Monocyte-derived transcriptome signature indicates antibody-dependent cellular phagocytosis as a potential mechanism of vaccine-induced protection against HIV-1
Figures
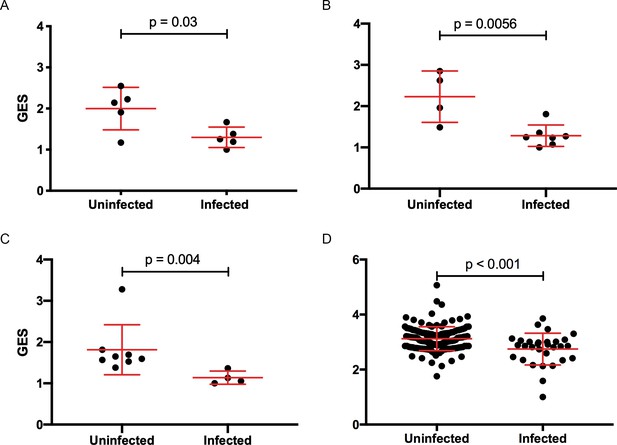
Composite gene expression scores (GES) are higher in the uninfected compared to infected groups.
GES computed from enriched genes in the geneset is higher in the uninfected compared to infected vaccinated NHP and humans. (A) Ad26/gp140 (09–11 NHP SIV challenge study, 58 enriched genes, N=10), (B) Ad26/gp140 (13–19 NHP SHIV challenge study, 58 enriched genes, N=11), (C) Ad26/Ad26+ gp140 (13–19 NHP SHIV challenge study, 68 enriched genes, N=12), and (D) ALVAC/gp 120 (RV144 human efficacy trial, 63 enriched genes, N=170). Statistical significance was calculated by either Mann-Whitney or unpaired t-test. NHP, non-human primate; SHIV, simian-human immunodeficiency virus; SIV, simian immunodeficiency virus.
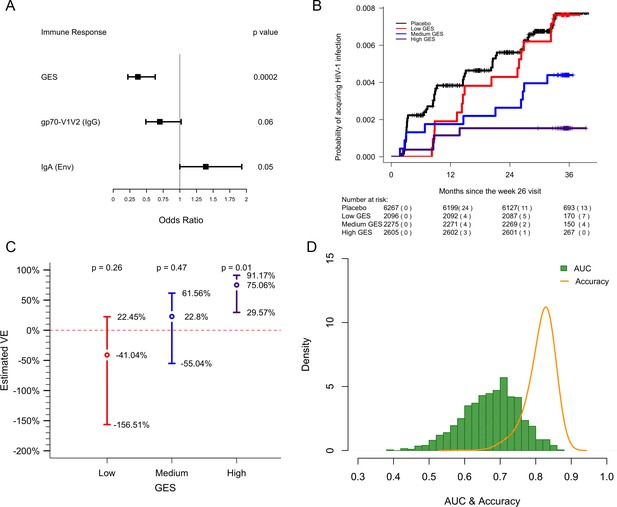
GES is a stronger correlate of reduced risk of infection in RV144.
A GES of the 63 enriched genes in the RV144 study was examined as a continuous variable (N=170). (A) GES is associated with lower odds of HIV acquisition compared to the other two primary correlates of risk. Variables were measured at week 26, 2 weeks post last vaccination. For each variable, the OR is reported per 1-SD increase. Transcriptome data was available only in a subset of the 246 donors. (B) Probability of acquiring HIV-1 is lower in individuals with higher GES. (C) Vaccine efficacy is increased significantly in individuals with high GES. (D) Distribution of AUC and accuracy plotted after repeating the process 1000 times showed that GES could predict HIV-1 infection with AUC of 0.67±0.08 and with accuracy of 0.81±0.04. GES, gene expression score.
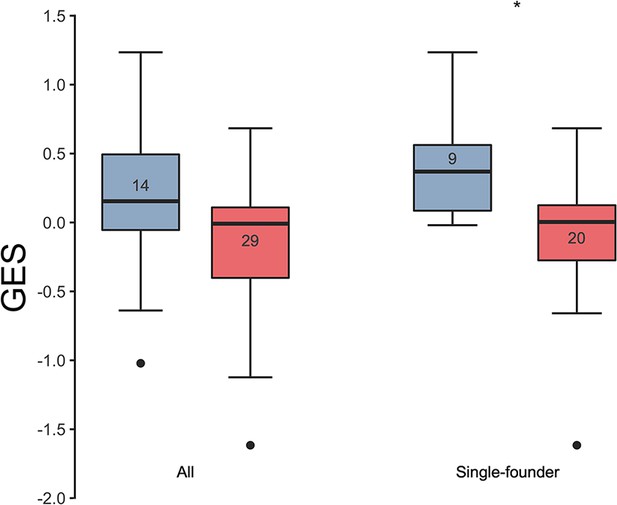
Association of the GES with HIV-1 breakthrough infections in a human vaccine trial.
