Giant ankyrin-B mediates transduction of axon guidance and collateral branch pruning factor sema 3A
Figures
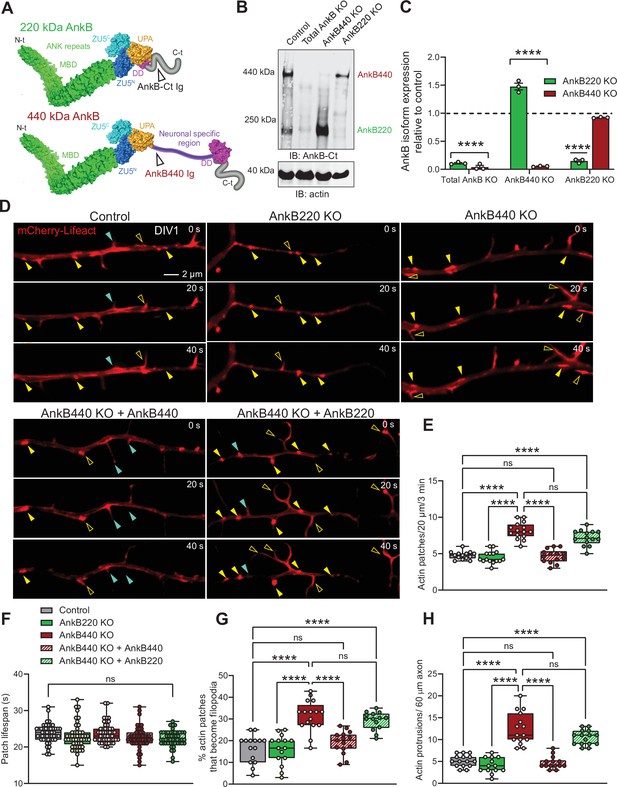
Ankyrin-B 440 suppresses the formation of axonal filopodia in vitro.
(A) Representation of the two major ankyrin-B isoforms in the mammalian brain and their functional domains. The localization of the epitopes recognized by the pan anti-ankyrin B (AnkB-Ct Ig) or the anti-440 kDa ankyrin-B (AnkB440 Ig) antibodies is indicated by arrowheads. (B) Western blot analysis of expression of ankyrin-B isoforms in the cortex of PND1 control mice and of mice individually lacking the 220 kDa ankyrin-B isoform (AnkB220 KO), the 440 kDa ankyrin-B isoform (AnkB440 KO), and both isoforms (Total AnkB KO). (C) Quantification of AnkB220 and AnkB440 levels normalized to actin in cortical lysates from PND1 mice of indicated genotypes relative to their levels in control brains. Data show mean ± SEM for three biological replicates per genotype representative of one of three independent experiments. Unpaired t test. ****p < 0.0001. (D) Images selected from timelapse sequences of DIV1 neurons expressing mCherry-Lifeact showing the formation of actin patches (asterisks), some of which do not form filopodia (closed yellow arrowheads) or lead to filopodia that may persist (open yellow arrowheads) or retract (closed teal arrowheads). Scale bar, 2 μm. (E) Quantification of actin patches observed in 20 μm of axons during a 3 minute acquisition period. Data was collected from n = 14 control, n = 15 AnkB220 KO, n = 15 AnkB440 KO, n = 14 AnkB440 KO+ AnkB440, and n = 14 AnkB440 KO+ AnkB220 axons from three independent experiments. (F) Quantification of actin patch lifespan derived from the analysis of n = 50 patches per each group collected from three independent experiments. (G) Quantification of proportion of actin patches that result in filopodia during the movie acquisition interval derived from analyses of n = 15 control, n = 15 AnkB220 KO, n = 16 AnkB440 KO, n = 15 AnkB440 KO+ AnkB440, and n = 14 AnkB440 KO+ AnkB220 axons from three independent experiments. (H) Quantification of filopodia per 60 μm of axons obtained from the analysis of n = 15 control, n = 13 AnkB220 KO, n = 15 AnkB440 KO, n = 14 AnkB440 KO+ AnkB440, and n = 15 AnkB440 KO+ AnkB220axons from three independent experiments. The box and whisker plots in E–H represent all data points collected arranged from minimum to maximum. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. nsp >0.05, ****p < 0.0001.
-
Figure 1—source data 1
Ankyrin-B 440 regulates the formation of axonal filopodia and branches.
- https://cdn.elifesciences.org/articles/69815/elife-69815-fig1-data1-v1.zip
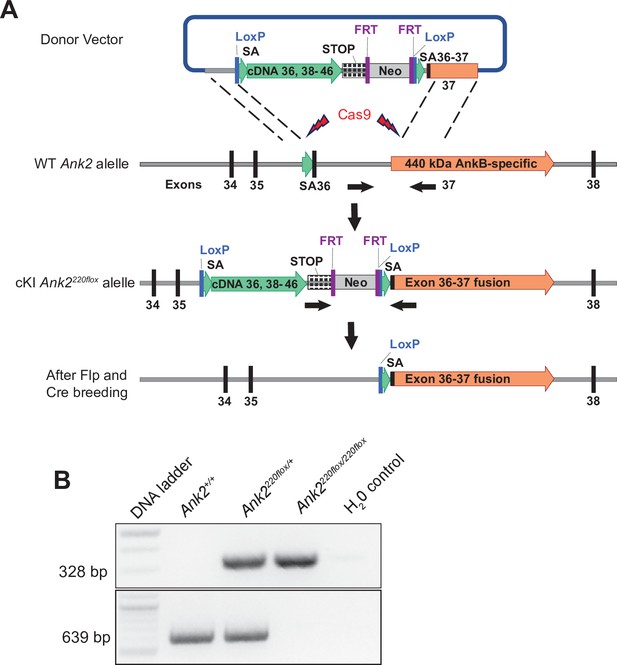
Development of conditional AnkB220 knockout mouse.
(A) Strategy used to generate a targeting Ank2 allele (cKI Ank2220flox). A donor vector containing a rescue cDNA cassette in which exons 36–37 of Ank2 were fused to prevent splicing of exon 37 unique to the AnkB440 transcript was introduced into mouse ES cells using CRISPR/Cas9. Transgenic mice bearing the cKI Ank2220flox allele were crossed to a mouse line expressing the flippase (FLP) recombinase to eliminate expression of the Neomycin (Neo) selection marker. Breeding to Nestin-Cre resulted in loss of AnkB220 while preserved AnkB440 levels. (B) Example of results of genotyping PCR from mouse genomic DNA to identify mice heterozygous or homozygous for the conditional Ank2220flox allele.
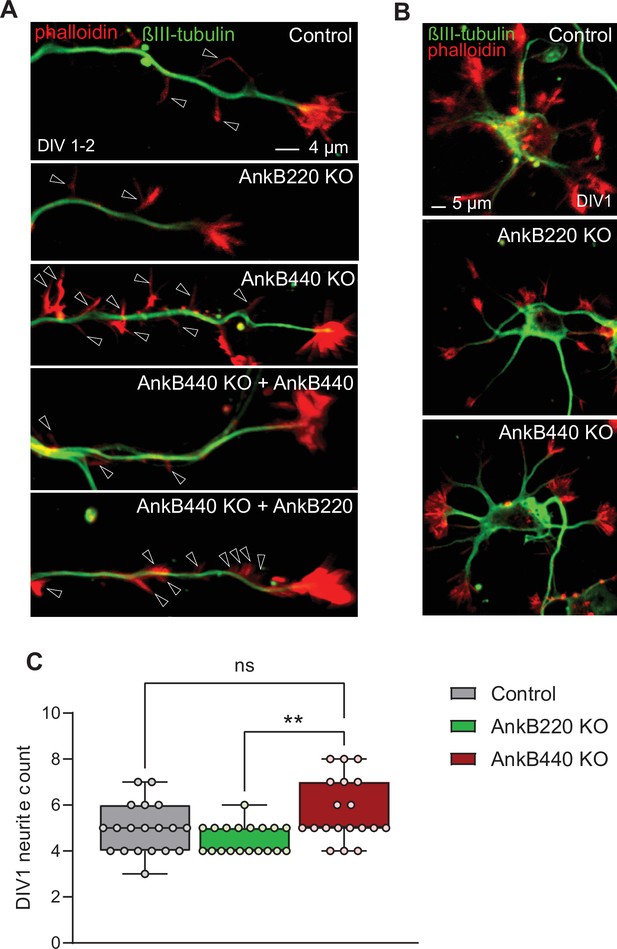
AnkB440 loss increases axon filopodia without early changes in total neurite count.
