PEAR, a flexible fluorescent reporter for the identification and enrichment of successfully prime edited cells
Figures
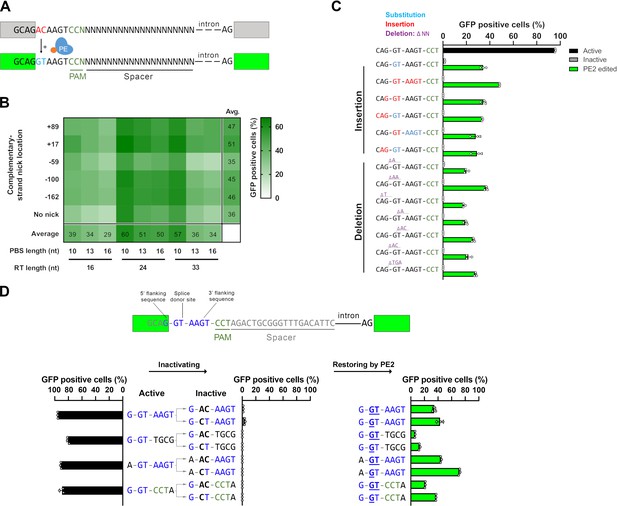
Principle of the prime editor activity reporter (PEAR) assay.
(A) Schematic of the PEAR. The mechanism of PEAR is based on the same concept as BEAR (see Figure 1—figure supplement 1A and B), and it contains the same inactive splice site, shown in (A). PE can revert the ‘G-AC-AAGT’ sequence to the canonical ‘G-GT-AAGT’ splice site. Prime editing occurs downstream of the cut site in the target; hence, this method enables us to position the spacer sequence within the intron, thus, the entire length of the spacer (shown as ‘N’-s) is freely adjustable in PEAR. The altered bases of the splice site are shown in red, the edited bases are shown in blue, and the PAM sequence is shown as dark green letters, the nCas9 is blue and the fused reverse transcriptase is orange. (B) Optimization of PBS, RT, and complementary DNA strand nicking on the PEAR-GFP plasmid. The heatmap shows the average percentage of GFP-positive cells of three replicates of transfections with the PEAR-GFP plasmid in combination with PE vectors which also contain the different pegRNAs and the sgRNAs for secondary nicking. The position of the second nick is given in relation to the first nick. Positive values indicate 3′, negative values indicate 5′ direction on the targeted DNA. When no second nick was introduced, it is indicated as ‘no nick’. (C, D) Flow cytometry measurements of HEK293T cells transfected with active (positive control) or with inactive PEAR plasmids either along with nCas9 for negative controls or with PE2 vectors (black, gray, or green columns, respectively). (C) In the PEAR system, GFP fluorescence can be restored from various inactive splice sequences not only by substitutions but also by insertions/deletions. Each inactive sequence (gray columns) was corrected (green columns) to the canonical splice site by PE2 with the optimal pegRNA (pegRNA 1) from (B). The edited sequences are shown next to the columns. Red indicates inserted, blue substituted, and purple deleted bases. (D) Prime editing can result in various active splice sequences, further demonstrating the sequence flexibility of the PEAR system. Based on Tálas et al., 2021, four additional active splice site variants were selected. To generate suitable inactive plasmids, splicing was disrupted by systematically replacing the 5′-GT splice donor site to 5′-AC and 5′-CT (negative controls with inactive sequences, gray columns). With the appropriate pegRNAs, PE2 was able to restore GFP fluorescence in every case (green columns). Letters highlighted in blue indicate the bases of the canonical splice donor site and the flanking sequences which influence splicing the most: 5′-G-GT-AAGT-3′; the altered bases of the inactive sequences are bold, and bases in the active sequences that are reverted by PE2 are underlined. (C, D) Columns represent means ± SD of three parallel transfections (white circles). For all measured values see Figure 1—source data 1.
-
Figure 1—source data 1
Measured values for Figure 1.
