Integrin α5β1 nano-presentation regulates collective keratinocyte migration independent of substrate rigidity
Figures
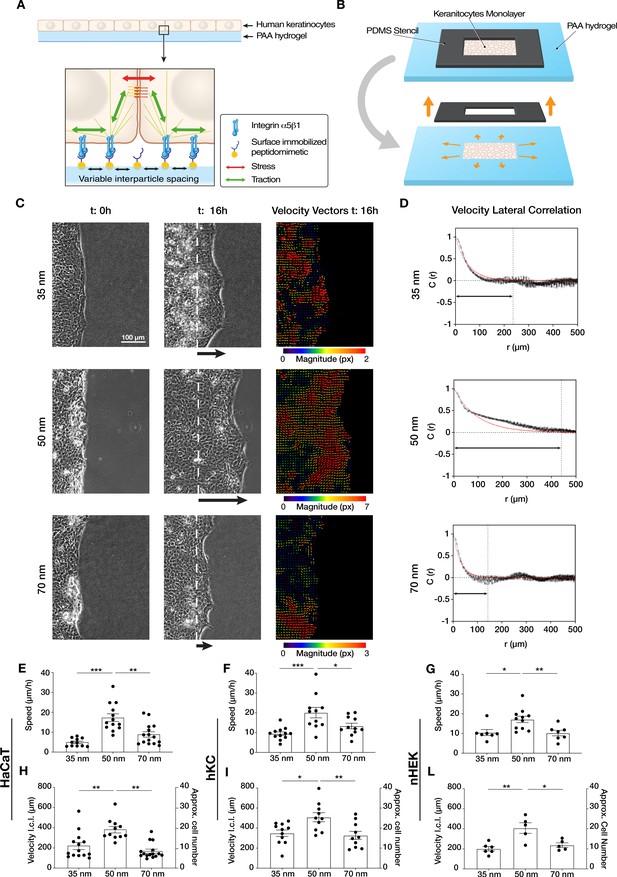
Efficient collective cell migration depends on integrin α5β1 ligand nanospacing.
(A) Keratinocyte monolayers coordinate their intra/intercellular tractions/stresses on polyacrylamide (PAA) hydrogels nanopatterned with integrin α5β1 peptidomimetic. (B) Schematic representation of the migration experiment setup. (C) Representative images of time-lapse experiments at t = 0 hr (start) and t = 16 hr (end), with corresponding velocity vectors plots in pixel (px) of keratinocyte (HaCaT) sheet migration on hydrogels with 35, 50, and 70 nm integrin α5β1 ligand lateral spacing. The dotted white lines illustrate the initial starting point of the monolayers immediately following stencil removal, and the arrows indicate the direction of movement. (D) Representative velocity lateral correlation length, C(r) where r = distance, curves of keratinocyte (HaCaT) velocity vectors at t = 16 hr. The quantification of migration speed and the lateral correlation length in HaCaT (E, H), human epidermis-derived keratinocytes (hKC) (F, I), and primary human epidermal keratinocytes (nHEK) (G, L) show an optimum at 50 nm integrin α5β1 ligand lateral spacing. Scatter plots show values with mean ± s.e.m. from at least three independent experiments. *p < 0.05; **p < 0.01; ***p < 0.001 using a Mann-Whitney test.
-
Figure 1—source data 1
Data points for graphs in Figure 1 and its supplements.
- https://cdn.elifesciences.org/articles/69861/elife-69861-fig1-data1-v1.xlsx
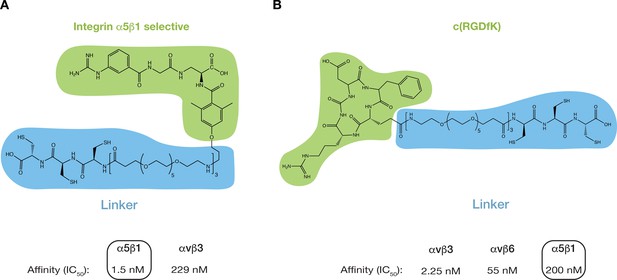
Chemical structures of integrin α5β1 selective peptidomimetic (A) and c(RGDfK) (B).
Highlighted in green are the integrin-selective moieties of the peptides and in blue the functionalization required for interaction with the gold nanoparticles. At the bottom are the integrin subtype-specific affinity values as reported elsewhere (Kapp et al., 2017; Rechenmacher et al., 2013).