(A) Images of DIV1-2 cortical neurons of indicated genotypes stained for phalloidin and βIII-tubulin. Open arrowheads indicate the presence of axonal filopodia. Scale bar, 4 μm. (B) Images of stage 1 DIV1 cortical neurons of indicated genotypes stained for phalloidin and βIII-tubulin. Scale bar, 5 μm. (C) Total neurite count at DIV1 computed from n = 19 neurons per genotype from three independent experiments. The box and whisker plots in C represent all data points collected arranged from minimum to maximum. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. nsp >0.05, **p < 0.01.
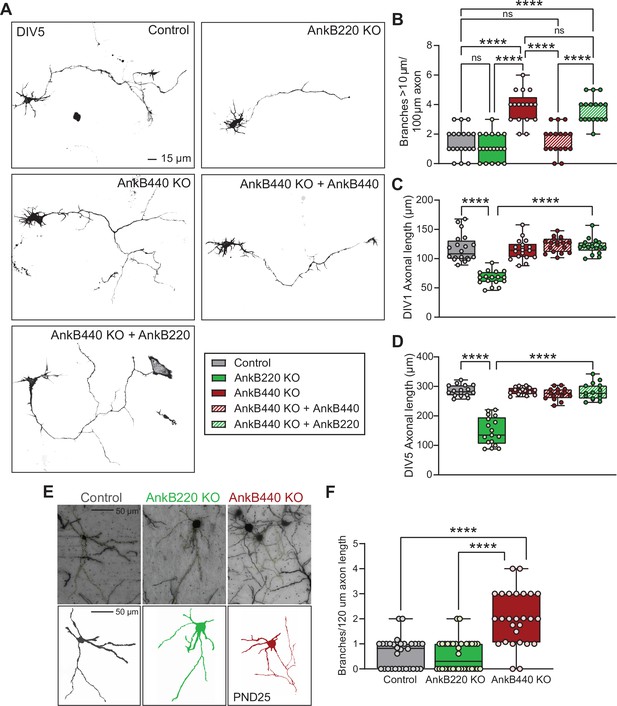
AnkB440 and AnkB220 isoforms exert different roles in axonal growth and branching.
(A) Representative images of mCherry-labeled DIV5 cortical neurons of indicated genotypes. Scale bar, 15 μm. (B) Quantification of axonal branches per 100 μm of axon of DIV5 (n = 19 control, n = 19 AnkB220 KO, n = 17 AnkB440 KO, n = 17 AnkB440 KO+ AnkB440, and n = 17 AnkB440 KO+ AnkB220) mCherry-labeled neurons from three independent experiments. (C) Quantification of axonal length of DIV1 (n = 20 control, n = 17 AnkB220 KO, n = 18 AnkB440 KO, n = 18 AnkB440 KO+ AnkB440, and n = 17 AnkB440 KO+ AnkB220) mCherry-labeled neurons from three independent experiments. (D) Quantification of axonal length of DIV5 (n = 16 control, n = 17 AnkB220 KO, n = 15 AnkB440 KO, n = 15 AnkB440 KO+ AnkB440, and n = 15 AnkB440 KO+ AnkB220) mCherry-labeled neurons from three independent experiments. (E) Images of Golgi-stained (top) projection neurons in layers II-III of the primary somatosensory cortex of PND25 mice and corresponding traces (bottom). Scale bar, 50 μm. (F) Quantification of axonal branches per 120 μm of axon (n = 30 control, n = 34 AnkB220 KO, and n = 27 AnkB440 KO) Golgi-stained neurons from n = 3 mice/genotype from one experiment. The box and whisker plots in B–D, F represent all data points collected arranged from minimum to maximum. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. nsp >0.05, ****p < 0.0001.
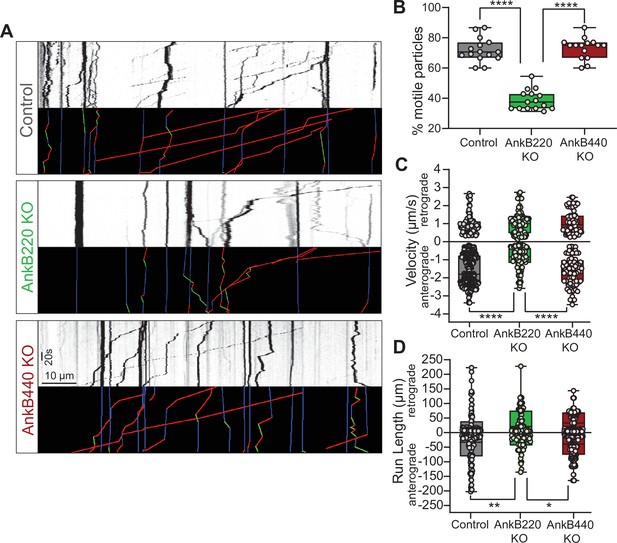
AnkB220, but not AnkB440, promotes axonal organelle transport.
(A) Kymographs showing motility of GFP-tagged LAMP1 late endosomes/lysosomes at the distal axon of DIV7 neurons from the indicated genotypes. Trajectories in the color-coded kymographs are shown in green for anterograde-moving, red for retrograde-moving, and blue for static cargo (Scale bar, 10 μm and 20 s). (B) Percent of motile GFP-LAMP1 cargo per axon computed from n = 15 axons per genotype. (C) Anterograde and retrograde velocity of GFP-LAMP1 cargo in axons of indicated genotypes (control: anterograde n = 85, retrograde n = 364; AnkB220 KO: anterograde n = 87, retrograde n = 101; AnkB440 KO: anterograde n = 73, retrograde n = 76). (D) Run length of GFP-LAMP1 cargo in axons of indicated genotypes (control: anterograde n = 103, retrograde n = 115; AnkB220 KO: anterograde n = 79, retrograde n = 65; AnkB440 KO: anterograde n = 144, retrograde n = 171). The box and whisker plots in B–D represent all data points collected from three independent experiments arranged from minimum to maximum. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. *p < 0.05, **p < 0.01, ****p < 0.0001.
AnkB220 promotes axonal organelle transport.
Timelapse images illustrate the motility of GFP-tagged LAMP1 late endosomes/lysosomes at the distal axon of DIV7 cortical neurons from the indicated genotypes. Movies are played at 15 frames/second. Scale bar, 10 μm.
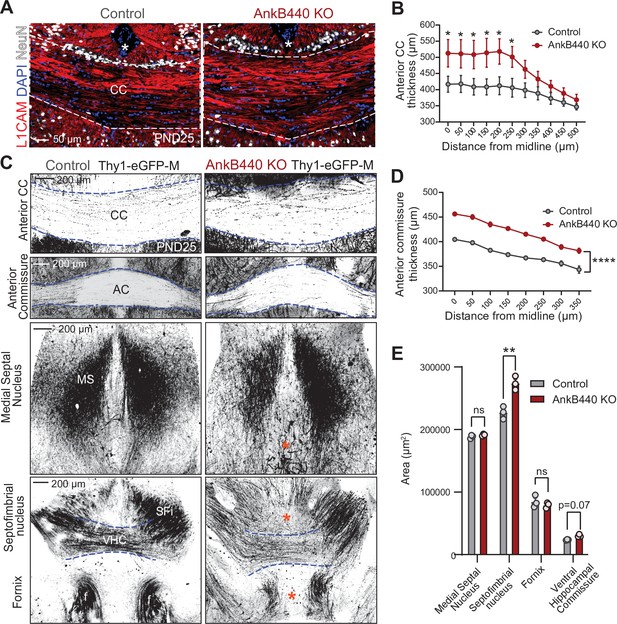
Loss of AnkB440 alters the formation of axonal tracts in mouse brains.
