, middle, and superficial portion of the pyramidal layer (SP). Black circles represent detected synaptic events. Note that the occurrence of s-EPSCs increases from top (superficial) to bottom (deep), whereas no apparent change in s-IPSC frequency can be identified among different locations. (C1) s-EPSC amplitude recorded in E12.5, E14.5, and E16.5 CA1PNs. On the left panel, violin plot of E12.5, E14.5, and E16.5 PNs s-EPSC amplitude (E12.5 vs E14.5, Padj: 0.3228; E12.5 vs E16.5, Padj:0.4012; E14.5 vs E16.5, Padj:0.3228). On the right panel, color-coded scatterplot of the same data plotted against the radial position of each cell. No linear correlation between s-EPSC amplitude and location (p: 0.4495,CI95% [–0.45; –0.44]) was found. (C2) s-EPSC frequency recorded in E12.5, E14.5, and E16.5 CA1PNs. On the left panel, violin plot of the three birthdate groups (E12.5 vs E14.5, Padj:0.3386, E12.5 vs E16.5, Padj:0.4845, E14.5 vs E16.5, Padj:0.4845). On the right panel, scatterplot of the same data against the radial position. S-EPSC frequency increases linearly with the cell location, the two being positively correlated (p: 0.017; CI95% [0.186; 0.710]). The deeper, the more frequent spontaneous excitatory events a cell receives. (D1) s-IPSC amplitude recorded in E12.5, E14.5 and E16.5 CA1PNs. On the left panel, violin plot of the three birthdate groups (E12.5 vs E14.5, Padj:0.8238; E12.5 vs E16.5, Padj>0.999; E14.5 vs E16.5, Padj>0.999). On the right panel, scatterplot of the same data against the radial position. No linear correlation between s-IPSC amplitude and location was found (p: 0.12, CI95% [–0.177; –0.492]). (D2) s-IPSC frequency recorded in E12.5, E14.5, and E16.5 CA1PNs. On the left panel, violin plot of the three birthdate groups (E12.5 vs E14.5, Padj:0.282; E12.5 vs E16.5, Padj:0.4526; E14.5 vs E16.5, Padj:0.1575). On the right panel, scatterplot of the same data against the radial position. No linear correlation between s-IPSC frequency and location was found (p: 0.067, CI95% [–0.019; –0.485]). (E1) Ratio between EPSC and IPSC (E/I) amplitude recorded in E12.5, E14.5, and E16.5 CA1PNs. On the left panel, violin plot of the three birthdate groups. E14.5 PNs display a lower ratio than E12.5 PNs (Padj: 0.010; CI95% [0.031; 0.210]) and E16.5 PNs (Padj: 0.006; CI95% [–0.193; –0.057]), suggesting that their overall synaptic drive leans more toward inhibition than the other two groups (E12.5 vs E16.5, Padj:0.2061). On the right panel, scatterplot of the same data against the radial position. No linear correlation between E/I amplitude ratio and location was found (p:0.4615, CI95% [–0.38; 0.33]). (E2) E/I frequency ratio recorded in E12.5, E14.5, and E16.5 CA1PNs. On the left panel, violin plot of the three birthdate groups (E12.5 vs E14.5, Padj: 0.61, E12.5 vs E16.5; Padj: 0.61; E14.5 vs E16.5, Padj: 0.59). On the right panel, scatterplot of the same data against the radial position. E/I ratio linearly correlates with the cell location, similarly to s-EPSC frequency (p: 0.0043; CI95% [0.126; 0.654]). Violin plots present medians (center), interquartile ranges (bounds), minima and maxima. Color-code: E12.5: light blue, E14.5: dark blue, E16.5: magenta. The gray shaded area in scatterplots represents the thickness of the stratum pyramidale. *p < 0.05.
