Sex-specific pubertal and metabolic regulation of Kiss1 neurons via Nhlh2
Figures
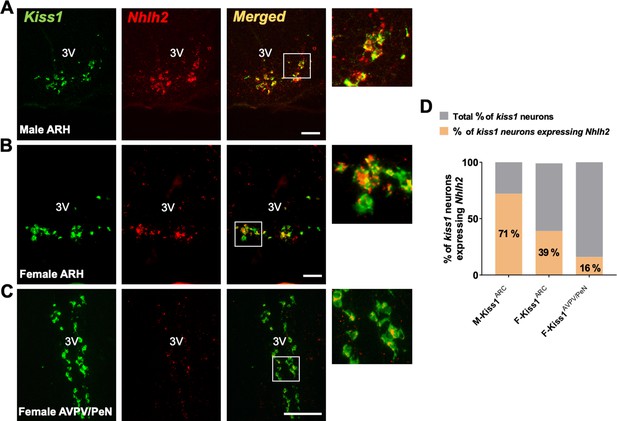
Nhlh2 is a marker of Kiss1 neurons.
(A) In situ hybridization (RNAscope) of Kiss1 and Nhlh2 in the ARH of adult male (A) and ovariectomized female mice (B), and in the AVPV/PeN of ovariectomized+estradiol treated female mice (C). (D) Percentage of Kiss1 neurons expressing Nhlh2. Scale bar=100 μm.
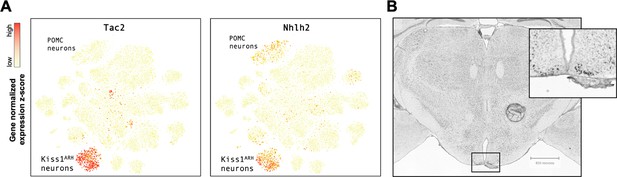
Nhlh2 is enriched in Kiss1 neurons of adult mice.
(A) Expression of marker genes shown by recoloring of neuron-only spectral tSNE plot of 13,079 neurons. (B) In situ hybridization of Nhlh2 in the brain of an adult male mouse by the Allen Brain Atlas. tSNE, t-distributed stochastic neighbor embedding.
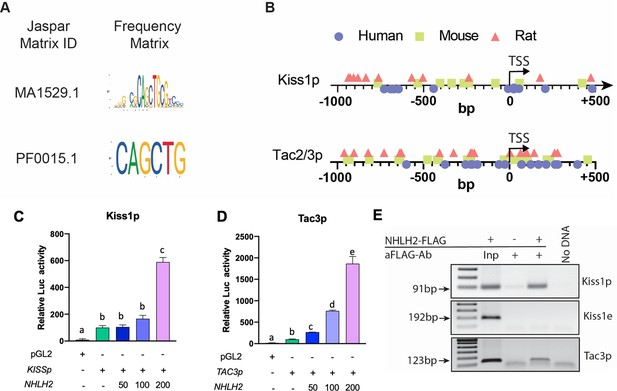
NHLH2 is recruited to and stimulates KISS1 and TAC3 promoter activity.
(A, B) Identification of NHLH2 biding sites in Kiss1 and Tac2/3. NHLH2 DNA recognition sites as identified in two Jaspar matrices (A). Distribution of NHLH2 binding sites in human, mouse, and rat Kiss1 promoters (Kiss1p) and TAC2/3 promoters (Tac2/3 p) (B). (C–E) Effect of NHLH2 on luciferase activity regulated by the human KISS1 (C) and TAC3 (D), promoter (p). Neuro-2a cells were transfected with luciferase reporter constructs containing the 5′ flanking region of the indicated genes. After 48 h, the cells were harvested and assayed for luciferase activity. Bars represent mean± SEM (n=three biological replicates per group). Groups with different letters are significantly different (P<0.05), as determined by one-way ANOVA followed by the Student-Newman-Keuls test. (E) Association of NHLH2 with the 5′ promoter region of the KISS1 (Kiss1p) and TAC3 (Tacr3p) genes as determined by PCR amplification of DNA immunoprecipitated with antibodies recognizing the FLAG/OctA epitope tagging NHLH2. As negative control, a region of the Kiss1 promoter outside of the area containing Nhlh2 E-box was assayed (Kiss1e). Inp=Input. Data presented as the mean± SEM.
-
Figure 2—source data 1
Association of NHLH2 with the 5' promoter regeion of KISS1.
- https://cdn.elifesciences.org/articles/69765/elife-69765-fig2-data1-v2.pdf
-
Figure 2—source data 2
Association of NHLH2 with a region of the Kiss1 promoter outside of the area containing Nhlh2 E-box was assayed (Kiss1e).