(A) Images of coronal sections of PND25 mouse brains taken through the anterior portion of the corpus callosum (CC) and stained for L1CAM to label axons, and DAPI and NeuN to respectively stain total and neuronal nuclei. An asterisk indicates the position of the brain midline. Scale bar, 50 μm. (B) Quantification of thickness of anterior region of the CC at different points from the brain midline from control (n = 4) and AnkB440 KO (n = 5) brains. (C) Images of different axon tracts of control and AnkB440 KO PN25 brains that also express the Thy1-GFP-M reporter. Scale bar, 200 μm. Blue lines demark the boundaries of indicated commissural tracts. Orange asterisks indicate mistargeted axons. AC (anterior commissure), MS (medial septal nucleus), VHC (ventral hippocampal commissure), SFi (septobimbrial nucleus). (D) Quantification of thickness of the AC at different points from the brain midline from control (n = 3) and AnkB440 KO (n = 3) brains. (E) Area of indicated tracts evaluated from control (n = 3) and AnkB440 KO (n = 3) Thy1-GFP-M-positive brains. Data in B, D, E represent mean ± SEM. Unpaired t test. nsp >0.05, *p < 0.05, **p < 0.01, ****p < 0.0001 (for all distances evaluated in D).
-
Figure 2—source data 1
Loss of AnkB440 alters axonal tracts in mouse brains.
- https://cdn.elifesciences.org/articles/69815/elife-69815-fig2-data1-v1.zip
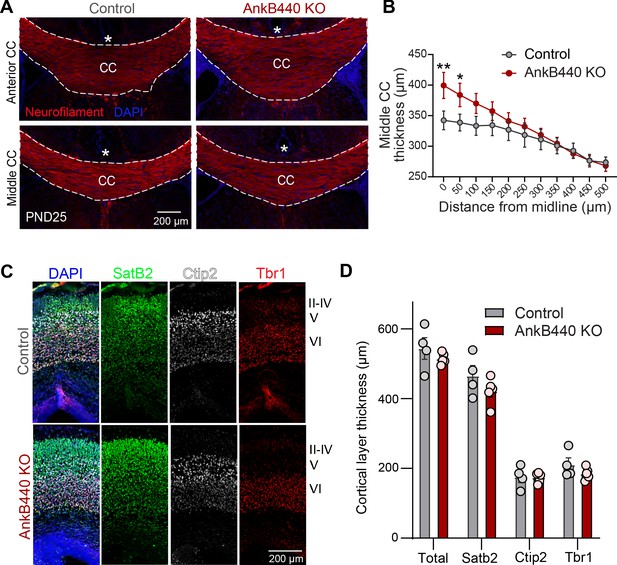
AnkB440 loss causes corpus callosum hyperplasia.
(A) Images of coronal sections of PND25 mouse brains taken through the anterior and middle portions of the corpus callosum (CC) and stained for neurofilament to label axons and DAPI to stain nuclei. An asterisk indicates the position of the brain midline. Scale bar, 200 μm. (B) Quantification of thickness of middle regions of the CC at different points from the brain midline from control (n = 4) and AnkB440 KO (n = 5) brains. (C) Images of PND0 brains stained for Satb2, Ctip2, and Tbr1 to label neocortical layers and DAPI to stain nuclei. Scale bar, 200 μm. (D) Quantification of total, Sabt2-, Ctip2-, and Tbr1-positive cortical layer thickness from PND0 control (n = 5–6) and AnkB440 KO (n = 4) brains. Data in B and D represent mean ± SEM. Unpaired t test. *p < 0.05, **p < 0.01.
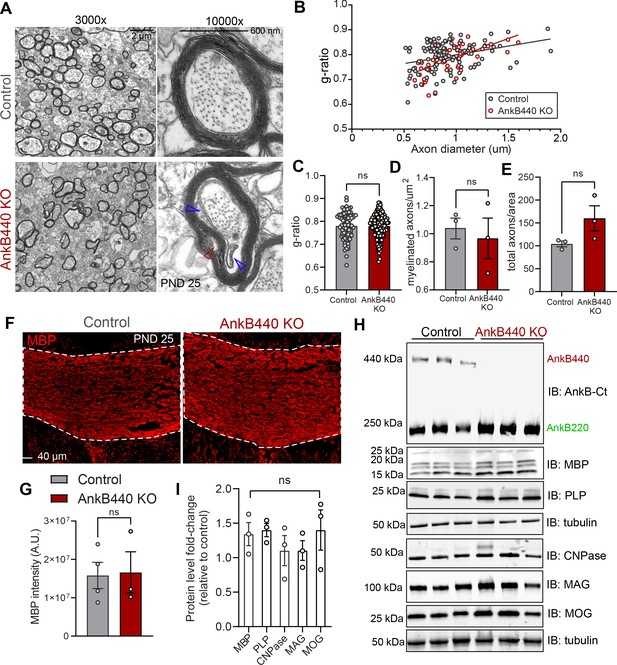
Loss of AnkB440 does not change levels of myelination of CC axons.
(A) TEM images of cross-sections through the CC near the brain midline from PND25 control and AnkB440 KO mice (n = 3 mice/genotype). Images were taken at 3000 x (scale bar, 2 μm) and 10,000 x (scale bar, 600 nm) magnification. Open red and blue arrowheads respectively indicate loose myelin wraps and enlarged inner tongues. (B) G-ratio versus axon diameter computed from control (n = 115) and AnkB440 KO (n = 63) axons from n = 3 mice/genotype. (C) Myelin g-ratio computed from (n = 84) and AnkB440 KO (n = 120) axons from n = 3 mice/genotype. (D) Myelinated axons per μm2 and (E) total axons per field of view (10,000 x images) computed from at least three independent images per mouse and n = 3 mice/genotype. (F) Images of coronal sections of PND25 mouse brains taken through the anterior portions of the CC (white lines) and stained for MBP. Scale bar, 40 μm. (G) MBP fluorescent intensity in the CC computed from n = 3 brains/genotype. (H) Western blot analysis of the expression of AnkB isoforms and multiple proteins (MBP, myelin-PLP, CNPase, MAG, MOG) associated with myelination in the cortex of PND25 control and AnkB440 KO mice (n = 3 lysates/genotype). (I) Fold-change in levels of myelination-associated proteins in cortical lysates from PND25 AnkB440 KO mice normalized to tubulin and computed relative to the expression of each protein in control brains. Data in C–E, G, and I show mean ± SEM for three biological replicates per genotype. Unpaired t test. nsp >0.05.
-
Figure 3—source data 1
Loss of AnkB440 does not alter levels of myelination of CC axons.
- https://cdn.elifesciences.org/articles/69815/elife-69815-fig3-data1-v1.zip
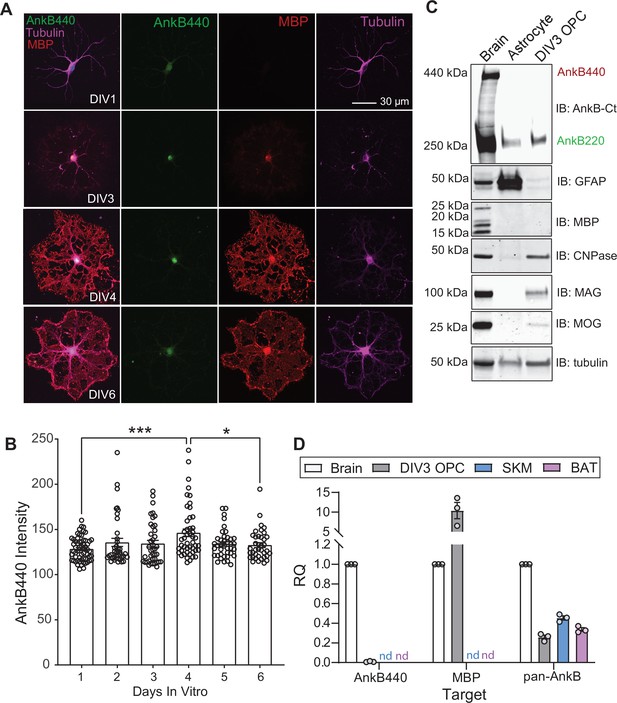
AnkB440 is not expressed at detectable levels in differentiated OPCs.