GES was significantly higher in the placebo (blue) compared to vaccine recipients (red) among the RV144 participants with single-founder breakthrough infection (p<0.05). Numbers indicate the number of participants plotted. Asterisks indicate significant pairwise differences by the Mann-Whitney test. GES, gene expression score.
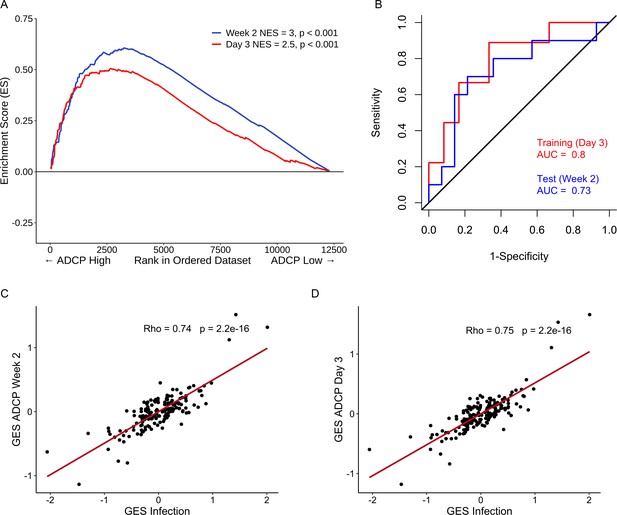
Strong relationship between functional ADCP responses in a human vaccine trial and the protective RV144 signature.
The geneset that associated with protection in an efficacy study was also enriched with higher magnitude of ADCP measured 2 weeks after vaccination in an immunogenicity trial that employed the RV144 vaccine regimen. NES from RNA-seq data at time points (A) 2 weeks (118 enriched genes) (N=24) and 3 days (93 enriched genes) (N=21) post the RV144 vaccine regimen in the RV306 trial are indicated. (B) The model built using ADCP GES from day 3 was able to predict ADCP responses measured 2 weeks after the last vaccination with an accuracy of 0.71. The ROC curve illustrates the discriminating ability of the classifier from the day 3 training data set (AUC=0.8, 95% CI: 0.6–0.99, p=0.01) and the week 2 testing data set (AUC=0.73, 95% CI: 0.5–0.95, p=0.03) to predict ADCP responses. (C) GES computed from the enriched genes associating with ADCP correlated strongly with the protective GES in the RV144 study (N=170) at time points 2 weeks (115 enriched genes) and (D) 3 days (91 enriched genes) post the RV144 vaccine regimen. ADCP, antibody-dependent cellular phagocytosis; CI, confidence interval; GES, gene expression score; NES, normalized enrichment score.
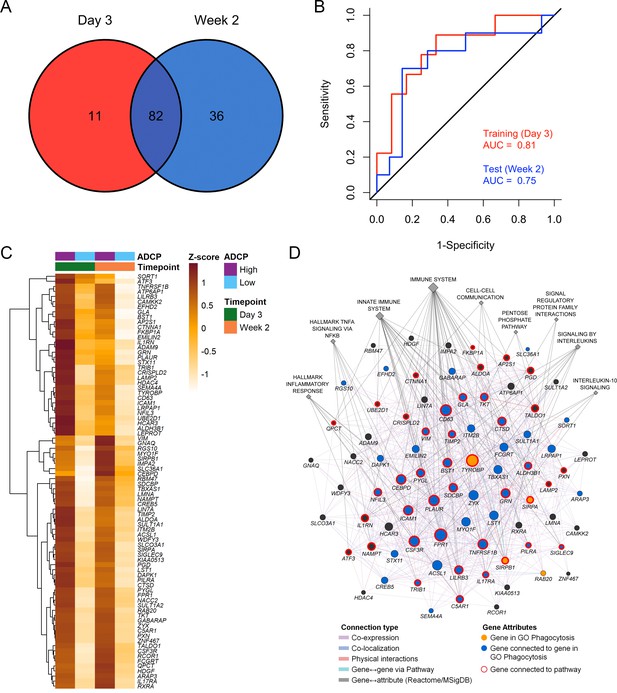
Overlapping enriched genes associating with ADCP responses.