- https://cdn.elifesciences.org/articles/69504/elife-69504-fig1-data1-v1.xlsx
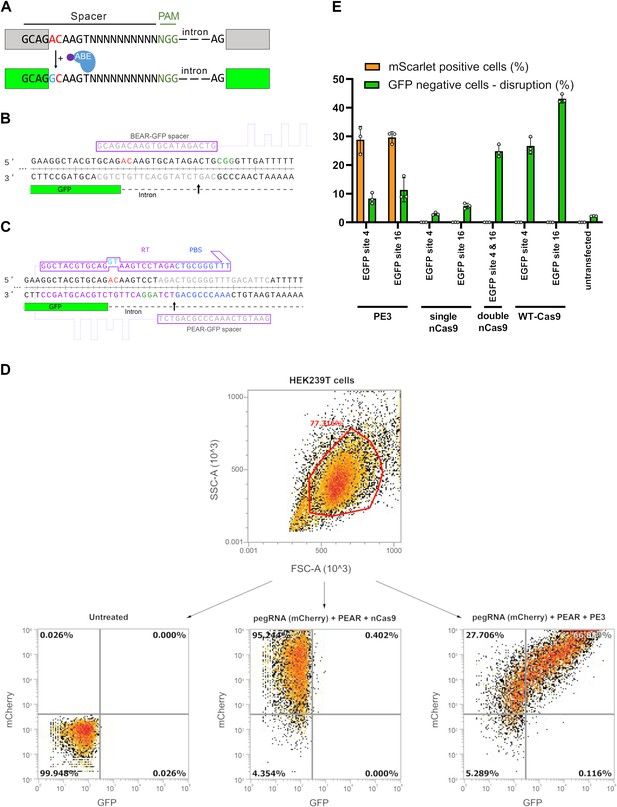
The differences between the base editor activity reporter (BEAR) and PEAR assays.
(A) Schematic representation of the BEAR. BEAR consists of a split GFP coding sequence separated by an intron, of which the 5′ splice donor site (G-GT-AAGT) is altered, resulting in an inactive splice site and a dysfunctional protein (gray). ABE converts the inactive splice site into a functional one. Here, the ‘G-AC-AAGT’ inactive splice site is illustrated, which can be modified by ABE to ‘G-GC-AAGT,’ which is a functional non-canonical splice site, and hence, restores GFP expression (green). In this assay, BEs act on the sense strand of the DNA. The altered bases of the splice site are shown in red, the edited base is shown in blue, and the variable bases in the sequence of the spacer are shown as ‘N’-s. The PAM sequence is dark green, nCas9 is blue, and the fused tadA deaminase is purple. (B) Detailed view of the 5′ splice site and the surrounding sequences of the BEAR-GFP plasmid. The 3′ end of the first exon of GFP is shown in green, the intron is shown as a dashed line. The spacer sequence and the target sequence are shown in gray, the PAM is green, and the inactive splice site is red. The Cas9 nick site is indicated by a black arrow. (C) Detailed view of the 5′ splice site and the surrounding sequences of the PEAR-GFP plasmid. The 3′ end of the first exon of GFP is shown in green, the intron is shown as a dashed line. The spacer sequence in the pegRNA and the target sequence in the DNA are shown in gray, the PAM is green, and the inactive splice site is red. The RT (purple) and the PBS (blue) sequences in the pegRNA and the targeted sequences are also colored. The Cas9 nick site is indicated by a black arrow. (D) A density plot of HEK293T cells when all live single cells (determined by FSC and SSC parameters) are either (1) untransfected (left plot), and thus show no fluorescence for mCherry or GFP, (2) co-transfected with a pegRNA-mCherry plasmid and the PEAR-GFP plasmid but with a nCas9, thus displaying mCherry but no GFP fluorescence, (3) or pegRNA-mCherry and PEAR-GFP plasmid is co-transfected with a prime editor expressing plasmid, causing the splice site to be edited, and thus displaying both mCherry and GFP fluorescence. (E) A HEK293T cell line containing an integrated EGFP expression cassette was co-transfected with the BEAR-mScarlet plasmid, a pegRNA and a second nicking sgRNA for BEAR-mScarlet, GFP targeting sgRNA(s) and either WT-Cas9, nCas9, or PE2. Cells were monitored for mScarlet and EGFP fluorescence (orange and green columns, respectively) 3 days after transfection. Columns represent means ± SD of three parallel transfections. For all measured values, see Figure 1—figure supplement 1—source data 1. PEAR, prime editor activity reporter.