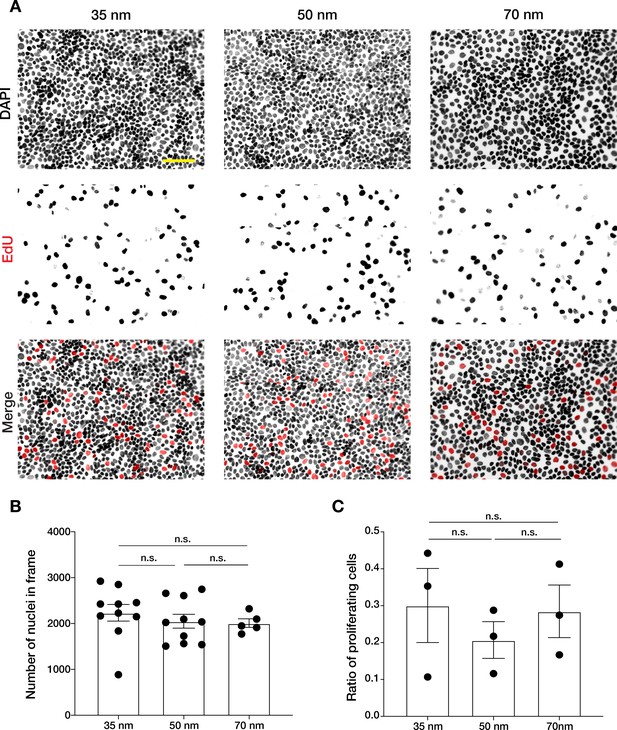
Keratinocyte monolayer density and proliferation are not affected by integrin α5β1 nanopatterning.
(A) Representative immunofluorescent images of keratinocyte nuclei stained with DAPI (top row), 5-ethynyl-2′-deoxyuridine (EdU) (middle row), and merge (bottom row, DAPI black, EdU red) in monolayers obtained on 35, 50, and 70 nm inter-ligand spacing after 6 hr EdU incorporation. Scale bar is 100 µm. Quantification of (B) number of nuclei per frame shows no significant differences in cell density or (C) proliferation ratio between the different conditions. Column bars show mean ± s.e.m. from at least three independent experiments. n.s. = not significant using an unpaired t-test.
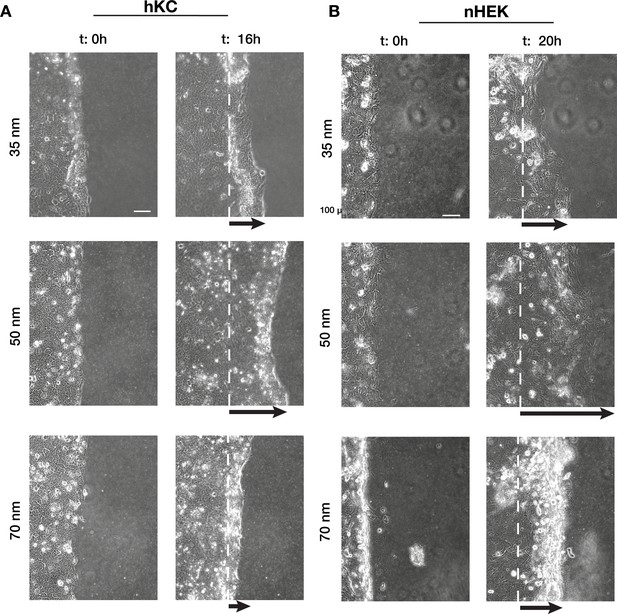
Human epidermis-derived keratinocyte (hKC) and primary human epidermal keratinocytes (nHEK) migratory behaviour is comparable with HaCaT.
Representative images of time-lapse experiments at t = 0 hr (start) and t = 16 hr/20 hr (end) of hKC (A) and nHEK (B) sheet migration on hydrogels with 35, 50, and 70 nm integrin α5β1 ligand lateral spacing. The dotted white lines illustrate the initial starting point of the monolayers immediately following stencil removal, and the arrows indicate the direction of movement. Scale bar is 100 µm.
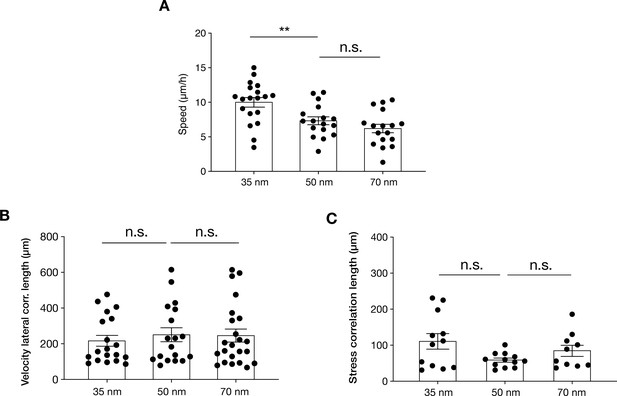
HaCaT keratinocyte collective behaviour on nanopatterned c(RGDfK) surfaces.