- https://cdn.elifesciences.org/articles/69765/elife-69765-fig2-data2-v2.pdf
-
Figure 2—source data 3
Association of NHLH2 with the 5' promoter regeion of TAC3.
- https://cdn.elifesciences.org/articles/69765/elife-69765-fig2-data3-v2.pdf
-
Figure 2—source data 4
Images of uncropped geles.
- https://cdn.elifesciences.org/articles/69765/elife-69765-fig2-data4-v2.pptx
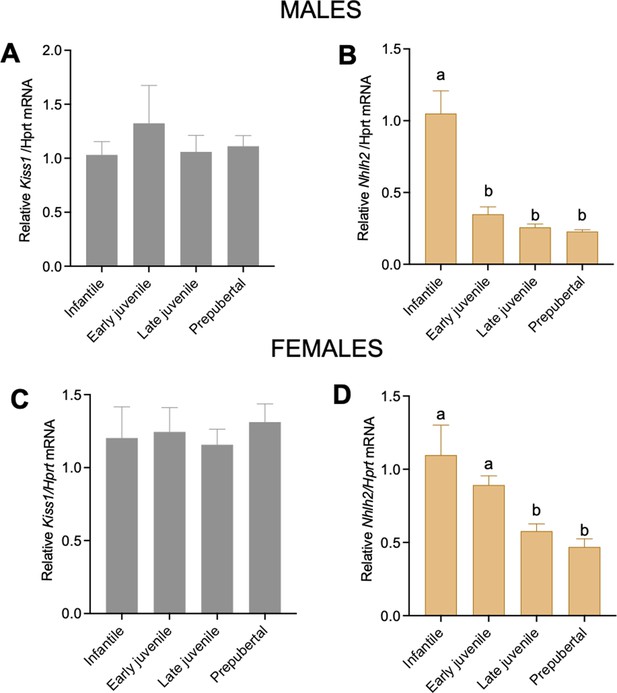
Gene expression of Kiss1 and Nhlh2 in the ARH during the prepubertal period.
Expression profile of Kiss1 and Nhlh2 in the MBH of WT male (n=5/group) (A, B) and female (n=6/group) (C, D) mice at different postnatal time points: infantile, early juvenile, late juvenile, and prepubertal, normalized to the housekeeping gene Hprt. Groups with different letters are significantly different (p<0.05), as determined by one-way ANOVA followed by the Student-Newman-Keuls test. Data presented as the mean± SEM.
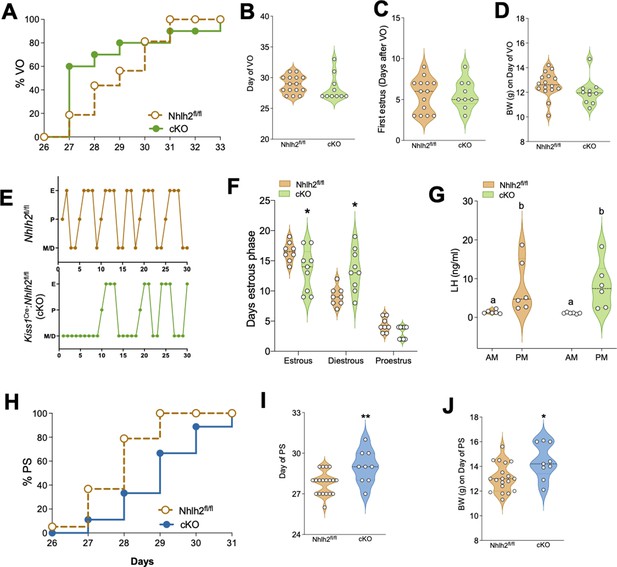
Kiss1Cre:Nhlh2fl/fl male mice present delayed puberty onset while females display irregular estrous cycles.