-
Figure 1—figure supplement 1—source data 1
Measured values for Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/69504/elife-69504-fig1-figsupp1-data1-v1.xlsx
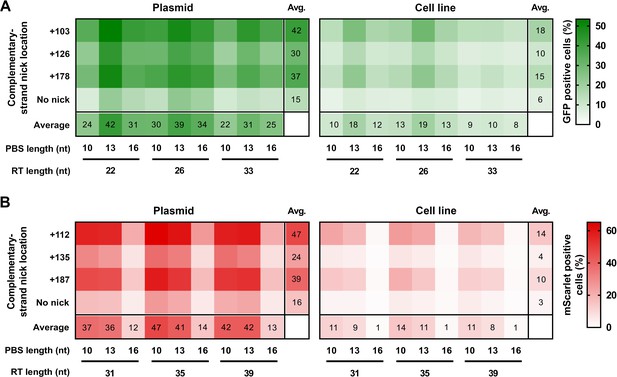
Prime editing with PEAR in a genomic and plasmid context.
PEs targeting BEAR sequences either located in plasmids (Plasmid) or incorporated into the genome (Cell line) were transfected into cells alongside different sgRNAs for different complementary strand nick locations. The heatmaps show the average percentage of GFP (A) and mScarlet (B) positive cells derived from three replicates. For all measured values see Figure 2—source data 1. BEAR, base editor activity reporter; PEAR, prime editor activity reporter.
-
Figure 2—source data 1
Measured values for Figure 2.
- https://cdn.elifesciences.org/articles/69504/elife-69504-fig2-data1-v1.xlsx
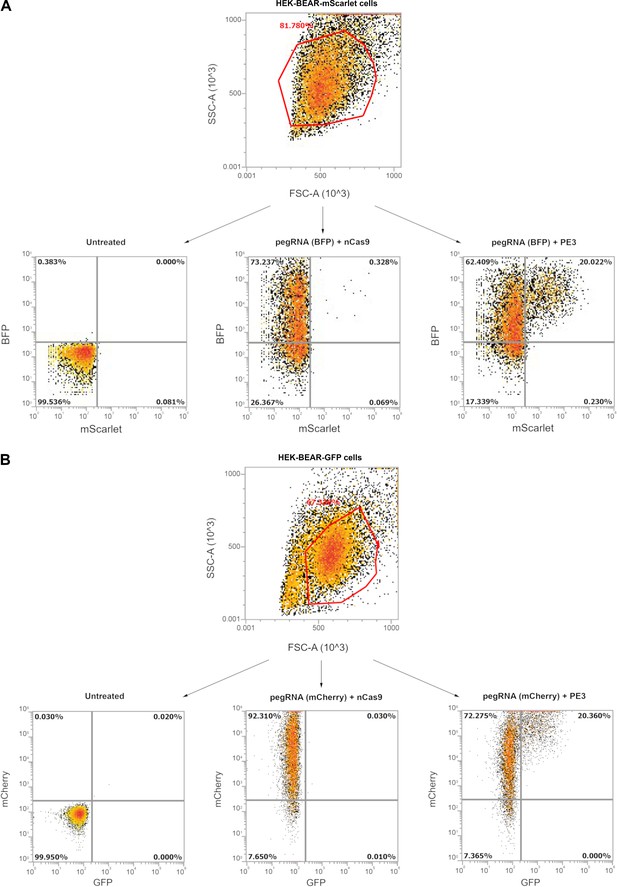
Gating examples from HEK-BEAR cell lines.
Gating examples are shown from Figure 2 where cells harboring a genomically integrated copy of BEAR-mScarlet (A) or BEAR-GFP (B) (HEK-BEAR-mScarlet and HEK-BEAR-GFP cell lines, respectively) sequences were subjected to various prime editing conditions. On the upper sections density plots of HEK-BEAR-mScarlet (A) or HEK-BEAR-GFP (B) cells are shown when all live single cells (determined by FSC and SSC parameters) are either (1) untransfected (left plot), and thus show no fluorescence for either fluorescent proteins; (2) co-transfected with a pegRNA-BFP (A) or a pegRNA-mCherry (B) plasmid but with a nCas9, thus displaying BFP but no mScarlet fluorescence in case of HEK-BEAR-mScarlet cells (A) or displaying mCherry but no GFP fluorescence in the case of HEK-BEAR-GFP cells (B); (3) the pegRNA plasmid is co-transfected with a prime editor expressing plasmid, causing the splice site to be edited, and thus displaying both BFP and mScarlet fluorescence in the case of HEK-BEAR-mScarlet cells (A) or displaying mCherry and GFP fluorescence in the case of HEK-BEAR-GFP cells (B). BEAR, base editor activity reporter.