-
Figure 3—source data 1
Source data for spontaneous synaptic currents of fate-mapped CA1 PNs.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig3-data1-v2.csv
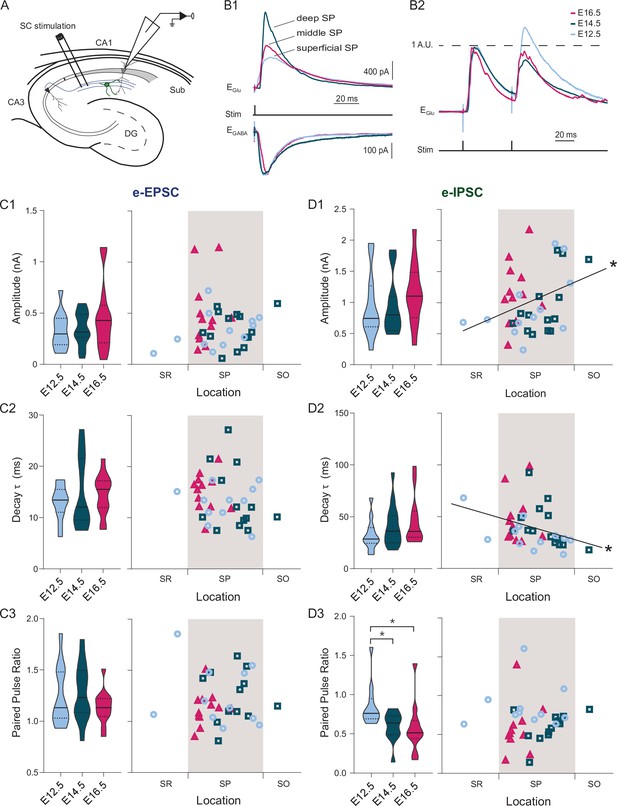
Both developmental and positional factors determine Schaffer collateral-evoked inhibition.
(A) Experimental paradigm in which synaptic responses to Schaffer collaterals (SC) stimulation are recorded from CA1PNs. DG: dentate gyrus, Sub: subiculum. (B1) Representative mean traces of evoked inhibitory (top) and excitatory (bottom) synaptic currents (e-IPSCs and e-EPSCs) recorded upon electric stimulation in the stratum radiatum from CA1PNs, located respectively in the deep, middle, and superficial portion of the pyramidal layer (SP). Note that the amplitude and kinetics of e-IPSCs vary from deep to superficial. (B2) Representative mean traces of paired pulse response recorded at glutamatergic receptor reversal potential (EGlu) and normalized on the first pulse amplitude. The current response to the second pulse in the E12.5PNS shown is proportionally larger than in E14.5 and E16.5 CA1PNs. (C1-3) e-EPSC amplitude (C1, E12.5 vs E14.5, Padj:0.617; E12.5 vs E16.5, Padj:0.5985; E14.5 vs E16.5, Padj:0.617), decay (C2, E12.5 vs E14.5, Padj:0.6165; E12.5 vs E16.5, Padj:0.4648; E14.5 vs E16.5, Padj:0.6165), and paired pulse ratio (C3, PPR; E12.5 vs E14.5, Padj:0.916; E12.5 vs E16.5, Padj:0.718; E14.5 vs E16.5, Padj:0.9165) recorded in E12.5, E14.5 and E16.5 CA1PNs. Left, violin plot of the three birthdate groups. Right, scatterplot of the same data against the radial position. No linear correlation with soma location was found in any of the three measures (e-EPSC amplitude, p:0.126; decay, 0.1315; PPR, 0.4412). (D1–D3) e-IPSC amplitude (D1, E12.5 vs E14.5, Padj:0.418; E12.5 vs E16.5, Padj:0.222; E14.5 vs E16.5, Padj:0.222), decay (D2, E12.5 vs E14.5, Padj:0.4018; E12.5 vs E16.5, Padj:0.375; E14.5 vs E16.5, Padj:0.4018), and PPR (D3) recorded in E12.5, E14.5 and E16.5 CA1PNs. Left, violin plot of the three birthdate groups. Right, scatterplot of the same data against the radial position. e-IPSC amplitude (p: 0.013; CI95% [0.048; 0.576]) and decay (p: 0.0051; CI95% [–0.599; –0.097]) display a positive and negative correlation with cell location, respectively (whereas PPR does not present any correlation, p:0.3473). Overall, deeper cells are subject to larger and faster SC-associated inhibitory currents than superficial ones. PPR is higher in E12.5 CA1PNs than in E14.5 (Padj: 0.0326; CI95% [0.015.; 0.342]) and E16.5 (Padj: 0.0177; CI95% [0.083; 0.452]), without any significant difference in E14.5 vs E16.5 (Padj:0.145). Violin plots present medians (center), interquartile ranges (bounds), minima and maxima. Color-code: E12.5 PNs: light blue, E14.5: dark blue, E16.5: magenta. The gray shaded area in scatterplots represents the thickness of the stratum pyramidale. *p < 0.05.