(A) Images of cultured OPCs at indicated DIVs, representative of three independent cultures stained for AnkB440, MBP and tubulin. Scale bar, 30 μm. (B) Quantification of AnkB440 fluorescent intensity in differentiated OPCs at indicated DIVs. Data represent mean ± SEM collected from n = 38–58 OPCs/DIV from three independent experiments. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. *p < 0.05, ***p < 0.001. (C) Western blot analysis of the expression of AnkB isoforms, GFAP, and multiple proteins (MBP, CNPase, MAG, and MOG) associated with myelination in total lysates from DIV25 mouse brain, DIV17 mouse astrocyte cultures, and DIV3 differentiated rat OPC cultures. (D) RQ levels of AnkB440, Mbp, and total AnkB transcripts in brain, DIV3 differentiated OPCs, skeletal muscle (SKM) and brown adipose tissue (BAT). GAPDH was used as internal control and RQ levels expressed relative to the transcript expression in brain. Data was computed using three biological replicates per group and three technical replicates per biological replicate. nd: not detected.
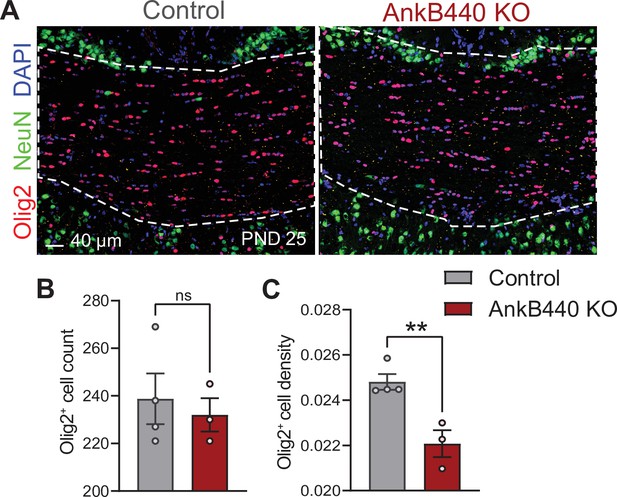
AnkB440 KO mice have normal number of oligodendrocytes in the corpus callosum.
(A) Images of coronal sections of PND25 mouse brains taken through the anterior portions of the CC (white discontinued lines) and stained for Olig2 to label oligodendrocyte and for DAPI and NeuN to respectively label total and neuronal nuclei. Scale bar, 40 μm. (B) Quantification of total number of Olig2+ cells. (C) Quantification of density of Olig2+ cells per CC area. Data in B, C show mean ± SEM of n = 4 control and n = 3 AnkB440 brains. Unpaired t test. nsp >0.05, **p < 0.01.
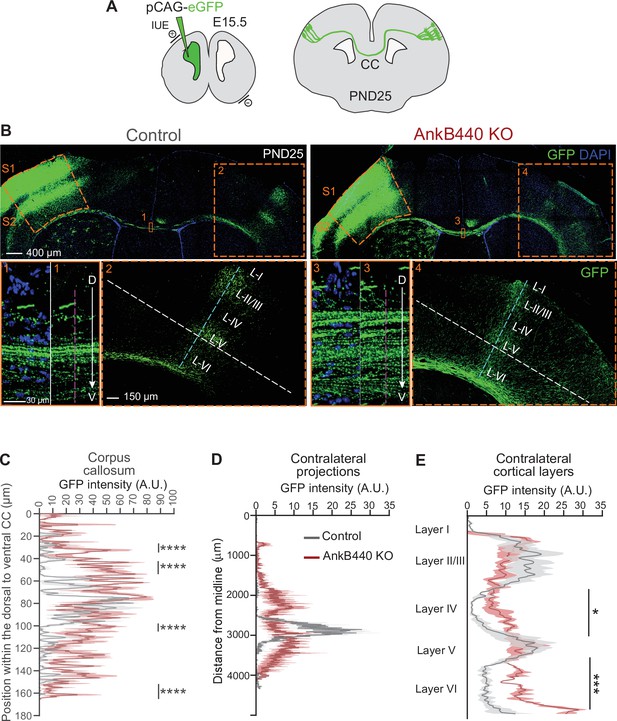
AnkB440 deficiency alters the establishment and refinement of callosal projections.
(A) Diagram illustrating the strategy used to label callosal projecting axons with GFP via IUE of layer II/III S1 neurons at E15.5 and to assess their contralateral targeting at PND25. (B) Coronal sections of PND25 control and AnkB440 KO brains electroporated with pCAG-GFP at E15.5. Scale bar, 400 μm. Insets 1 and 3 (scale bar, 30 μm) show high-magnification images of the boxed CC regions. Insets 2 and 4 (scale bar, 150 μm) show high magnification images of the boxed contralateral cortical regions. The dorsoventral (D–V) order of S1 axons within the CC was grossly disrupted in AnkB440 KO mice (inset 3). (C) Quantification of the dorsoventral distribution of GFP signal in the CC at PND25 as a function of the distance from the most dorsal (D) portion of the CC, measured through the region demarked by a discontinuous pink line in insets 1 and 3. (D) GFP labeling reveals disruption of homotopic S1 callosal projection targeting to the S1/S2 border in AnkB440 KO brains (inset 4), and more contralateral axon branches aberrantly innervating all cortical layers. (D) Quantification of GFP fluorescence of contralateral axon projections as a function of their distance from the brain midline measured through the region demarked by a discontinuous white line in insets 2 and 4. (E) Quantification of GFP fluorescence of axon projections in the contralateral cortical layers measured through the region demarked by a discontinuous teal line in insets 2 and 4. Data in C–E represent mean ± SEM (shadow) of n = 3–4 controls and n = 3 AnkB440 KO brains. Unpaired t test. *p < 0.05, ***p < 0.001, ****p < 0.0001.
-
Figure 4—source data 1
Loss of AnkB440 does not alter levels of myelination of CC axons.
- https://cdn.elifesciences.org/articles/69815/elife-69815-fig4-data1-v1.zip
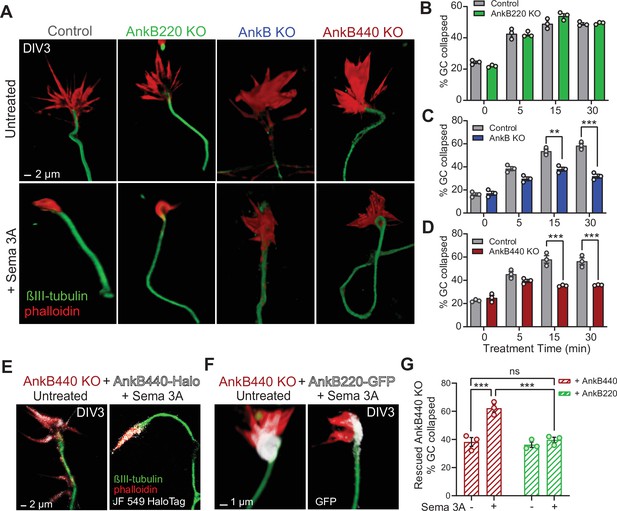
AnkB440 is required for Sema 3A-induced GC collapse.