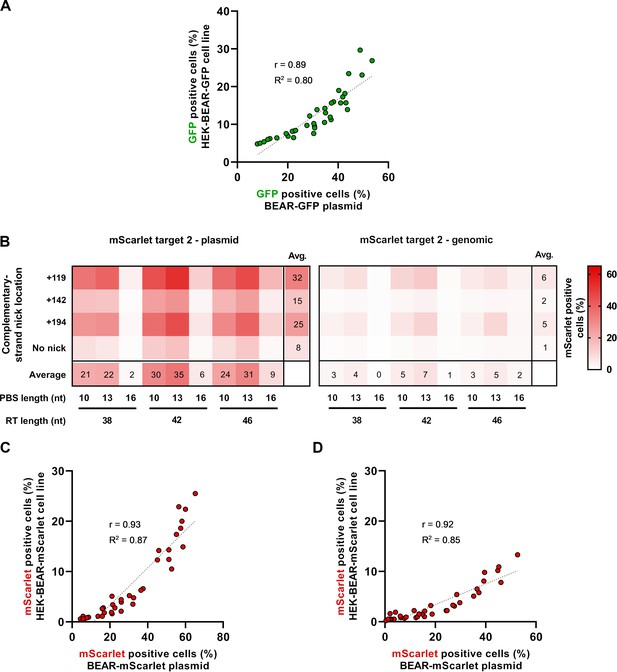
Prime editing of the BEAR-mScarlet plasmid with target 2.
(A) Scatter plot of GFP-positive cells measured either when the BEAR-GFP plasmid or the HEK-BEAR-GFP cell line (Figure 2A) was edited by PE3 with various pegRNA and secondary nicking sgRNA combinations (Pearson’s r=0.89). (B) The heatmaps show the average percentage of mScarlet positive cells derived from three replicates where PE vectors targeting inactive splice sequences located on plasmids (Plasmid) or incorporated into the genome (Cell line) were transfected into cells alongside different sgRNAs for different complementary strand nick locations. The position of the second nick is indicated on the left side of the heatmaps with positive numbers or when no second nick was introduced, it is indicated as ‘no nick’. (C–D) Scatter plot of mScarlet positive cells measured either when the BEAR-mScarlet plasmid or the HEK-BEAR-mScarlet cell line was edited by PE3 targeting mScarlet target 1 (C) data are from Figure 2B or target 2 (D) data are from (B) with various pegRNAs and secondary nicking sgRNA combinations (Pearson’s r=0.92 for (C) and r=0.93 for (D)). For all measured values, see Figure 2—figure supplement 2—source data 1. BEAR, base editor activity reporter.
-
Figure 2—figure supplement 2—source data 1
Measured values for Figure 2—figure supplement 2.
- https://cdn.elifesciences.org/articles/69504/elife-69504-fig2-figsupp2-data1-v1.xlsx
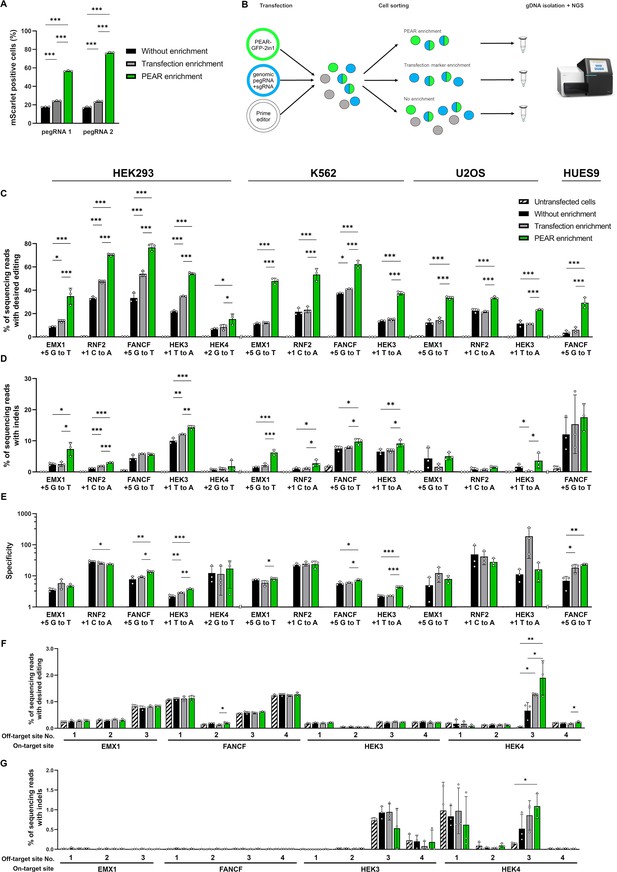
PEAR enriches prime edited cells.
