Genetic depletion studies inform receptor usage by virulent hantaviruses in human endothelial cells
Figures
Suitability of TIME cells as a model to study hantavirus entry and the generation of knockout cells.
(a) Upper panels, total flow cytometry plots of HUVEC and TIME cells stained for endothelial cell markers PECAM and von Willebrand factor (vWF). Medium and lower panels, surface flow cytometry plots of HUVEC and TIME cells stained for PCDH1, β3 integrin, DAF, β1 integrin. (b) Surface flow cytometry of wild-type (WT) and knockout (KO) TIME cells stained as above. Histograms of WT cells are shown in gray; single- and double-KO cells are shown in color. (c) Western blot analysis of WT TIME cells and KO cells ± cDNA. β-Actin was used as a loading control.
-
Figure 1—source data 1
Original blot of WT TIME cells and KO cells ± cDNA.
- https://cdn.elifesciences.org/articles/69708/elife-69708-fig1-data1-v1.zip
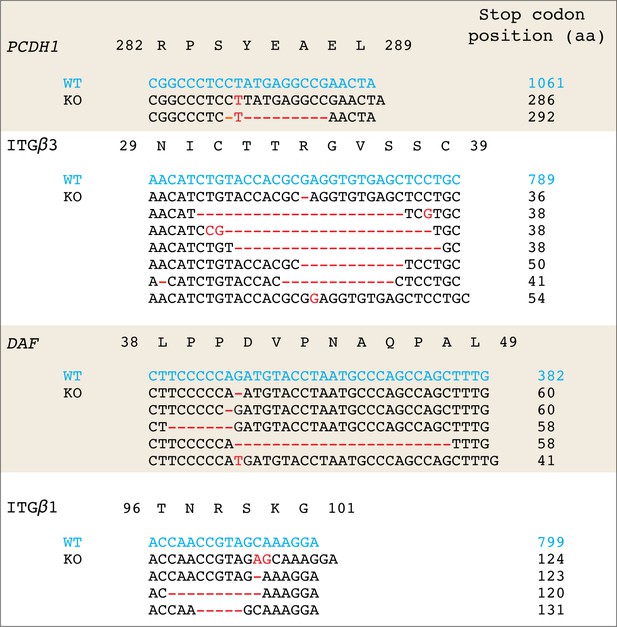
Sanger sequences retrieved from the targeted genomic loci for each knockout cell population.
Sequences of interests were amplified by PCR and then TA-cloned into the pGEM-T vector. For each knockout cell population, 15–20 clones were subjected to Sanger sequencing. All sequenced clones showed an indel at the targeted site, resulting in a frameshift that brought one or more stop codons into frame.
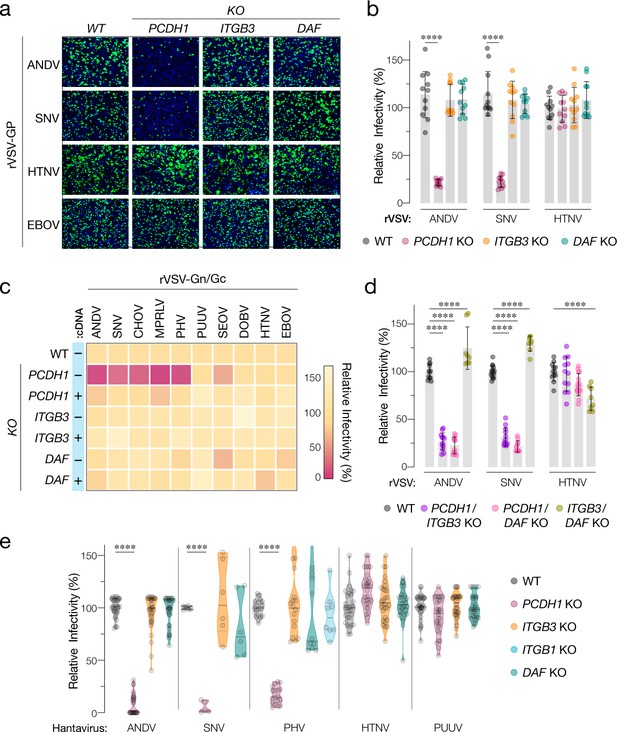
Hantavirus receptor requirement in endothelial cells.