-
Figure 4—source data 1
Source data for evoked synaptic currents of fate-mapped CA1 PNs.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig4-data1-v2.csv
-
Figure 4—source data 2
Table summarizing linear regression analyses for PSCs excluding E12.5PNs in the SR.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig4-data2-v2.csv
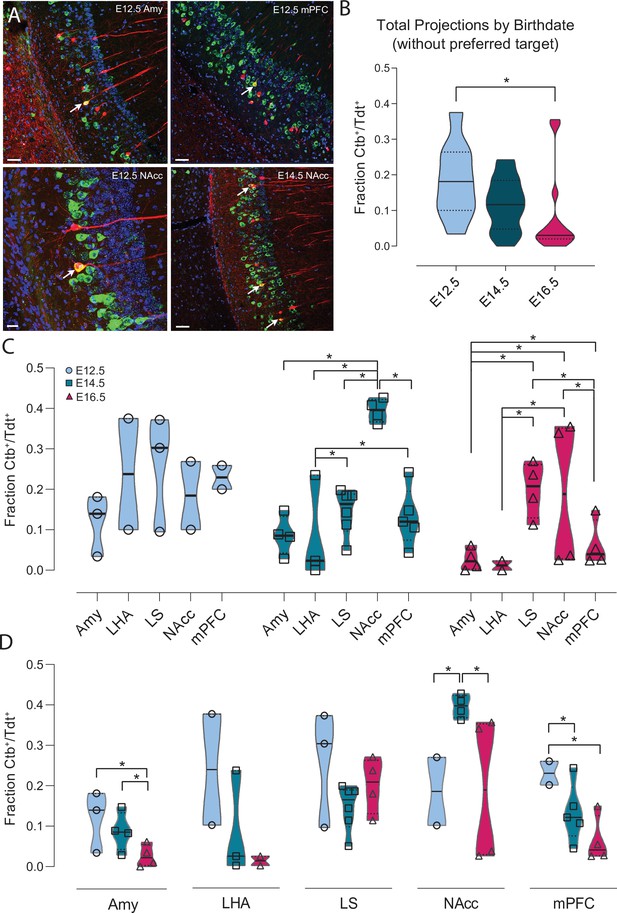
Ventral CA1 PNs with different birthdates exhibit distinct output connectivity.
(A) Examples of cholera toxin subunit b (Ctb) retrograde labeling (green) in ventral CA1 after injection in amygdala in Neurog2CreER-Tdt mouse induced at E12.5 (top left), medial prefrontal cortex in E12.5 mouse (top right), Nucleus Accumbens (NAcc) in E12.5 mouse (bottom left) and NAcc in E14.5 mouse (bottom right). Colabeled Ctb+/Tdt+ cells are indicated by a white arrow. Scalebars: top left, top right and bottom right, 50 µm; bottom left, 20 µm. (B) Quantification of the total Ctb+/Tdt+ cell fraction for the three birthdates, excluding the preferred region for each (E12.5 = lateral septum (LS), E14.5 = NAcc, E16.5 = LS). Overall, significantly more Ctb+/Tdt+ were found in E12.5 PNs than E16.5 (Padj: 0.0129, CI95% [0.013; 0.19]) while no significant difference was observed when comparing E12.5 with E14.5 (Padj: 0.070) or E16.5 PNs (Padj: 0.146). (C) Fraction of Ctb+/Tdt+ cells in E12.5, E14.5 and E16.5 PNs by target region. Note how E12.5 neurons project more homogeneously to all structures probed, while marked inter-regional differences appear among E14.5 and E16.5 neurons. (D) Fraction of Ctb+/Tdt+ cells in amygdala (Amy), lateral hypothalamic area (LHA), lateral septum (LS), nucleus accumbens (NAcc), and medial prefrontal cortex (mPFC) by time of neurogenesis. E12.5 PNs project more prominently than other birthdate groups to Amy and mPFC, while NAcc is preferentially targeted by E14.5.
-
Figure 5—source data 1
Table summarizing fate-mapped Ctb+-Tdt+ PN co-labeling data.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig5-data1-v2.csv
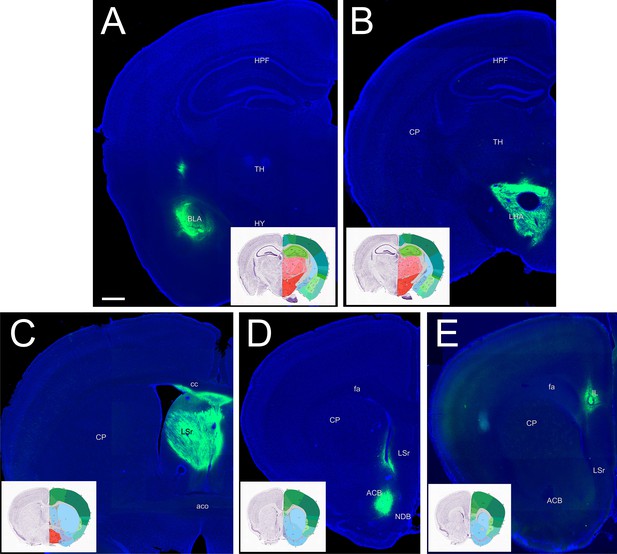
Representative sections of injections sites.