(A) Quantification of keratinocyte sheet migration speed, velocity lateral correlation length (B), and stress correlation length (C) on 35, 50, and 70 nm inter-receptor spacing. Scatter plots show individual values and mean ± s.e.m. from at least three independent experiments. **p < 0.01, n.s = not significant using a Mann-Whitney test.
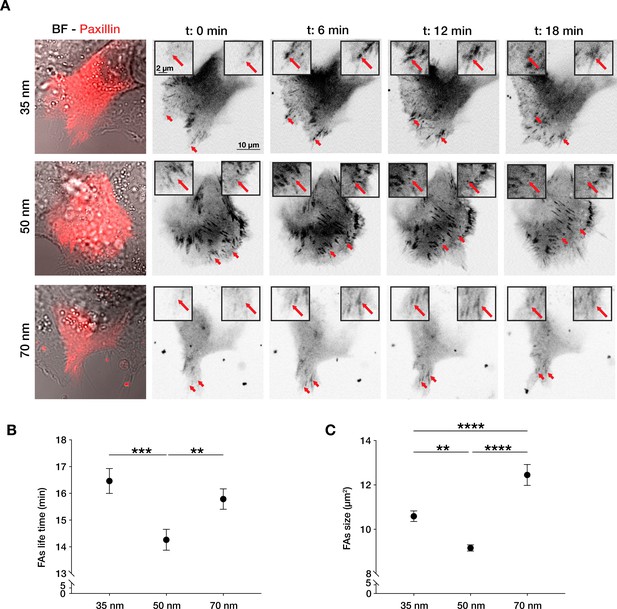
Integrin α5β1 lateral spacing governs focal adhesion dynamics and size.
(A) Representative still frames from time sequence over 18 min from transiently transfected keratinocytes with mCherry-α-paxillin migrating on 35, 50, or 70 nm integrin α5β1 ligand lateral spacing hydrogels. The red arrows and insets highlight the development of the same focal adhesion over culture time. Left column images are of BF = bright-field overlaid with mCherry-α-paxillin (red) at t = 0. Other images are of mCherry-α-paxillin in B&W over culture time (t = 0, 6, 12, 18 min). (B, C) Quantification of focal adhesions (FAs) lifetime and size (area, in µm2) on the three different spacing surfaces. The data points show mean values ± s.e.m. of >10 cells/condition, from at least three independent experiments. **p < 0.01; ***p < 0.001; ****p < 0.0001 using a Mann-Whitney test.
-
Figure 2—source data 1
Data points for graphs in Figure 2 and its supplements.
- https://cdn.elifesciences.org/articles/69861/elife-69861-fig2-data1-v1.xlsx
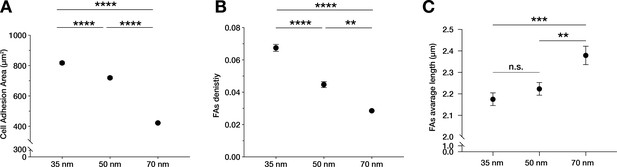
Cell adhesion area and focal adhesion density and length depends on ligand spacing.
Quantification of cell area in contact with the nanopatterned hydrogels within the monolayer (A), focal adhesions (FAs) density (B), and FAs length (C) on 35, 50, and 70 nm integrin α5β1 ligand lateral spacing. The data points show mean values ± s.e.m. of >10 cells/condition, from at least three independent experiments. **p < 0.01; ***p < 0.001, ****p < 0.0001, n.s. = not significant using a Mann-Whitney test.