(a) Representative images of eGFP-positive rVSV-infected wild-type (WT) and PCDH1, ITGB3, and DAF knockout (KO) TIME cells. Nuclei were stained with Hoechst (blue). (b) WT and KO cells were exposed to the indicated rVSV-Gn/Gc. n=11 for each cell line from three independent experiments. WT versus KO cells, two-way ANOVA with Dunnett’s test; ***p<0.0001. Other comparisons were not statistically significant (p>0.05). (c) WT and KO cells lacking (–cDNA) or expressing the corresponding cDNA (+cDNA) were exposed to rVSVs bearing the indicated hantavirus glycoproteins. Viral infectivities are shown in the heatmap. Averages are from three independent experiments (d) WT and double-KO cells were exposed to rVSV-Gn/Gc. PCDH1/ITGB3 KO, n = 12; PCDH1/DAF KO, n = 12; and ITGB3/DAF KO, n=9 from three independent experiments. WT versus KO cells, two-way ANOVA with Dunnett’s test; ****p<0.0001. Other comparisons were not statistically significant. (e) Cells were exposed to authentic hantaviruses and infected cells were manually enumerated by immunofluorescence microscopy for ANDV, HTNV, and PUUV (each point represents infectivity of the average of positive cells per field relative to WT). Data are from two independent experiments. PHV- and SNV-infected cells were detected and enumerated by automated imaging following immunofluorescence staining. For PHV: WT and PCDH1 KO n=18, ITGB3 KO n = 16 from four independent experiments; DAF KO, n=10, ITGB1 KO n=8, from three independent experiments. For SNV: WT, ITGB3 KO, DAF KO n=6; PCDH1 KO n = 5 from three independent experiments. Averages ± SD are shown. WT versus KO cells, two-way ANOVA with Tukey’s test; ***p<0.0001. Other comparisons were not statistically significant (p>0.05).
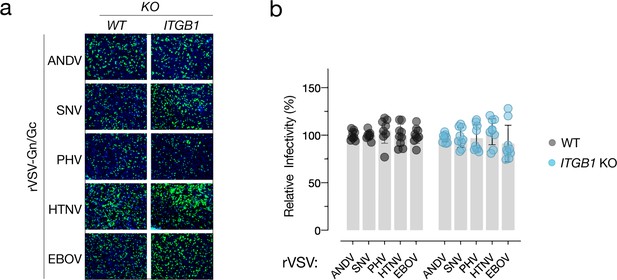
Dispensability of β1 integrin in TIME cells.
(a) Representative images of eGFP-positive rVSV-infected wild-type (WT) and ITGB1 knockout (KO) TIME cells. Nuclei were stained with Hoechst (blue). (b) WT and KO cells were exposed to the rVSV bearing the ANDV, SNV, PHV, HTNV, and EBOV glycoproteins. n=12 for each cell line from three independent experiments. WT versus KO cells, two-way ANOVA. Comparisons were not statistically significant (p>0.05).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | PCDH1 | GenBank | Gene ID: 5097 | |
| Gene (Homo sapiens) | ITGB3 | GenBank | Gene ID: 3690 | |
| Gene (Homo sapiens) | DAF | GenBank | Gene ID: 1604 | |
| Gene (Homo sapiens) | ITGB1 | GenBank | Gene ID: 3688 | |
| Strain, strain background (virus) | rVSV eGFP ANDV Gn/Gc | Kleinfelter et al., 2015 | ||
| Strain, strain background (virus) | rVSV eGFP SNV Gn/Gc | Kleinfelter et al., 2015 | ||
| Strain, strain background (virus) | rVSV eGFP HTNV Gn/Gc | Kleinfelter et al., 2015 | ||
| Strain, strain background (virus) | rVSV eGFP SEOV Gn/Gc | Jangra et al., 2018 | ||
| Strain, strain background (virus) | rVSV eGFP DOBV Gn/Gc | Slough et al., 2019 | ||
| Strain, strain background (virus) | rVSV eGFP MPRLV Gn/Gc | Jangra et al., 2018 | ||
| Strain, strain background (virus) | rVSV eGFP PHV Gn/Gc | Jangra et al., 2018 | ||
| Strain, strain background (virus) | rVSV mNeongreen-P PUUV-Gn/Gc | Kerkman et al., 2019, This study | Laboratory of K. Chandran | |
| Strain, strain background (virus) | rVSV mNeongreen-P CHOV Gn/Gc | This study | GenBank # KT983772.1 | Laboratory of K. Chandran/VSV antigenome plasmid (Whelan et al., 1995), plasmids expressing T7 polymerase and VSV N, P, M, G, and L (Witko et al., 2006) |