Representative sections including the injection site of Cholera toxin subunit B retrograde tracer, coupled to Alexa647 (here shown in green, DAPI: blue). (A) Amygdala; (B) Lateral Hypothalamic Area (LHA); (C) Lateral Septum (LS), (D) Nucleus Accumbens shell (NAcc); (E) medial prefrontal cortex (mPFC). Scalebar: 500 µm. Inserts are from the Allen Brain Atlas (https://mouse.brain-map.org/experiment/thumbnails/100048576?image_type=atlas); Images A, B, C, D, E correspond to Atlas sections #74, 72, 53, 43, 40 of 132, respectively. ACB: nucleus accumbens; BLA: basolateral amygdalar nucleus; CP: caudate putamen; HPF: hippocampal formation; HY: hypothalamus; IL: infralimbic area; LHA: lateral hypothalamic area; LSr: lateral septal nucleus; NDB: diagonal band nucleus; TH: thalamus; aco: anterior commissure, olfactory limb; cc: corpus callosum; fa: corpus callosum, anterior forceps.
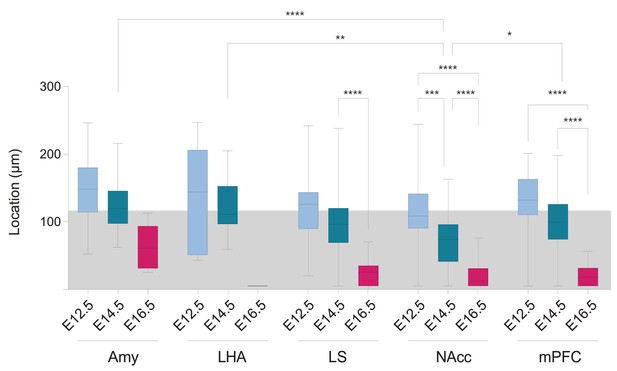
Cell location of fate-mapped Ctb+/Tdt+ PNs in ventral CA1.
The distribution of Ctb+/Tdt+ cell location is shown for all injected sites. Amygdala (Amy); Lateral Hypothalamic Area (LHA); Lateral Septum (LS), Nucleus Accumbens shell (NAcc); medial prefrontal cortex (mPFC). The gray shaded area represents the thickness of the stratum pyramidale (SP). Overall, the distributions are reminiscent of the radial organization of CA1 PNs according to their birthdate, with earlier-born cells located in deeper locations of SP, and conversely. This is most evident in NAcc- and mPFC-projecting PNs. Please notice that NAcc-projecting E14.5 PNs are shifted toward more superficial positions than E14.5 PNs targeting other regions (NAcc-E14.5 vs. LHA-E14.5, p<0.0054; mPFC-E14.5 vs. NAcc-E14.5, p: 0.0147; Amy-E14.5 vs. NAcc-E14.5, p<0.0001), except for the LS (LS-E14.5 vs. NAcc-E14.5, p: 0.954). Boxplots present medians (center), interquartile ranges (bounds), minima and maxima. Color-code: E12.5 PNs: light blue, E14.5: dark blue, E16.5: magenta. Kruskal-Wallis, main effect: p<0.0001. For pairwise comparisons: *p < 0.05, **p < 0.01, ***p < 0.001, **** p < 0.0001.
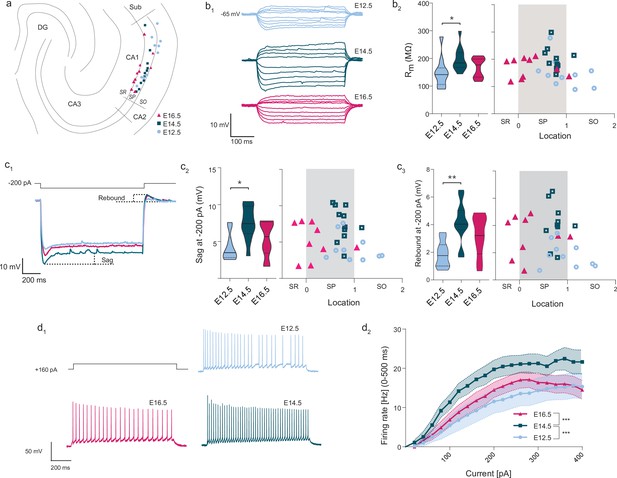
Intrinsic excitability varies according to embryonic birthdate.