(A) There were 82 overlapping genes between the day 3 (N=21) and week 2 (N=24) ADCP enriched genes in the RV306 study. (B) The model using GES obtained from the 82 genes was also able to predict ADCP responses measured 2 weeks after vaccination with an accuracy of 0.71. The ROC curve illustrates the discriminating ability of the classifier from the day 3 training data set (AUC=0.81, 95% CI: 0.62–1, p=0.007) and the week 2 testing data set (AUC=0.75, 95% CI: 0.53–0.97, p=0.02) to predict ADCP responses. (C) Heatmap showing the hierarchical clustering of gene expression of the 82 genes (day 3 and week 2 after 4th vaccination) when stratified by magnitude of ADCP responses measured 2 weeks after the 4th immunization. (D) The list of 82 ADCP enriched genes was uploaded in GeneMANIA. Edges represent physical interactions, co-expression, co-localization, and shared pathways. Circles depict the 82 genes, gold circles are the four genes that belong to the gene ontology Phagocytosis pathway, blue circles are genes that are directly connected to them, diamonds indicate related pathways, and the color of the edge indicates the type of connection. ADCP, antibody-dependent cellular phagocytosis; CI, confidence interval; GES, gene expression score; ROC, receiver operator characteristic.
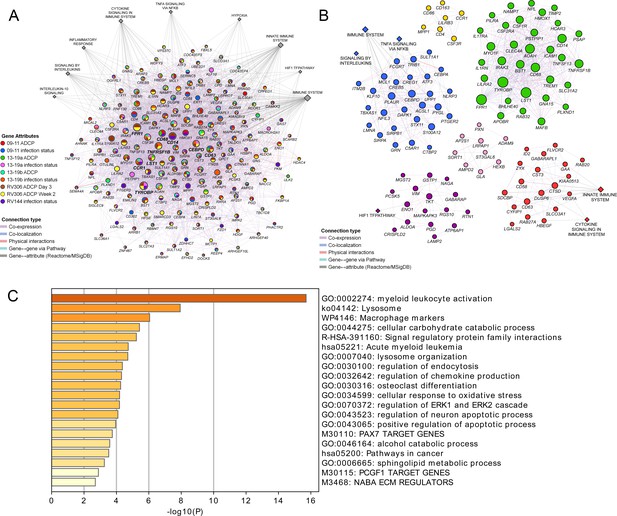
Pathway analyses of the enriched genes in the different vaccine studies.
A meta-analysis of pathways including enriched genes with reduced infection or higher ADCP was performed. (A) Genes that were enriched in at least one of the nine ADCP or infection status analyses (178) were used as input for GeneMANIA in Cytoscape. The connections between the different genes and the top MSigDB and Reactome pathways are shown. Each gene is represented by a circle and size is proportional to the number of connections with other genes or pathways. The color of each node indicates the enrichment status in the different studies. (B) Clustering of the enriched genes from the different studies. The color of each node represents the membership in a cluster and size is proportional to the number of connections with other genes or pathways. (C) Pathway enrichment analysis results of the 63 enriched genes that associated with reduced infection in the RV144 study. ADCP, antibody-dependent cellular phagocytosis.
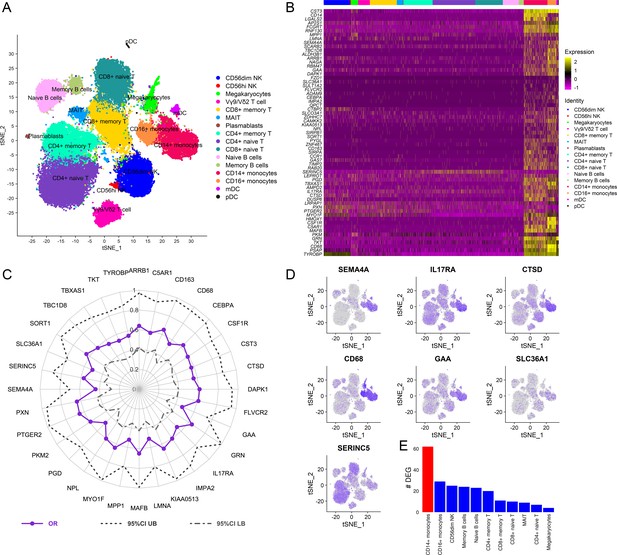
Cellular origin of the RV144 signature.
Single-cell CITE-seq in vaccinated participants (N=12) who received the RV144 vaccine regimen (day 3 after last vaccination) identified expression of the genes in the signature in cells from the myeloid lineage. (A) Clustering based on cell surface expression of CITE-seq data. (B) Heatmap of the mRNA expression of the 63 genes from the RV144 signature from single cells. Columns represent single cells from different protein cell subsets and rows the mRNA gene expression. (C) Radar plot showing significant genes in the signature that associated with decreased risk of infection in RV144 (p<0.05, q<0.1) (N=170). (D) Feature plots of the expression of the most protective genes show that SEMA4A, IL17RA, CTSD, CD68, and GAA were mainly expressed in monocytes. (E) CD14+ monocytes had the highest number of differentially expressed genes (DEGs) when comparing high versus low ADCP (2 weeks after vaccination) from single-cell CITE-seq vaccinated participants who received the RV144 vaccine regimen (day 3 after last vaccination). ADCP, antibody-dependent cellular phagocytosis.