Female Kiss1Cre:Nhlh2fl/fl mice present normal puberty onset as assessed by daily monitoring of vaginal opening (VO) and documented by cumulative percent of animals at VO (A), mean age of VO (B) Nhlh2fl/fl (n=15) and Kiss1Cre:Nhlh2fl/fl (n=10) and first estrus (C) Nhlh2fl/fl (n=14) and Kiss1Cre:Nhlh2fl/fl (n=9). These females have normal body weight (BW) at the age of VO (D). Estrous cyclicity, assessed by daily vaginal cytology for 30 days is irregular in Kiss1Cre:Nhlh2fl/fl mice (E, F), presenting longer time in diestrus compared to control, Nhlh2fl/fl (n=8) and Kiss1Cre:Nhlh2fl/fl (n=9). *p<0.05 two-way ANOVA followed by Bonferroni test. (G) LH measurements in the morning (AM [10 a.m.]) and afternoon (PM [7:30 p.m.]) after lights off of female mice subjected to an LH surge induction protocol. n=6/group. Groups with different letters are significantly different. Two-way ANOVA followed by Bonferroni post hoc test. (H, I) Pubertal development of male Kiss1Cre:Nhlh2fl/fl mice and control littermates determined by preputial separation. *p<0.05 by Student's t-test. (J) BW of males at the age of preputial separation. Nhlh2fl/fl (n=19) and Kiss1Cre:Nhlh2fl/fl (n=9). *p<0.05 by Student's t-test. Data presented as median (dotted line)+distribution of the data and its probability density (violin plot). cKO=Kiss1Cre:Nhlh2fl/fl. LH, luteinizing hormone.
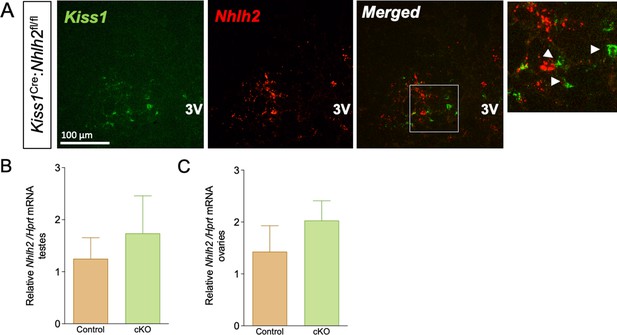
Validation of the Kiss1Cre:Nhlh2fl/fl mouse model.
Representative images depicting the absence of co-expression of Kiss1 (green) and Nhlh2 (red) mRNA in the ARH of adult Kiss1Cre:Nhlh2fl/fl male mice (A). White arrows indicate examples of Kiss1 neurons showing the absence of Nhlh2 expression in them. Expression of Nhlh2 in testes (B) and ovaries (C) from adult Kiss1Cre:Nhlh2fl/fl mice and their littermate controls (n=4–5/group). cKO=Kiss1Cre:Nhlh2fl/fl.
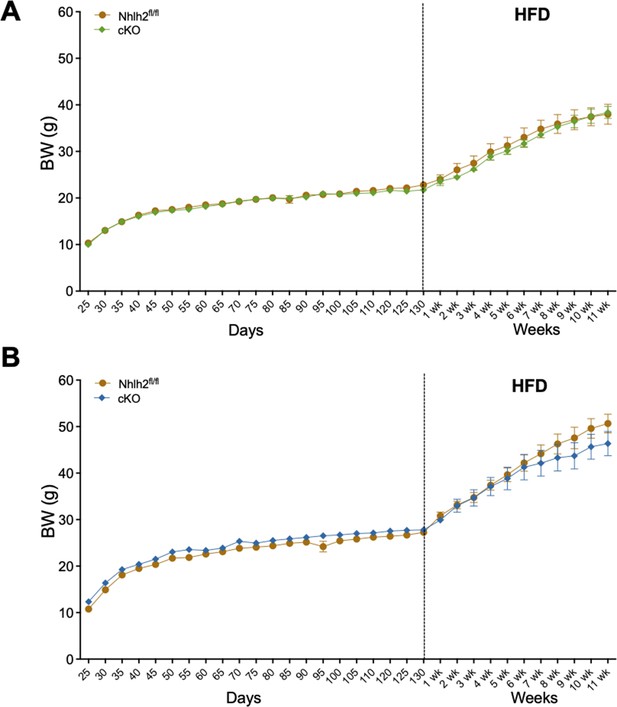
Kiss1Cre:Nhlh2fl/fl mice display normal body weight (BW).
BW of Kiss1Cre:Nhlh2fl/fl male and female mice and their respective littermate controls were monitored for 130 days under regular chow diet followed by 11 weeks on a 60% high-fat diet (HFD). cKO=Kiss1Cre:Nhlh2fl/fl.
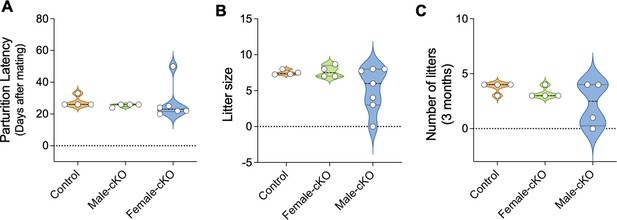
Kiss1Cre:Nhlh2fl/fl mice are fertile.