(A) Flow cytometry measurements of prime edited (i.e., mScarlet positive) cells in the HEK-BEAR-mScarlet cell line. The PEAR-GFP plasmid was co-transfected with PE protein-coding plasmid, two pegRNAs targeting either the plasmid or the genomic PEAR sequences and an sgRNA (for complementary strand nicking at position +112). To edit the PEAR-GFP plasmid, two efficient pegRNAs were selected from a previous experiment (Figure 1B) (pegRNA 1 and pegRNA 2 are those shown in Figure 1B with an RT of 24 and a PBS of 10 nucleotides, and an RT of 33 and a PBS of 10 nucleotides, respectively). The mScarlet positive cell count was gated for either all live single cells (no enrichment, black bars); for BFP-positive cells (transfection enrichment, gray bars); and for GFP-positive cells (PEAR enrichment, green bars). (B) Schematic workflow of PEAR enrichment experiments. Various cell lines were co-transfected with PEAR-GFP-2in1, a pegRNA, and a complementary nicking sgRNA targeting a genomic sequence (the latter two carrying a BFP expression cassette to monitor transfection efficiency), and the PE2-encoding plasmid. Cells expressing GFP, BFP, or both are represented as green, blue or half green/half blue circles, respectively. Cells not expressing these fluorescent proteins are shown in gray. Three days after transfection, cells were sorted into three fractions: single living cells, cells expressing BFP, and cells expressing GFP. After purification of genomic DNA and PCR amplification of the targeted amplicon, the percentage of editing was determined by NGS from each sorted sample. (C–E) The PEAR-GFP-2in1 plasmid and endogenous genomic targets were co-edited in HEK293T, K562, U2OS, and HUES9 cells. Cells were enriched and analyzed as described in (B). Results from cells without enrichment are shown in black, transfection enrichment in gray, PEAR enrichment in green, and untransfected cells in striped black and white. Precise prime editing (C) and unwanted indel formation (D) were quantified from the same samples. Specificity (prime editing%/indel%) was calculated separately for each sample (E). Columns represent means ± SD of three parallel transfections (white circles). When indel% was below the detection limit of NGS, specificity was calculated with 0.05% indel to avoid falsely high specificity values. Differences between samples were tested using one-way ANOVA. Only statistically significant differences are shown, differences to untransfected cells are not shown. *p<0.05, **p<0.01, ***p<0.001. (F, G) Off-target prime editing (F) and indel formation (G) were analyzed for known off-targets of target sites from (C) and (D) in HEK293T cells. Columns represent means ± SD of three parallel transfections (white circles). Differences between samples were tested using one-way ANOVA. Only statistically significant differences are shown. *p<0.05, **p<0.01, ***p<0.001. For all measured values, applied statistic tests and exact p values see Figure 3—source data 1. BEAR, base editor activity reporter; PEAR, prime editor activity reporter.
-
Figure 3—source data 1
Measured values and detailed statistics for Figure 3.