GES of the most significant genes is a correlate of reduced risk of infection in RV144.
A GES of the 32 enriched genes that were also significantly associated with decreased risk of acquisition in a univariate analysis in the RV144 study was examined as a continuous variable (N=170). (A) GES is associated with lower odds of HIV acquisition compared to the other two primary correlates of risk. Variables were measured at week 26, 2 weeks post last vaccination. For each variable, the OR is reported per 1-SD increase. Transcriptome data was available only in a subset of the 246 donors. (B) Probability of acquiring HIV-1 is lower in individuals with higher GES. (C) Vaccine efficacy is increased significantly in individuals with high GES. (D) Distribution of AUC and accuracy plotted after repeating the process 1000 times showed that GES could predict HIV-1 infection with AUC of 0.69±0.08 and with accuracy of 0.81±0.04. GES, gene expression score; OR, odds ratio.
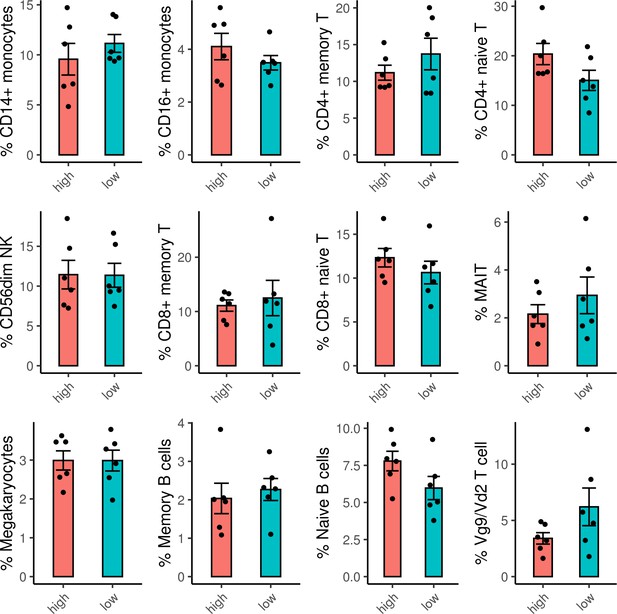
Frequencies of cell subsets do not differ between ADCP high and low samples.
Frequencies of each cell subset were calculated for each sample. For the cell subsets with average frequencies >1 % of the sample, there were no significant differences between the frequencies of the cell subsets in the ADCP high versus low samples. ADCP, antibody-dependent cellular phagocytosis.
Tables
Gene signature associates with vaccine protection in multiple trials.
| Study | Vaccine regimen | Species | Partial protection | N | Method | Protective signature |
|---|---|---|---|---|---|---|
| 09–11 | Ad26/gp140 | NHP | Y | 10 | RNA-seq | Y |
| 13–19 | Ad26/gp140 | NHP | Y | 11 | RNA-seq | Y |
| 13–19 | A26/Ad26+ gp140 | NHP | Y | 12 | RNA-seq | Y |
| 13–19 | Ad26/MVA+ gp140 | NHP | Y | 9 | RNA-seq | N |
| ALVAC-SIV/gp120 | NHP | Y | 27 | Microarray | Y | |
| DNA-SIV/ALVAC+ gp120 | NHP | Y | 12 | Microarray | Y | |
| RV144 | ALVAC/gp120 | Human | Y | 170 | Microarray | Y |
| HVTN 505 | DNA/rAd5 | Human | N | 42 | RNA-seq | N |
Additional files
-
Supplementary file 1
Additional tables supporting the findings in this study.
(a) Number of enriched genes from the geneset in different comparisons from multiple studies. (b) Number of enriched genes from the geneset in different comparisons from multiple studies. (c) Relationship between genes associating with ADCP in this study and previously identified pathways. (d) Enriched ADCP genes that belong to the gene ontology phagocytosis pathway.
- https://cdn.elifesciences.org/articles/69577/elife-69577-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69577/elife-69577-transrepform1-v2.docx
-
Source code 1
Code to generate figures from "Monocyte-derived transcriptome signature indicates antibody-dependent cellular phagocytosis as a potential mechanism of vaccine-induced protection against HIV-1".
- https://cdn.elifesciences.org/articles/69577/elife-69577-supp2-v2.zip