(A) Anatomical location of neurobiotin-filled CA1PNs recorded from acute horizontal slices in current-clamp experiments. DG: dentate gyrus, Sub: subiculum, SR: stratum radiatum, SP: stratum pyramidale, SO: stratum oriens. (B1) Representative membrane responses to a series of hyper- and depolarizing current steps recorded from fate-mapped CA1PNs. Note the larger deflection of membrane potential in the E14.5 neuron. (B2) Membrane input resistance (Rm) of fate-mapped CA1PNs. Left, violin plot of the three birthdate groups. Right, scatterplot of the same data against the radial position. Rm is higher in E14.5 than E12.5 cells (Padj: 0.025; CI95% [–89.71; –11.15]; E12.5 vs E16.5, E16.5 vs E14.5, Padj:0.348). No linear correlation was found with cell location (p:0.139). (C1) Sag potential response and rebound of fate-mapped CA1PNs, following –200 pA current injections. In this example, E14.5 CA1PNs display a greater sag response than E12.5 and E16.5 PNs. (C2) Sag response. Left, violin plot of the three birthdate groups. Right, scatterplot of the same data against the radial position. Sag response is significantly higher in E14.5 than E12.5 cells (Padj: 0.0026; CI95% [1.74; 6.05]; E12.5 vs E16.5, E16.5 vs E14.5, Padj:0.128). No linear correlation was found with cell location (p:0.27). (C3) Rebound potential. Left, violin plot of the three birthdate groups. Right, scatterplot of the same data against the radial position. Sag response is significantly higher in E14.5 than E12.5 cells (Padj: P:0.0063; CI95% [–3.82;–1.29]; E12.5 vs E16.5, Padj:0.054; E16.5 vs E14.5, Padj:0.223). No linear correlation was found with cell location (p:0.139). (D1) Examples of firing responses to a depolarizing current step recorded from fate-mapped CA1PNs. Note that the number of action potential fired by E16.5 and E12.5 PNs is lower than E14.5 PNs. (D2) Input-output curves of fate-mapped CA1PNs. E14.5 PNs have a higher firing rate than E12.5 PNs (Padj < 0.0001; CI95% [5.21; 11.14]) and E16 (Padj < 0.0001; CI95% [–9.91; –4.13]), suggesting that they are more excitable (E16.5 vs. E12.5, Padj:0.645). Effect of current injection F (2, 540) = 25.65, p < 0.0001; effect of birthdate F (19, 540) = 10.71, p < 0.0001; interaction F (38, 540) = 0.2355, p > 0.9999, ordinary two-way ANOVA with Tukey’s post hoc test. Data are represented as means ± standard errors of the means. Violin plots present medians (center), interquartile ranges (bounds), minima and maxima. Color-code: E12.5: light blue, E14.5: dark blue, E16.5: magenta. The gray shaded area in scatterplots represents the thickness of the stratum pyramidale. *p < 0.05; **p < 0.01; ***p < 0.001.
-
Figure 6—source data 1
Source data for intrinsic membrane properties of fate-mapped CA1 PNs.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig6-data1-v2.csv
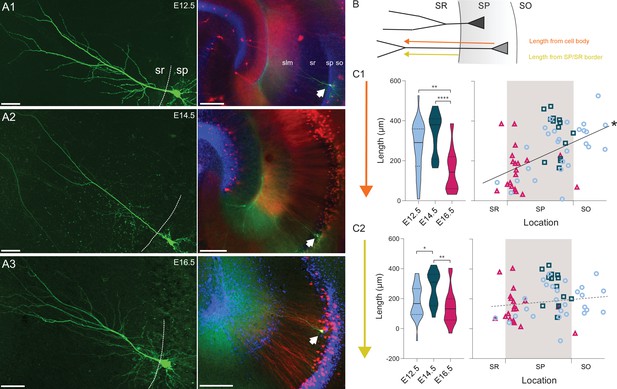
Embryonic origin is a determinant of dendritic morphology.