(A) Images of the distal portion of axons of DIV3 cortical neurons untreated and treated with Sema 3 A and stained with phalloidin and βIII-tubulin. Scale bar, 2 μm. (B–D) Percent of GC collapsed before and after Sema 3 A treatment of control, AnkB220 (B), total AnkB KO (C) and AnkB440 KO (D) cortical neurons. Data in B-D represent mean ± SEM collected from an average of n = 120–200 GCs/treatment/genotype per each experiment. Each dot in the graph represents one of three independent experiments. Unpaired t test. **p < 0.01, ***p < 0.001. (E) Images of DIV3 AnkB440 KO cortical neurons expressing Halo-tagged AnkB440 untreated and treated with Sema 3 A and stained with Janelia Fluor 549 HaloTag ligand to visualize AnkB440. Scale bar, 2 μm. (F) Images of DIV3 AnkB440 KO cortical neurons expressing GFP-tagged AnkB220 untreated and treated with Sema 3 A and stained for GFP to visualize AnkB220. Scale bar, 1 μm. (G) Percent of GC collapsed before and after Sema 3 A treatment of AnkB440 KO neurons rescued with AnkB440 or AnkB220 cDNAs. Data represent mean ± SEM collected from an average of n = 150 GCs/treatment condition/transfection from three independent experiments. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. nsp >0.05, ***p < 0.001.
-
Figure 5—source data 1
AnkB440 promotes Sema 3A-induced GC collapse.
- https://cdn.elifesciences.org/articles/69815/elife-69815-fig5-data1-v1.zip
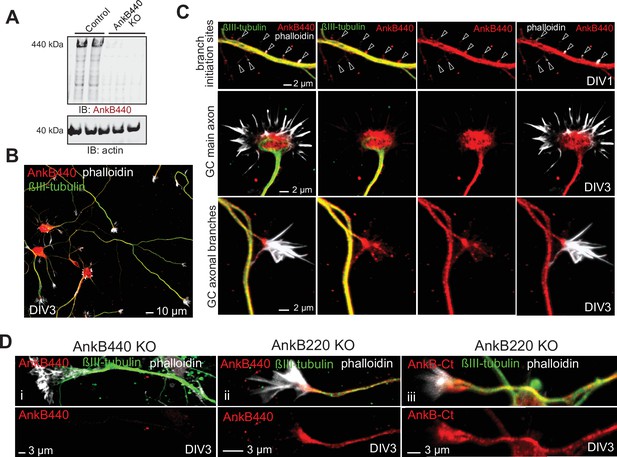
AnkB440 is abundant in axonal growth cones.
(A) Western blot analysis of expression of AnkB440 in total cortical lysates of PND1 control and AnkB440 KO mice show specificity of the antibody against the AnkB440 isoform. (B) Images of DIV3 control cortical neurons stained with the AnkB440-specific antibody, phalloidin (to label F-actin), and βIII-tubulin indicate the broad distribution of AnkB440 in axons. Scale bar, 10 μm. (C) Higher magnification images show that AnkB440 is abundantly expressed at sites of nascent axonal filopodia (arrowheads, already present before microtubule invasion), and at the growth cone (GC) of the main axon and of collateral axon branches. Scale bar, 2 μm. AnkB440 staining colocalizes with both F-actin- and microtubule-enriched axonal structures. (D) Staining of DIV3 AnkB440 KO cortical neurons show specificity of the antibody against the AnkB440 isoform (i). Staining of AnkB220 KO neurons with AnkB440-specific (ii) and AnkB-Ct (iii) antibodies show the localization of AnkB440 in the axon and at the GC. Scale bar, 3 μm.
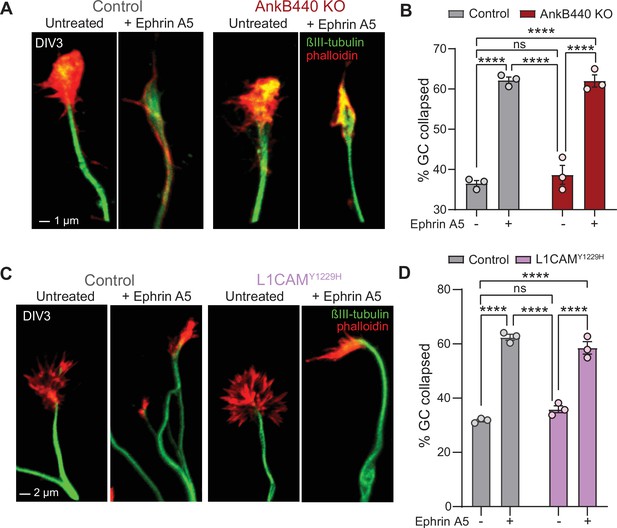
AnkB440 does not enable Ephrin A5-induced GC collapse.
(A) Images of the distal portion of axons of DIV3 cortical control and AnkB440 KO neurons untreated and treated with Ephrin A5 and stained with phalloidin and βIII-tubulin. Scale bar, 1 μm. (B) Percent of GC collapsed before and after Ephrin A5 treatment of control and AnkB440 cortical neurons. Data represent mean ± SEM collected from an average of n = 80 GCs/treatment/genotype per experiment. (C) Images of the distal portion of axons of DIV3 cortical control and Y1229H L1CAM mutant neurons untreated and treated with Ephrin A5 and stained with phalloidin and βIII-tubulin. Scale bar, 2 μm. (D) Percent of GC collapsed before and after Ephrin A5 treatment of control and Y1229H L1CAM neurons. Data in B and D represent mean ± SEM collected from an average of n = 80 GCs/treatment/genotype/experiment. Each dot in the graphs represents one of three independent experiments. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. nsp >0.05, ****p < 0.0001.
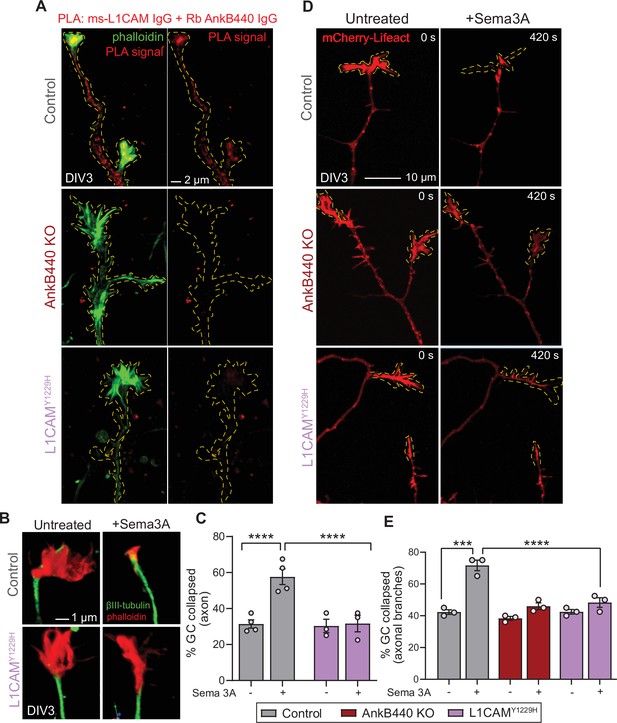
The AnkB440-L1CAM complex promotes GC collapse induced by Sema 3 A.
(A) Images show PLA signal between AnkB440 and L1CAM in axons and GCs of DIV3 cortical neurons from the indicated genotypes. Phalloidin staining was used to identify GCs. Scale bar, 2 μm. (B) Images of the distal portion of the main axon untreated and treated with Sema 3 A and stained with phalloidin and βIII-tubulin. Scale bar, 1 μm. (C) Percent of axon GCs that collapse before and after Sema 3 A treatment. (D) Images from timelapse sequences of DIV3 cortical neuron expressing mCherry-Lifeact before and after Sema 3 A treatment. Scale bar, 10 μm. (E) Percent of collateral axon branches GCs that collapse before and after Sema 3 A treatment. Data in C,E represent mean ± SEM collected from an average of n = 80 GCs/treatment/genotype/experiment. Each dot represents one out of three or four independent experiments. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. ***p < 0.001, ****p < 0.001.