| Strain, strain background (virus) | rVSV-EBOV/Mayinga GP (EBOV/H.sap-tc/COD/76/ Yambuku-Mayinga) | Wong et al., 2010 | ||
| Strain, strain background (Hantavirus) | ANDV isolate Chile-9717869 | N/A | ||
| Strain, strain background (Hantavirus) | HTNV isolate 76–118 | N/A | ||
| Strain, strain background (Hantavirus) | PUUV isolate Sotkamo | N/A | ||
| Strain, strain background (Hantavirus) | SNV isolate SN77734 | Botten et al., 2000 | ||
| Strain, strain background (Hantavirus) | PHV | N/A | Laboratory of K. Chandran | |
| Cell line (H. sapiens) | 293T | ATCC | Cat. # CRL-3216 | |
| Cell line (H. sapiens) | HUVEC | Lonza | Cat. # C2517A | Primary cell |
| Cell line (H. sapiens) | TIME (endothelial cell line) | ATCC | Cat. # CRL-4025 | |
| Cell line (C. aethiops) | Vero | CCL-81 | Cat. # CCL-81 | |
| Genetic reagent (H. sapiens) | TIME PCDH1 KO (endothelial cell line) | This study | Laboratory of K. Chandran/Lentiviral transduction/lentiCRISPR v2-sgRNA PCDH1 | |
| Genetic reagent (H. sapiens) | TIME ITGB3 KO (endothelial cell line) | This study | Laboratory of K. Chandran/Lentiviral transduction/lentiCRISPR v2-sgRNA ITGB3 | |
| Genetic reagent (H. sapiens) | TIME ITGB1 KO (endothelial cell line) | This study | Laboratory of K. Chandran/Lentiviral transduction/lentiCRISPR v2-sgRNA ITGB3 | |
| Genetic reagent (H. sapiens) | TIME DAF KO (endothelial cell line) | This study | Laboratory of K. Chandran/Lentiviral transduction/lentiCRISPR v2-sgRNA DAF | |
| Genetic reagent (H. sapiens) | TIME PCDH1/ITGB3 KO (endothelial cell line) | This study | Laboratory of K. Chandran/TIME PCDH1 KO + Lentiviral transduction/lentiCRISPR v2-sgRNA ITGB3 | |
| Genetic reagent (H. sapiens) | TIME PCDH1/DAF KO (endothelial cell line) | This study | Laboratory of K. Chandran / TIME PCDH1 KO + Lentiviral transduction/ lentiCRISPR v2-sgRNA DAF | |
| Genetic reagent (H. sapiens) | TIME ITGB3/DAF KO (endothelial cell line) | This study | Laboratory of K. Chandran / TIME ITGB3 KO + Lentiviral transduction/ lentiCRISPR v2-sgRNA DAF | |
| Genetic reagent (H. sapiens) | TIME PCDH1 KO + PCDH1 (endothelial cell line) | This study | Canonical isoform Q08174-1 (Uniprot) | Laboratory of K. Chandran / Retroviral transduction/ pBabe-PCDH1 |
| Genetic reagent (H. sapiens) | TIME ITGB3 KO + ITGB3 (endothelial cell line) | This study | Canonical isoform P05106-1 (Uniprot) | Laboratory of K. Chandran / Retroviral transduction/ pBabe-ITGB3 |
| Genetic reagent (H. sapiens) | TIME DAF KO + DAF (endothelial cell line) | This study | Canonical isoform P08174-1 (Uniprot) | Laboratory of K. Chandran / Retroviral transduction/ pBabe-DAF |
| Antibody | AF480-α-Human-vWF (Rabbit monoclonal) | Abcam | Cat. # ab195028 | Flow 1:250 |
| Antibody | PeCy7-α-Human-PECAM (Mouse monoclonal) | BD | Cat. # 563651 | Flow 1:1000 |
| Antibody | AF 647-α-Human- β3-Integrin (Mouse monoclonal) | Biolegends | Cat. # 336407 | Flow 1:200 |
| Antibody | PE-α-Human-DAF (Mouse monoclonal) | BD | Cat. # 555694 | Flow 1:1600 |
| Antibody | APC-α-Human-β1-Integrin (Mouse monoclonal) | BD | Cat. # 59883 | Flow 1:20 |
| Antibody | α–Human-PCDH1 mAb 3305 (human monoclonal) | Jangra et al., 2018 | Flow 1:200 | |
| Antibody | Convalescent serum (human polyclonal) | Stoltz et al., 2007 | IF: 1:40 | |
| Antibody | AF488-α–-Human IgG (Goat polyclonal) | Invitrogen | Cat. # A-11013 | Flow 1:200 |