(A1-3) On the left, representative examples of CA1 PNs dendritic arborization in neurobiotin-filled cells. The dashed line is drawn at the border between stratum pyramidale (SP) and stratum radiatum (SR). On the right, the same cell as on the left is shown at lower magnification and indicated by a white arrow. Red and blue channels represent Tdtomato and DAPI, respectively. (A1) E12.5 PN, (A2) E14.5 PN, (A3) E16.5 PN. Note that the main dendritic branch bifurcates more distally in E14.5 cells. Scalebars: left, 50 µm; right, 200 µm. so: stratum oriens; sp: stratum pyramidale; sr: stratum radiatum; slm: stratum lacunosum-moleculare. (B) Scheme illustrating the primary dendrite length measured from soma or from the border between SP and SR to the first major dendritic bifurcation. SO: stratum oriens. (C1) Primary dendrite length from soma to bifurcation in fate-mapped CA1PNs. Left, violin plot of the three birthdate groups. Right, scatterplot of the same data against the radial position. E16.5PNs display a reduced dendrite length, in respect to E12.5 (p: 0.0001; CI95% [37.74; 190.36]) and E14.5 PNs (p < 0.0001; CI95% [92.07; 262.82]; E12.5 vs E14.5, p: 0.031. Sidak’s alpha for significance: 0.0169). The dendrite length from soma is markedly correlated with the soma location (p < 0.0001; CI95% [0.318; 0.683]), likely due to the contribution of the dendritic segment within the SP. (C2) Primary dendrite length measured between the SP/SR border and the dendrite bifurcation in fate-mapped CA1PNs. Left, violin plot of the three birthdate groups. Right, scatterplot of the same data against the radial position. The dendritic length is larger in E14.5 than E12.5 PNs (Padj: 0.0056; CI95% [–172.99; –6.14]) and E16 cells (Padj: 0.0004; CI95% [36.48; 217.12], E12.5 vs E16.5, p: 0.116. Sidak’s alpha for significance: 0.0169). No linear correlation was found with the cell location (P:0.109). Violin plots present medians (center), interquartile ranges (bounds), minima and maxima. Color-code: E12.5: light blue, E14.5: dark blue, E16.5: magenta. The gray shaded area in scatterplots represents the thickness of the stratum pyramidale. *p < 0.05. **p < 0.01. ****p < 0.0001.
-
Figure 7—source data 1
Source data for morphometric properties of fate-mapped CA1 PNs.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig7-data1-v2.csv
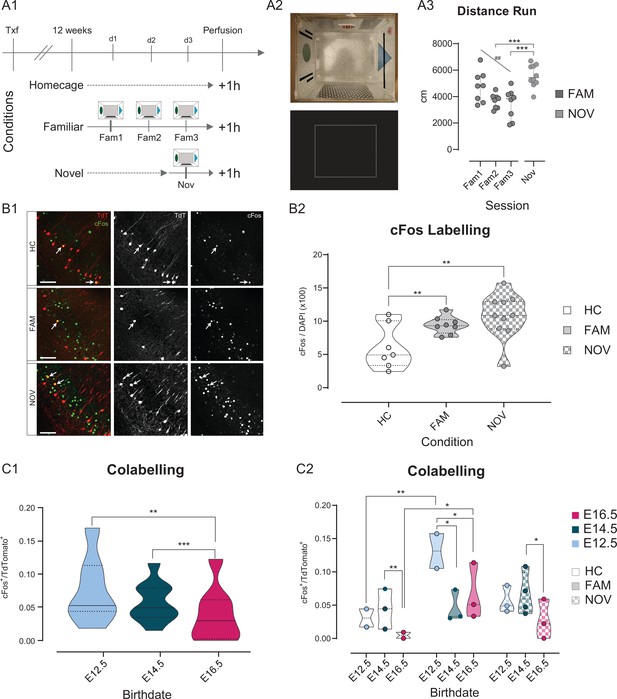
Fate-mapped CA1PNs differ in cFos expression following exploratory behavior.