-
Figure 6—source data 1
The AnkB440-L1CAM complex promotes Sema 3A-induced GC collapse.
- https://cdn.elifesciences.org/articles/69815/elife-69815-fig6-data1-v1.zip
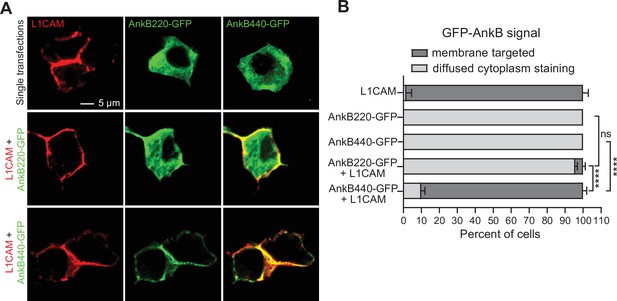
L1CAM recruits AnkB440 to the cell membrane.
(A) Images of HEK 293T/17 cells independently transfected with plasmids encoding L1CAM, AnkB220-GFP, and AnB440-GFP, and L1CAM co-transfected with each of the two GFP-tagged AnkB isoforms. Scale bar, 5 μm. (B) Percent of cells in which L1CAM, AnkB220-GFP, or AnkB440-GFP are targeted to the plasma membrane or found diffused throughout the cytoplasm for each experiment. Data represent mean ± SEM collected from an average of n = 15 cells/transfection/experiment from three independent experiments. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. nsp >0.05, ****p < 0.0001.
The AnkB440-L1CAM complex promotes GC collapse induced by Sema 3 A.
Timelapse images illustrate changes in F-actin dynamics at the GC of collateral axon branches and fillopodia of DIV3 cortical neurons in response to Sema 3 A. Movies are played at 15 frames/second. Scale bar, 5 μm.
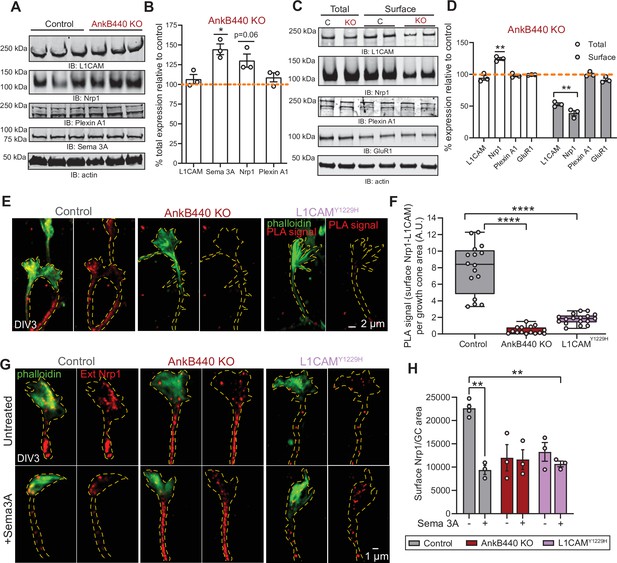
AnkB440 stabilizes the Sema 3 A receptor complex L1CAM-Nrp1 at the cell surface of GCs.
(A) Western blot analysis of the expression of Sema 3 A, L1CAM and the Sema 3 A receptors Nrp1 and Plexin A1 in the cortex of PND1 control and AnkB440 KO mice. (B) Quantification of protein levels normalized to actin in cortical lysates from PND1 mice of indicated genotypes relative to the normalized levels of each protein in control brains. Data show mean ± SEM for three biological replicates per genotype for one experiment. Unpaired t test. *p < 0.05. (C) Western blot analysis of total and surface levels of indicated proteins in DIV3 control and AnkB440 KO cortical neurons. (D) Quantification of total and surface protein levels normalized to actin relative to the normalized levels of each protein in control cortical neurons. Data show mean ± SEM for three biological replicates per genotype for one experiment. Unpaired t test. **p < 0.01. (E) Images show PLA signal between L1CAM and Nrp1 at the cell surface of the axon and GCs of DIV3 cortical neurons from the indicated genotypes. This assay used an Nrp1 antibody that selectively recognizes an extracellular epitope. Phalloidin staining was used to identify GCs. Scale bar, 2 μm. (F) Quantification of PLA signal at the GC surface relative to GC area collected from an average n = 15 GCs/genotype. The box and whisker plots represent all data points arranged from minimum to maximum. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. ****p < 0.0001. (G) Images show Nrp1 localization at the surface of GCs of DIV3 neurons untreated and treated with Sema 3 A, which induces the internalization of surface Nrp1. Scale bar, 1 μm. (H) Quantification of surface Nrp1 levels at GCs relative to GC area at the basal state and upon Sema 3A-induced Nrp1 internalization. Data represent mean ± SEM collected from an average of n = 90 GCs/treatment/genotype/experiment. Each dot represents one out of three independent experiments. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. **p < 0.01.
-
Figure 7—source data 1
AnkB440 stabilizes the L1CAM-Nrp1 complex at the cell surface of GCs.
- https://cdn.elifesciences.org/articles/69815/elife-69815-fig7-data1-v1.zip
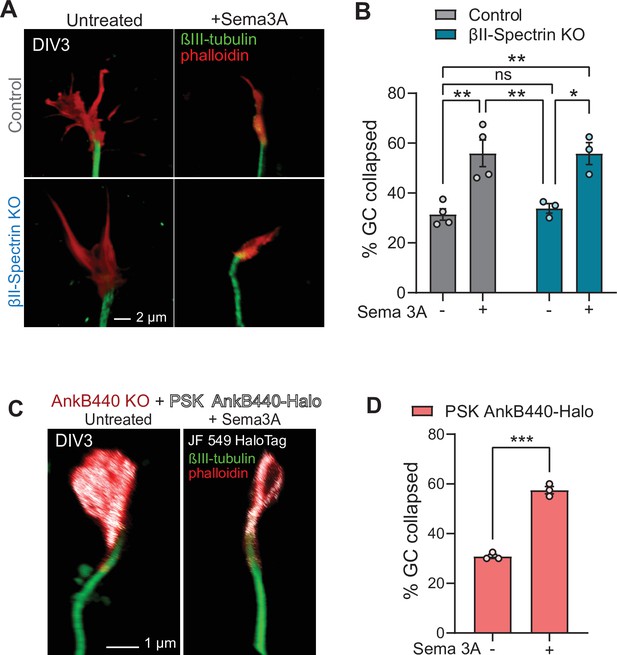
AnkB440 does not require βII-spectrin or binding to microtubules to transduce Sema 3 A signaling.
(A) Images of the distal portion of the axon of control and βII-pectrin KO DIV3 cortical neurons untreated and treated with Sema 3 A and stained with phalloidin and βIII-tubulin. Scale bar, 2 μm. (B) Percent of axon GCs that collapse before and after Sema 3 A treatment. (C) Images of the distal portion of the axon of DIV3 AnkB440 KO cortical neurons expressing mutant PSK AnkB440-Halo untreated and treated with Sema 3 A. Scale bar, 1 μm. (B) Percent of axon GCs that collapse before and after Sema 3 A treatment. Data in B and D represent mean ± SEM collected from an average of n = 70–90 GCs/treatment/genotype/experiment. Each dot represents one out of three or four independent experiments. Data in B was analyzed by one-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. Data in D was analyzed by unpaired t test. nsp >0.05, *p < 0.05, **p < 0.01, ***p < 0.001.