| Antibody | AF594-α–Human IgG (Goat polyclonal) | Invitrogen | Cat. # A-11014 | IF 1:500 |
| Antibody | AF594-α-Mouse IgG (Goat polyclonal) | Invitrogen | Cat. # A32742 | IF 1:500 |
| Antibody | AF488-α-Rabbit IgG (Goat polyclonal) | Invitrogen | Cat. # A-11008 | IF 1:500 |
| Antibody | α–Human PCDH1 (Mouse monoclonal) | Santa Cruz | Cat. # sc-81816 | WB 1:200 |
| Antibody | α–Human β3 Integrin (Rabbit polyclonal) | Cell Signaling | Cat. # 4702 | WB 1:300 |
| Antibody | α–Human-DAF (Mouse monoclonal) | Santa Cruz | Cat. # NaM16-4D3 | WB 1:200 |
| Antibody | α–Human β Actin (Mouse monoclonal) | Santa Cruz | Cat. # sc-47778 | WB 1:300 |
| Antibody | IRDye 680LT α-Mouse (Goat polyclonal) | LI-COR | Cat. # 926–68020 | WB 1:10,000 |
| Antibody | IRDye 680LT Goat α-Rabbit IgG 680 (Goat polyclonal) | LI-COR | Cat. # 926–68021 | WB 1:10,000 |
| Antibody | α-hantavirus nucleocapsid B5D9 (Mouse monoclonal) | Progen | Cat. # B5D9-C | IF 1:50 |
| Antibody | α-HTNV nucleoprotein NR-12152 (Rabbit polyclonal) | BEI resources | Cat. # NR-12152 | IF 1:500 |
| Recombinant DNA reagent | pBabe-puro (plasmid) | Morgenstern and Land, 1990 | Addgene plasmid # 1764 | |
| Recombinant DNA reagent | pBabe-PCDH1 (plasmid) | Jangra et al., 2018 | ||
| Recombinant DNA reagent | pBabe-ITGB3 (plasmid) | This study | Laboratory of K. Chandran | |
| Recombinant DNA reagent | pBabe-DAF (plasmid) | This study | Laboratory of K. Chandran | |
| Recombinant DNA reagent | lentiCRISPR v2 (plasmid) | Sanjana et al., 2014; Shalem et al., 2014 | ||
| Recombinant DNA reagent | lentiCRISPR v2-sgRNA PCDH1 (plasmid) | Jangra et al., 2018 | Laboratory of K. Chandran/5′-GTTTGAGCGGCCCTCCTATGAGG-3′ /PAM sequence is underlined but not included in oligos | |
| Recombinant DNA reagent | lentiCRISPR v2-sgRNA DAF (plasmid) | This study | Laboratory of K. Chandran / 5′-CCCCCAGATGTACCTAATGCCCA-3′ / PAM sequence is underlined but not included in oligos | |
| Recombinant DNA reagent | lentiCRISPR v2-sgRNA ITGB3 (plasmid) | This study | Laboratory of K. Chandran / 5′-CCACGCGAGGTGTGAGCTCCTGC-3′ /PAM sequence is underlined but not included in oligos | |
| Recombinant DNA reagent | lentiCRISPR v2-sgRNA ITGB1 (plasmid) | This study | Laboratory of K. Chandran/5′-AATGTAACCAACCGTAGCAAAGG-3′ /PAM sequence is underlined but not included in oligos | |
| Sequence-based reagent | SNV degenerate primers | This study | Laboratory of S. Bradfute/SNV F: CAgCTgTgTCTgCATTggAgAC SNV R: TARAgYCCgATggATTTCCAATCA | |
| Sequence-based reagent | SNV probe | This study | Laboratory of S. Bradfute/ TMGB1: F-TCAAAACCTgTTgATCCA NFQ MGB | |
| Commercial assay or kit | pGEM-T Vector | Promega | Cat. # A3600 | |
| Commercial assay or kit | VIC/TAMRA-dye probe | Applied Biosystems | Cat. # 4310881E | |
| Commercial assay or kit | TaqMan Fast Advanced Master Mix | Applied Biosystems | Cat. # 4444556 | |
| Commercial assay or kit | Foxp3/Transcription Factor Staining | Tonbo | Cat. # TNB-0607-KIT | |
| Commercial assay or kit | Zombie NIR Fixable Viability Kit | BioLegend | Cat. # 423105 | Flow 1:4000 |
| Software, algorithm | Cytation 5 Cell Imaging Multi-Mode Reader | Biotek |
| gRNA target site location | Modified sequence in pBabe- PCDH1/ ITGB3/ DAF | |
|---|---|---|
| PCDH1 | GTTTGAGCGGCCCTCCTATGAGG | aTTcGAGaGaCCtagtTATGAGG |
| ITGB3 | CCACGCGAGGTGTGAGCTCCTGC | CtACtaGAGGcGTatcaagCTGt |
| DAF | CCCCCAGATGTACCTAATGCCCA | CCgCCtGAcGTcCCaAAcGCgCA |