(A1-3) Experimental paradigm and validation for detecting cFos expression upon exploration of an arena. (A1) Schematic representation of the behavioral conditions, with 3 cohorts of tamoxifen-induced Neurog2CreER-Tdt mice: homecage (HC), no exploration, familiar (FAM), repeated exploration (20’) on three consecutive days, and novel (NOV), only one exploration (20’). (A2) Top, view from above of the exploration box containing visual, tactile, and olfactory (butanal) cues. In addition, a white noise was played in the experimental room. Bottom, representative occupancy heat map. (A3) Quantification of the distance run by animals during exploration of the arena. Upon repeated exposure (Fam1-3), mice explored progressively less the arena (RM one-way ANOVA, main effect = F(1,14): 11.52, p: 0.0044, Slope:–660.1 ± 194.5, CI95% of slope [–242.95;–1077.2]), suggesting that novelty decreases over sessions. Also, NOV did not differ significantly from Fam1 (Padj: 0.26, CI95% [–323.80; 1921.7]), where mice explored for the first time, and was higher than Fam3 (Padj: 0.0006, CI95% [973.4; 3264.8]), corresponding to the last exploration (one-way ANOVA, main effect = F(3,30): 9.6745, p:0.0001). (B1-2) The expression of cFos in ventral CA1 is higher upon exploration of a novel environment, than after repeated exposures. (B1) Immunohistochemistry anti-cFos, performed on three E14.5 tamoxifen-induced mice. Top, HC; middle, FAM; bottom, NOV condition. Each shows the merged Tdt, cFos and DAPI image (left), Tdt (center), cFos (right). Note the increase in cFos signal from HC to FAM and NOV. Insert: magnification on two cells, one strongly cFos+, the other co-expressing Tdt and cFos. Scalebar: 100 µm. (B2) Fraction of cFos+ cells per animal. In both FAM (Padj: 0.0006, CI95% [–5.78; –1.99]) and NOV (Padj: 0.0006; CI95% [–7.86; –2.54]) animals, cFos immunoreactivity is increased compared to HC (Fam vs Nov, Padj:0.111). (C1-2) Quantification of the co-expression of cFos and Tdt in fate-mapped CA1PNs. (C1) Fraction of cFos+/Tdt+ cells by birthdate group, regardless of the condition. E16 cells display fewer co-labelled cells than E12 (Padj: 0.0022, CI95% [0.0097; 0.0483]) and E14 (Padj: 0.0009, CI95% [0.0137; 0.0459]; E12.5 vs E14.5, Padj:0.464). (C2) Fraction of cFos+/Tdt+ cells by birthdate and per condition. Each animal is represented as a datapoint indicating the fraction of cFos+/Tdt+ cells. Note that within the FAM condition, cFos+/Tdt+ cells are more abundant in E12-FAM than E14-FAM (Padj: 0.008, CI95% [0.0284; 0.135]) and E16-FAM (Padj: 0.0325, CI95% [0.0236;0.136]). Within E12 birthdate group as well, E12-FAM fraction of co-labeled cells is higher than E12-HC (Padj < 0.0001, CI95% [–0.151; –0.049]), but not than E12-NOV (Padj < 0.066, CI95% [0.0149; 0.123]). In addition, E14-NOV is greater than E16-NOV (Padj < 0.015, CI95% [0.0179; 0.0797]). Violin plots present medians (center), interquartile ranges (bounds), minima and maxima. Color-code: E12: light blue, E14: dark blue, E16: magenta. *p < 0.05. ##, **p < 0.01. ***p < 0.001.
-
Figure 8—source data 1
Source data of mouse locomotory activity for each behavioral session.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig8-data1-v2.csv
-
Figure 8—source data 2
Table summarizing DAPI, cFos+ and cFos+-Tdt+ labeling data following exploration.
- https://cdn.elifesciences.org/articles/69270/elife-69270-fig8-data2-v2.csv
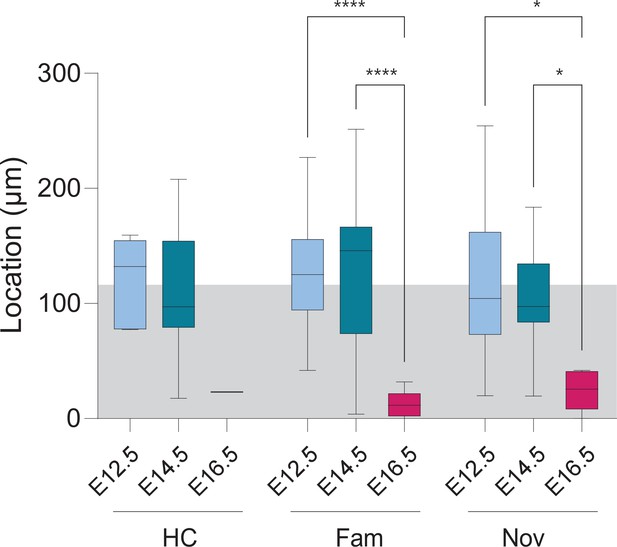
Cell location of fate-mapped cFos+/Tdt+ PNs in ventral CA1.