Male and female Kiss1Cre:Nhlh2fl/fl mice were mated with sexually experienced WT mates for 3 months. The time to deliver pups, that is, parturition latency (A), total number of pups in 3 months (B), and number of litters in 3 months (C) were monitored, n=4–7/group. Two-way ANOVA followed by Bonferroni post hoc test. Values are presented as median (middle line)±max/min (violin plot). cKO=Kiss1Cre:Nhlh2fl/fl. WT, wild-type.
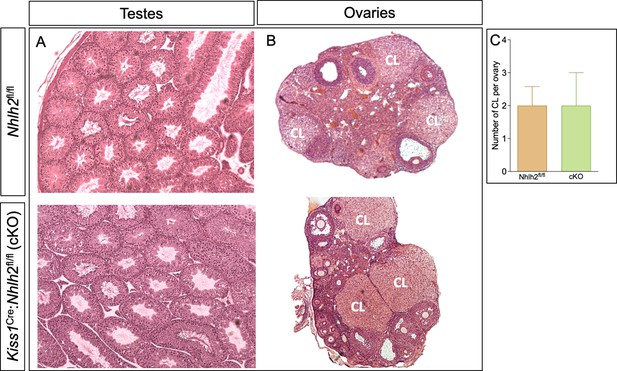
Histological analysis of testes and ovaries from Kiss1Cre:Nhlh2fl/fl mice.
Testicular (A) and ovarian (B) histology of Kiss1Cre:Nhlh2fl/fl male and female mice. (C) Number of corpora lutea compared with their respective littermate controls (n=3/group). cKO=Kiss1Cre:Nhlh2fl/fl.
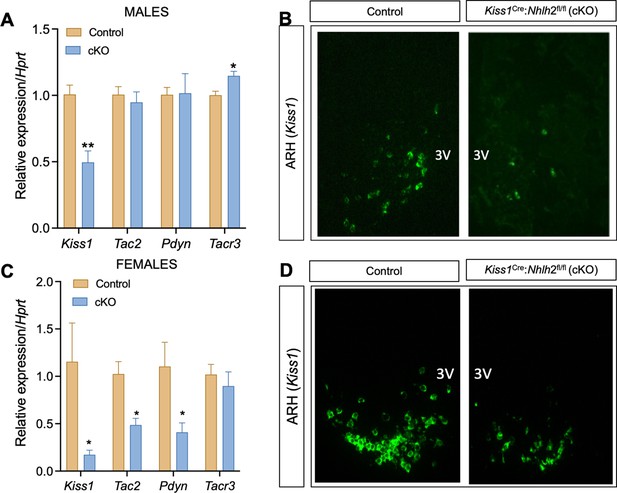
Expression of KNDy genes in the ARH of Kiss1Cre:Nhlh2fl/fl mice.
The expression of the KNDy genes (Kiss1, Tac2, and Pdyn) and the receptor for NKB (Tacr3) were assessed in the MBH of adult male (A) and female (C) Kiss1Cre:Nhlh2fl/fl mice and their control (Kiss1Cre/+) littermates (n=4/group; *p<0.05, **p<0.01. Two-way ANOVA followed by Bonferroni test). Representative images of in situ hybridization (RNAscope) depicting the expression of Kiss1 expression in the MBH of Kiss1Cre:Nhlh2fl/fl male (B) and female (D) mice compared to controls. Data presented as the mean± SEM. cKO=Kiss1Cre:Nhlh2fl/fl.
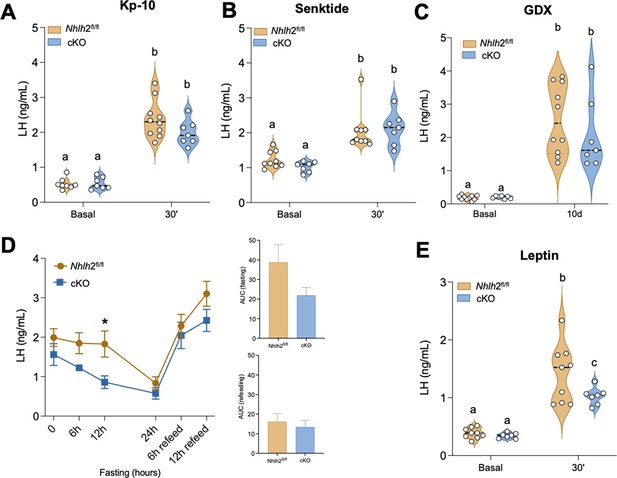
Functional assessment of Kiss1 neurons in Kiss1Cre:Nhlh2fl/fl male mice under fed and fasting conditions.