- https://cdn.elifesciences.org/articles/69504/elife-69504-fig3-data1-v1.xlsx
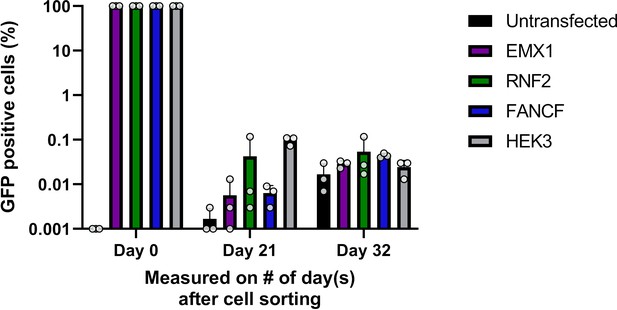
Stable integration of PEAR plasmids.
PEAR enriched populations (sorted for GFP, day 0) of HEK293T cells from the experiments in Figure 3C were passaged for 3–4 weeks. GFP-positive cells were measured by flow cytometry to determine whether fluorescence was due to stable integration of the plasmid. Columns represent means ± SD of three parallel transfections. For all measured values, see Figure 3—figure supplement 1—source data 1. PEAR, prime editor activity reporter.
-
Figure 3—figure supplement 1—source data 1
Measured values for Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/69504/elife-69504-fig3-figsupp1-data1-v1.xlsx
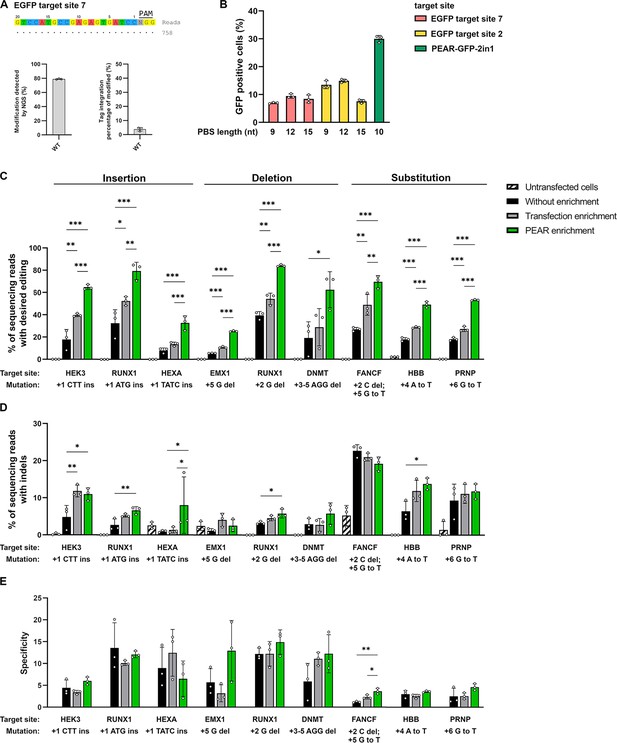
Enrichment with a PEAR reporter with no off-target activity.
(A) Potential off-target cleavage of WT-SpCas9 was assessed by GUIDE-seq on EGFP site 7. Read counts are shown for the on-target sequence only, as no off-targets were identified. On the bar charts, the on-target genome modification (indel+tag integration) and the tag integration frequency of the modified cells are shown that were analyzed by NGS. The other analyzed site (EGFP site 2) has previously been proven to have no detectable off-target sites by GUIDE-seq (Figure 5f, Kulcsár et al., 2020). (B) Flow cytometry measurements of HEK293T cells transfected with different 2in1 PEAR plasmids, and the PE2 encoding plasmid. Target sites EGFP site 7 and EGFP site 2 (pink and yellow columns, respectively) were candidates for the construction of a PEAR-GFP-2in1 plasmid with no genome-wide off-target activity. For the plasmid targeting pegRNA, three different PBS lengths were tested with each target in combination with the 24-nucleotide long RT-template identified as the most efficient in Figure 1B. The PEAR-GFP-2in1 plasmid used in previous experiments is shown in green. Columns represent means ± SD of three parallel transfections (white circles). (C–E) PEAR can enrich all types of editing (insertions, deletions, and substitutions) in HEK293T cells. Results from cells without enrichment are shown in black, transfection enrichment in gray, PEAR enrichment in green, and untransfected cells in striped black and white. Prime editing (C) and indel formation (D) were quantified from the same samples. (E) Specificity (prime editing%/indel%) was calculated separately for each sample. Columns represent means ± SD of three parallel transfections (white circles). When indel% was below the detection limit of NGS, specificity was calculated with 0.05% indel to avoid falsely high specificity values. Differences between samples were tested using one-way ANOVA. Only statistically significant differences are shown, differences to untransfected cells are not shown. *p<0.05, **p<0.01, ***p<0.001. For all measured values, applied statistic tests and exact p values see Figure 4—source data 1. PEAR, prime editor activity reporter.