The distribution of cFos+/Tdt+ cell location is shown for all conditions and birthdate group. Home-cage condition (HC); Familiar condition (Fam); Novel condition (Nov). The gray shaded area represents the thickness of the stratum pyramidale (SP). Overall, the distributions are reminiscent of the radial organization of CA1PNs according to their birthdate, with earlier-born cells located in deeper locations of SP, and conversely. This is significant in Fam (Fam-E12.5 vs. Fam-E16.5, p<0.0001; Fam-E14.5 vs. Fam-E16.5, p<0.0001; Fam-E12.5 vs. Fam-E14.5, p>0.9999) and Nov conditions (Nov-E12.5 vs. Nov-E16.5, p<0.0155; Nov-E14.5 vs. Nov-E16.5, p<0.0344; Nov-E12.5 vs. Nov-E14.5, p>0.9999). Kruskal-Wallis, main effect: p<0.0001. For pairwise comparisons: *p < 0.05, ****p < 0.0001.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | RjOrl:SWISS | Mouse Genome Informatics | MGI:6430821 | |
| Gene (Mus musculus) | Neurog2tm1(cre/Esr1*)And | Mouse Genome Informatics | MGI:2652037 | |
| Gene (Mus musculus) | Gt(ROSA)26Sortm14(CAG-tdTomato)Hze | Mouse Genome Informatics | J:155,793 | |
| Antibody | Anti-parvalbumin (goat polyclonal) | Swant | RRID:AB_10000345 | (1:1000) |
| Antibody | Anti-cholera toxin beta subunit (rabbit polyclonal) | Invitrogren | RRID:AB_779810 | (1:1000) |
| Antibody | Anti-cFos (rabbit polyclonal) | Abcam | RRID:AB_190289 | (1:5000) |
| Antibody | Anti-CBD28k (rabbit polyclonal) | Swant | RRID:AB_10000340 | (1:1000) |
| Antibody | AlexaFluor647-conjugate Anti-rabbit (donkey polyclonal) | Jackson Immunoresearch | RRID:AB_2492288 | (1:500) |
| Antibody | AlexaFluor647-conjugate Anti-rabbit (goat polyclonal) | Jackson Immunoresearch | RRID:AB_2338072 | (1:500) |
| Antibody | AlexaFluor488-conjugate Anti-goat (donkey polyclonal) | Jackson Immunoresearch | RRID:AB_2336933 | (1:500) |
| Chemical compound, drug | AlexaFluor647-conjugate Cholera Toxin subunit b | ThermoFisher | Catalog ID:C34778 | (0.1%) |
| Chemical compound, drug | Neurobiotin | Vectorlabs | Catalog ID:SP-112 | (0.5%) |
| Chemical compound, drug | Streptavidin-AlexaFluor488 | Thermofisher | Catalog ID:S11223 | (1:1000) |
| Software, algorithm | Patchmaster | Heka | RRID:SCR_000034 | |
| Software, algorithm | MATLAB | MathWorks | RRID:SCR_001622 | |
| Software, algorithm | GraphPad Prism | GraphPad | RRID:SCR_015807 | |
| Software, algorithm | MiniAnalysis | Synaptosoft | RRID:SCR_002184 | |
| Software, algorithm | CellProfiler | CellProfiler | RRID:SCR_007358 | |
| Software, algorithm | ZEN | Zeiss | RRID:SCR_013672 | |
| Software, algorithm | EthoVision XT | Noldus | RRID:SCR_000441 | |
| Software, algorithm | Fiji | Fiji | RRID:SCR_002285 | |
| Other | DAPI stain | Vector Laboratories | Catalog ID:H-1200 | Mounting medium |
Stereotaxic coordinates of CA1PNs target regions.
| Target region | AP | ML | DV |
|---|---|---|---|
| NAcc | + 1.8/ + 2.0 | −0.6/–0.45 | –4.0 |
| Amy | –1.4 | –3.3 | –4.3 |
| mPFC | + 2.0 | –0.35 | –1.8 |
| LHA | 1.34/–1.45 | −0.65/–1.0 | −4.8/–4.0 |
| LS | + 0.6/0.7 | −0.4/–0.35 | –2.6/2.55 |
-
NAcc, Nucleus Accumbens (shell), Amy: Amygdala; mPFC, medial prefrontal cortex; LHA, lateral hypothalamic area, LS: lateral septum.
Summary of adult Neurog2CreER-Tdt mice used in post-exploration cFos analysis.
| N° animals | HC | FAM | NOV | Total |
|---|---|---|---|---|
| E12.5 | 2 | 2 | 3 | 7 |
| E14.5 | 3 | 3 | 4 | 10 |
| E16.5 | 2 | 3 | 3 | 8 |
| Total | 7 | 8 | 10 | 25 |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69270/elife-69270-transrepform-v2.docx
-
Supplementary file 1
Statistics table.
- https://cdn.elifesciences.org/articles/69270/elife-69270-supp1-v2.csv