Central (icv) administration of Kp-10 (50 pmol/mouse in 5 μl) (Nhlh2fl/fl [n=10] and Kiss1Cre:Nhlh2fl/fl [n=7]) (A) or senktide (600 pmol/mouse in 5 μl) Nhlh2fl/fl (n=9) and Kiss1Cre:Nhlh2fl/fl (n=7) (B) was performed in adult male Kiss1Cre:Nhlh2fl/fl mice and controls. Blood samples were collected before and 30 min after treatment. Groups with different letters are significantly different two-way ANOVA followed by Bonferroni test. In (C), the ability of Kiss1Cre:Nhlh2fl/fl male mice to generate a compensatory LH rise after gonadectomy was assessed before surgery and 10 days post-gonadectomy. Nhlh2fl/fl (n=10) and Kiss1Cre:Nhlh2fl/fl (n=7). Two-way ANOVA followed by Bonferroni test. (D) Response of LH in gonadectomized male mice during 24 hr of fasting and 12 hr of refeeding (Nhlh2fl/fl [n=10] and Kiss1Cre:Nhlh2fl/fl [n=7]); *p<0.05 compared with the control group at 12 hr. Two-way ANOVA followed by Fisher’s test. The area under the curve (AUC) was determined during fasting and refeeding periods. (E) Response of mice to overnight fasting followed by leptin administration. Blood samples were collected before and 30 min after leptin (2 μg/mouse in 5 μl) treatment. Nhlh2fl/fl (n=9) and Kiss1Cre:Nhlh2fl/fl (n=6); Groups with different letters are significantly different. Two-way ANOVA followed by Bonferroni post hoc test. Data presented as median (dotted line)+distribution of the data and its probability density (violin plot) or the mean± SEM (in (D)). cKO=Kiss1Cre:Nhlh2fl/fl. LH, luteinizing hormone.
Tables
Primers used for ChIP.
| Gene | Accession # | Primers | Sequence | Amplicon (bp) |
|---|---|---|---|---|
| Kiss1 | NM_181692.1 | rKiss1p CHIP-F | TCGGGCAGCCAGATAGAGGAAGC | 91 |
| rKiss1p CHIP-R | TTGAGGGCCGAGGGAGAAGAG | |||
| Kiss1 | NM_181692.1 | rKiss1e CHIP-F | CCAGCCCGGGGGATGGGGTGTAA | 192 |
| rKiss1e CHIP-R | AGGGGCCAGCGCAGCAGCACAAT | |||
| Tac3 | NM_019162.2 | rTAC2p CHIP-F | ACGTGCGTGTCTGGGTATGTGA | 123 |
| rTAC2p CHIP-R | GGAGGGTTTGGGGGAGTCG | |||
| rPCSK1p CHIP-R | CCTTCGAGACAGCATTCACA |
-
F, forward; R, reverse.
Primers used for real-time RT-PCR.
| Gene | Accession # | Primers | Sequence |
|---|---|---|---|
| Hprt | NM_013556.2 | Hprt-F | CCTGCTGGATTACATTAAAGCGCTG |
| Hprt-R | GTCAAGGGCATATCCAACAACAAAC | ||
| Kiss1 | AF472576.1 | Kiss1-F | GCTGCTGCTTCTCCTCTGTG |
| Kiss1-R | TCTGCATACCGCGATTCCTT | ||
| Tac1 | NM_009311.2 | Tac1-F | ATGAAAATCCTCGTGGCCGT |
| Tac1-R | GTTCTGCATCGCGCTTCTTT | ||
| Pdyn | NM_018863.4 | Pdyn-F | ACAGGGGGAGACTCTCATCT |
| Pdyn-R | GGGGATGAATGACCTGCTTACT | ||
| Tacr3 | NM_021382.6 | Tacr3-F | GCCATTGCAGTGGACAGGTAT |
| Tacr3-R | ACGGCCTGGCATGACTTTTA | ||
| Nhlh2 | NM_178777.3 | Nhlh2-F | ACCAGAAGAGCCAAGAAGCCA |
| Nhlh2-R | GCGGGTGTATGGTTGTTCACTTAG |
-
F, forward; R, reverse.
Additional files
-
Supplementary file 1
Jaspar Analysis of Nhlh2 binding sites.
- https://cdn.elifesciences.org/articles/69765/elife-69765-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69765/elife-69765-transrepform-v2.docx