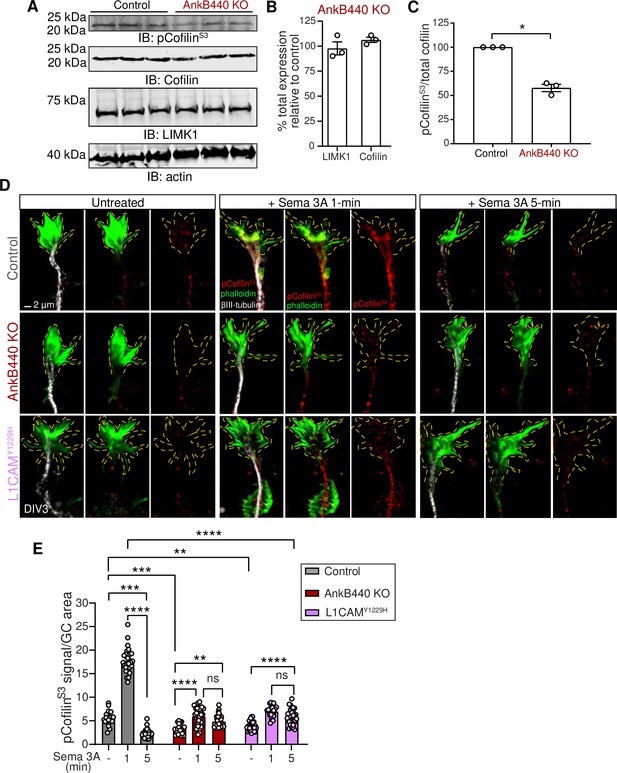
AnkB440 and its interaction with L1CAM are required for F-actin disassembly during GC collapse upon Sema 3 A treatment.
(A) Western blot analysis of the expression of cofilin, phospho-cofilin (Ser3) (pCofilinSer3) and LIMK1 in the cortex of PND1 control and AnkB440 KO mice. Actin is a loading control. (B) Quantification of total levels of cofilin and LIMK1 normalized to actin in cortical lysates from PND1 AnkB440 KO mice relative to the normalized levels of each protein in control brains. (C) Quantification of levels of pCofilinSer3 relative to total cofilin in cortical lysates from PND1 mice. Data in B and C represent mean ± SEM for three biological replicates per genotype. Unpaired t test. *p < 0.05. (D) Images of the distal portion of the main axon of DIV3 cortical neurons untreated and treated with Sema 3 A for 1 and 5 minutes and stained with phalloidin, βIII-tubulin, and pCofilinSer3. Dotted lines indicate GCs. Scale bar, 2 μm. (E) Quantification of pCofilinS3 signal at GCs relative to GC area at the basal state and upon Sema 3 A treatment for 1 and 5 min. Data represent mean ± SEM from an average of n = 25–40 GCs/treatment/genotype collected from three independent experiment. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. nsp >0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
-
Figure 8—source data 1
AnkB440 interaction with L1CAM promotes F-actin disassembly during Sema 3A-induced GC collapse.
- https://cdn.elifesciences.org/articles/69815/elife-69815-fig8-data1-v1.zip
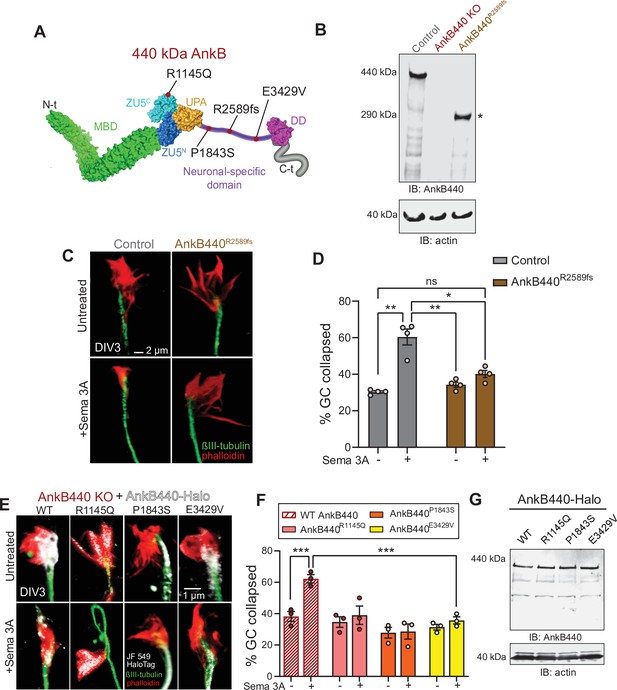
ASD-linked AnkB440 variants affect the transduction of Sema 3 A cues during GC collapse.
(A) Red dots indicate the position within AnkB440 functional domains of the ASD-linked ANK2 variants evaluated. (B) Western blot analysis of expression of AnkB440 in cortical lysates of PND1 control mice of the indicated genotypes assessed with an AnkB440-specific antibody. Brains of AnkB440R2589fs mice express a truncated 290 kDa AnkB440 fragment (asterisk). (C) Images of the distal portion of the axon of control and AnkB440R2589fs DIV3 neurons untreated and treated with Sema 3 A and stained with phalloidin and βIII-tubulin. Scale bar, 2 μm. (D) Percent of axon GCs that collapse before and after Sema 3 A treatment. Data represent mean ± SEM collected from an average of n = 130 GCs/treatment/genotype. Each dot represents one out of four independent experiments. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. nsp >0.05, *p < 0.05, **p < 0.01. (E) Images of the distal portion of the axon of DIV3 AnkB440 KO cortical neurons rescued with the indicated AnkB440-Halo plasmids untreated and treated with Sema 3 A. Scale bar, 1 μm. (F) Percent of axon GCs that collapse before and after Sema 3 A treatment. Data represent mean ± SEM collected from an average of n = 70 GCs/treatment/genotype. Each dot represents one out of three independent experiments. One-way ANOVA with Tukey’s post hoc analysis test for multiple comparisons. ***p < 0.001. (G) Western blot analysis of expression of AnkB440-Halo plasmids in HEK293T cells. Data is representative of three independent experiments.
-
Figure 9—source data 1
ASD-linked AnkB440 variants affect the transduction of Sema 3 A cues during GC collapse.
- https://cdn.elifesciences.org/articles/69815/elife-69815-fig9-data1-v1.zip
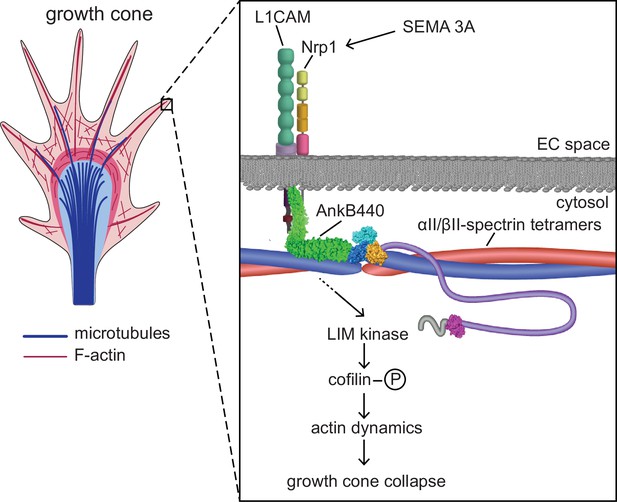
Proposed mechanism of AnkB440-mediated growth cone collapse.