-
Figure 4—source data 1
Measured values and detailed statistics for Figure 4.
- https://cdn.elifesciences.org/articles/69504/elife-69504-fig4-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HEK293T | ATCC | CRL-3216 | |
| Genetic reagent (H. sapiens) | HEK293.EGFP | Kulcsár et al., 2017 | ||
| Cell line (H. sapiens) | U2OS | ATCC | HTB-96 | |
| Cell line (H. sapiens) | K562 | ATCC | CCL-243 | |
| Genetic reagent (H. sapiens) | HEK-BEAR-GFP | Tálas et al., 2021 | ||
| Genetic reagent (H. sapiens) | HEK-BEAR-mScarlet | Tálas et al., 2021 | ||
| Cell line (H. sapiens) | HUES9 | Dr. Douglas Melton | ||
| Chemical compound, drug | Turbofect | Thermo Fischer Scientific Inc | R0531 | |
| Commercial assay or kit | Qubit dsDNA HS Assay Kit | Thermo Fischer Scientific Inc | Q32854 | |
| Commercial assay or kit | SF and SE Cell Line 4D-Nucleofector X Kit S | Lonza | V4XC-2032V4XC-1032 | |
| Recombinant DNA reagent | Plasmids | This study | see Supplementary file 2 | |
| Sequence-based reagents | DNA oligonucleotides | This study | see Supplementary file 3 | |
| Chemical compound, drug | Q5 High-Fidelity DNA Polymerase | New England Biolabs Inc | M0491L | |
| Chemical compound, drug | HiFi Assembly Master Mix | New England Biolabs Inc | E2621X | |
| Chemical compound, drug | KAPA Universal qPCR Master Mix | KAPA Biosystems | KK4602 | |
| Strain, strain background (Escherichia coli) | NEB5-alpha competent cells | New England Biolabs Inc | C2987I | |
| Recombinant DNA reagent | pCMV-PE2 | Addgene | (#132775) | |
| Recombinant DNA reagent | pAT9624-BEAR-cloning | Addgene | (#162986) | |
| Recombinant DNA reagent | pAT9651-BEAR-GFP | Addgene | (#162989) | |
| Recombinant DNA reagent | pAT9752-BEAR-mScarlet | Addgene | (#162991) | |
| Recombinant DNA reagent | pAT9658-sgRNA-mCherry | Addgene | (#162987) | |
| Recombinant DNA reagent | pAT9679-sgRNA-BFP | Addgene | (#162988) | |
| Recombinant DNA reagent | pX330-Flag-wtSpCas9-H840A | Addgene | (#80453) | |
| Recombinant DNA reagent | pX330-Flag-dSpCas9 | Addgene | (#92113) | |
| Recombinant DNA reagent | pX330-Flag-wtSpCas9 | Addgene | (#92353) |
Additional files
-
Supplementary file 1
Primers and PCR condition to detect PEAR plasmid integration.
Forward and reverse PCR primers, PCR product sizes, and a detailed PCR protocol suitable for detecting integrated PEAR plasmids are provided.
- https://cdn.elifesciences.org/articles/69504/elife-69504-supp1-v1.docx
-
Supplementary file 2
List of plasmids used in this study.
This file contains all of the plasmids used in the study with references and additional comments.
- https://cdn.elifesciences.org/articles/69504/elife-69504-supp2-v1.docx
-
Supplementary file 3
List of oligonucleotides used in this study.
This file contains the oligonucleotide sequences used for molecular cloning or PCR.
- https://cdn.elifesciences.org/articles/69504/elife-69504-supp3-v1.docx
-
Supplementary file 4
Sample indexing for NGS.
This file contains the necessary information to identify the NGS samples used in this study.
- https://cdn.elifesciences.org/articles/69504/elife-69504-supp4-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69504/elife-69504-transrepform1-v1.docx