AnkB440 binds the cytoplasmic domain of L1CAM to stabilize the L1CAM-Nrp1 holoreceptor complex at the cell surface. Transduction of Sema 3 A signals via the AnkB440-L1CAM-Nrp1 complex modulates F-actin dynamics through LIMK phosphorylation of cofilin to facilitate local F-actin disassembly and the collapse of GCs from the axon and collateral branches independently of βII-spectrin, or of AnkB440 interaction with microtubules. Source data. Source data contain the original files of the full raw unedited gels or blots and figures with the uncropped gels or blots with the relevant bands clearly labelled. Source data files also include numerical data that are represented as graphs in the figures.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Ank2440flox | PMID:31285321 | MGI ID: 6784039. Pending RRID assignment | Dr. Vann Bennett, Duke University |
| Genetic reagent (M. musculus) | Ank2Exon22-Neo | PMID:25533844Jackson Laboratory | RRID:IMSR_JAX:027916, Stock No: 027916 | Dr. Vann Bennett, Duke University |
| Genetic reagent (M. musculus) | Ank2220flox | This paper | See methods- Generation of AnkB220 knockout mice | |
| Genetic reagent (M. musculus) | Ank2R2589fs | PMID:31285321 | Dr. Vann Bennett, Duke University | |
| Genetic reagent (M. musculus) | L1CAMY1229H | PMID:31285321 | Dr. Vann Bennett, Duke University | |
| Genetic reagent (M. musculus) | Sptbn1flox | PMID:31209033Jackson Laboratory | RRID:IMSR_JAX:020288, Stock No: 020288 | Dr. Matthew N. Rasband, Baylor College of Medicine |
| Genetic reagent (M. musculus) | Nestin-Cre | PMID:31209033Jackson Laboratory | RRID:IMSR_JAX:003771, Stock No: 003771 | Dr. Rudiger Klein, Max-Planck Institute of Neurobiology |
| Genetic reagent (M. musculus) | Thy1-GFP line M | PMID:11086982Jackson Laboratory | RRID:IMSR_JAX:007788, Stock No: 007788 | Dr. Joshua R Sanes, Harvard University |
| Cell line (Homo sapiens) | HEK 293T/17 | American Type Culture Collection | Cat# CRL-11268,RRID:CVCL_1926 | |
| Antibody | anti-pan ankyrin-B (Rabbit polyclonal) | PMID:31209033, 31285321 | IF (1:500), WB (1:5000) | |
| Antibody | anti-440 kDa ankyrin-B (Rabbit polyclonal) | PMID:31285321 | IF (1:500), WB (1:5000) | |
| Antibody | anti-Sema 3 A (Rabbit polyclonal) | Abcam | Cat# ab23393, RRID:AB_447408 | WB (1:500) |
| Antibody | anti-Nrp1 (Rabbit polyclonal) | Abcam | Cat# ab25998, RRID:AB_448950 | WB (1:1000) |
| Antibody | anti-Mbp (Rat monoclonal) | Abcam | Cat# ab7349, RRID:AB_305869 | IF (1:100), WB (1:500) |
| Antibody | anti-L1CAM (mouse monoclonal) | Abcam | Cat# ab24345, RRID:AB_448025 | WB (1:200) |
| Antibody | anti-Satb2 (Mouse monoclonal) | Abcam | Cat# ab51502, RRID:AB_882455 | IF (1:200) |
| Antibody | anti-Tbr1 (Rabbit polyclonal) | Abcam | Cat# ab31940, RRID:AB_2200219 | IF (1:200) |
| Antibody | anti-Ctip2 (Rat monoclonal) | Abcam | Cat# ab18465, RRID:AB_2064130 | IF (1:500) |
| Antibody | anti-Plexin A1 extracellular (Rabbit polyclonal) | Alomone | Cat# APR-081, RRID:AB_2756765 | WB (1:200) |
| Antibody | anti-Nrp1 extracellular (Rabbit polyclonal) | Alomone | Cat# ANR-063, RRID:AB_2756695 | IF(1:50) |
| Antibody | anti-mGluR1, clone N355/1 (Mouse polyclonal) | Antibodies Incorporated | Cat# 75–327, RRID:AB_2315840 | WB (1:500) |
| Antibody | anti-phospho-Cofilin (Ser3) (Rabbit polyclonal) | Cell Signaling Technology | Cat# 3311, RRID:AB_330,238 | WB (1:1000) |
| Antibody | anti-LIMK1 (Rabbit polyclonal) | Cell Signaling Technology | Cat# 3842, RRID:AB_2281332 | WB (1:1000) |
| Antibody | anti-MAG (Rabbit monoclonal) | Cell Signaling Technology | Cat# 9043, RRID:AB_2665480 | WB (1:1000) |
| Antibody | anti-MOG (Rabbit monoclonal) | Cell Signaling Technology | Cat# 96457, RRID:AB_2800265 | WB (1:1000) |
| Antibody | anti-GFAP (mouse monoclonal) | Cell Signaling Technology | Cat# 3670, RRID:AB_561049 | WB (1:1000) |
| Antibody | anti-cofilin (Mouse monoclonal) | Proteintech | Cat# 66057–1-Ig, RRID:AB_11043339 | WB (1:1000) |
| Antibody | anti-Olig2 (Rabbit polyclonal) | Proteintech | Cat# 13999–1-AP, RRID:AB_2157541 | IF (1:200) |
| Antibody | Anti-pan neurofilament (mouse monoclonal) | Biolegend | Cat# 837904, RRID:AB_2566782 | IF (1:200) |
| Antibody | anti-actin (mouse monoclonal) | Millipore-Sigma | Cat# MAB1501, RRID:AB_2223041 | WB (1:2000) |
| Antibody | anti-CNPase(mouse monoclonal) | Millipore-Sigma | Cat# C5922, RRID:AB_476854 | WB (1:500) |
| Antibody | anti-NeuN (guinea pig polyclonal) | Millipore-Sigma | Cat# ABN90P, RRID:AB_2341095 | IF (1:1000) |
| Antibody | anti-L1CAM (rat monoclonal) | Millipore-Sigma | Cat# MAB5272, RRID:AB_2133200 | IF (1:200) |
| Antibody | anti- βIII-tubulin (mouse monoclonal) | Millipore-Sigma | Cat# MAB1637, RRID:AB_2341095 | IF (1:200) |
| Antibody | anti-myelin PLP (rabbit polyclonal) | Novus Biologicals | Cat# NBP1-87781, RRID:AB_11026674 | WB (1:1000) |
| Antibody | anti-pan tubulin (sheep polyclonal) | Cytoskeleton | Cat# ATN02, RRID:AB_10709401 | IF (1:200) |
| Antibody | anti-GFP (chicken polyclonal) | Antibodies Incorporated | Cat# GFP-1020, RRID:AB_10000240 | IF (1:1000) |
| Recombinant DNA reagent | pBa-AnkB440-Halo | PMID:31285321 | pBa-Halo backbone | |
| Recombinant DNA reagent | pBa-AnkB440P1843S-Halo pBa-AnkB440E3429V-Halo; | PMID:31285321 | pBa-Halo backbone | |
| Recombinant DNA reagent | pBa-AnkB440R1145Q-Halo | This paper | pBa-Halo backbone. See methods- plasmids | |
| Recombinant DNA reagent | pBa-AnkB440-PSK-Halo | PMID:32640013 | pBa-Halo backbone | |
| Recombinant DNA reagent | pCAG-AnkB220-GFP | This paper | pCAG-eGFP-N1 backbone. See methods- plasmids | |
| Recombinant DNA reagent | pBa-AnkB440-GFP | This paper | pBa-Halo backbone. See methods- plasmids | |
| Commercial assay or kit | GeneArt Site-Directed Mutagenesis System | ThermoFisher Scientific | Cat. #: A13282 | |
| Commercial assay or kit | In-Fusion Snap Assembly Starter Bundle | Takara | Cat. #: 638,945 | |
| Commercial assay or kit | DUOLINK PLA in situ detection kit-red | Millipore-Sigma | Cat. #: DUO92008-100RXN | |
| Commercial assay or kit | Duolink In Situ PLA Probe Anti-Mouse PLUS | Millipore-Sigma | Cat. #: DUO92001-100RXN | |
| Commercial assay or kit | Duolink In Situ PLA Probe Anti-Rabbit MINUS | Millipore-Sigma | Cat. #: DUO92005-100RXN | |
| chemical compound, drug | Alexa Fluor- 488,–568, –633 Phalloidin | ThermoFisher Scientific | Cat. #: A12379, A12380, A22284 | IF (1:100) |
| Peptide, recombinant protein | Semaphorin 3 A | Peprotech | Cat. #: 150–17 H | 250 ng/ml |
| Peptide, recombinant protein | Ephrin A5 | R&D Systems | Cat. #: 374-EA | 1 µg/ml |