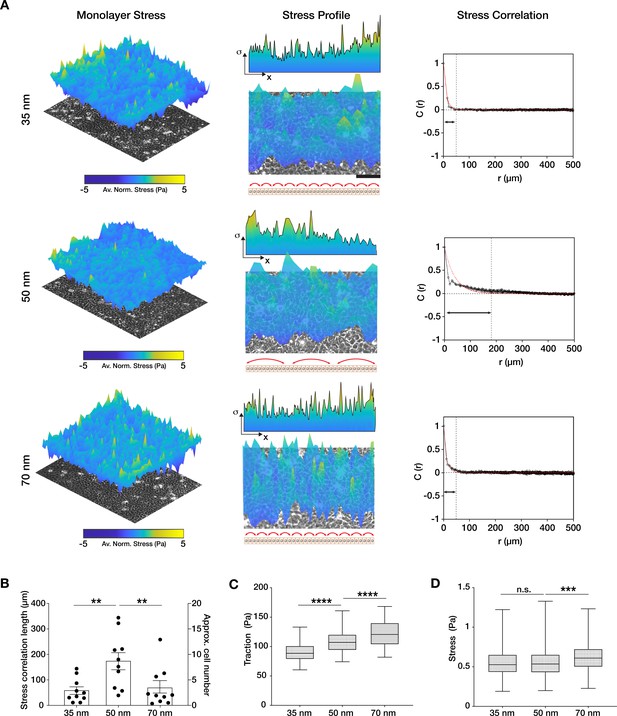
Traction force generation depends on integrin α5β1 ligand nanospacing.
(A) Monolayer stress plots and profiles, and spatial correlation curves from keratinocyte monolayers at the different integrin α5β1 ligand lateral spacings (35, 50, 70 nm). Stress correlation can be identified by inter-peak distance of the stress profile as schematically represented by the red arrows. Scale bar is 100 µm; σ = stress. The quantification of stress correlation lengths (B) shows an optimal intercellular force coordination at 50 nm inter-ligand spacing. (C) Traction forces and stresses (D) quantified in the three spacing conditions. Scatter and box and whiskers plots show values and mean ± s.e.m. from at least four independent experiments. n.s. = not significant; **p < 0.01; ***p < 0.001; ****p < 0.0001 using a Mann-Whitney test.
-
Figure 3—source data 1
Data points for graphs in Figure 3 and its supplements.
- https://cdn.elifesciences.org/articles/69861/elife-69861-fig3-data1-v1.xlsx
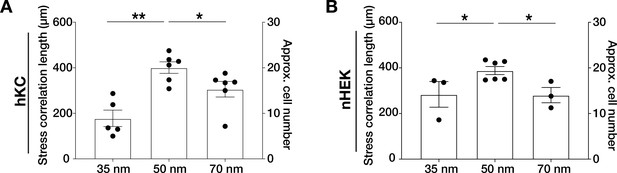
Quantification of stress correlation lengths in human epidermis-derived keratinocytes (hKC) (A) and primary human epidermal keratinocytes (nHEK) (B) monolayers on integrin α5β1 ligand lateral spacings (35, 50, 70 nm).
The data confirmed the optimal intercellular force coordination at 50 nm inter-ligand spacing. Scatter and box and whiskers plots show values and mean ± s.e.m. from at least four independent experiments. n.s. = not significant; *p < 0.05; **p < 0.01 using a Mann-Whitney test.
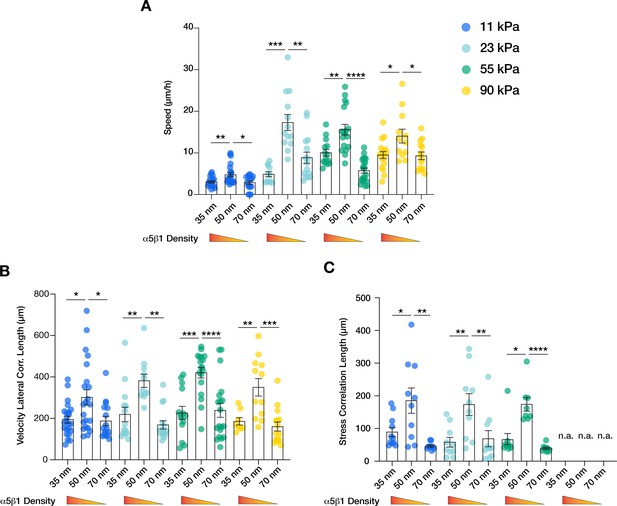
α5β1 ligand spacing outweighs substrate stiffness in collective cell migration.
Quantification of the migratory speed (A), velocity lateral correlation length (B), and stress correlation length (C) of keratinocyte sheets on different substrate rigidities (11 kPa – blue, 23 kPa – teal, 55 kPa – green, 90 kPa – yellow) and integrin α5β1 ligand lateral spacing (35, 50, 70 nm). Scatter plots show values and mean ± s.e.m. from at least three independent experiments. n.a. = not applicable; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 using a Mann-Whitney test.
-
Figure 4—source data 1
Data points for graphs in Figure 4.
- https://cdn.elifesciences.org/articles/69861/elife-69861-fig4-data1-v1.xlsx
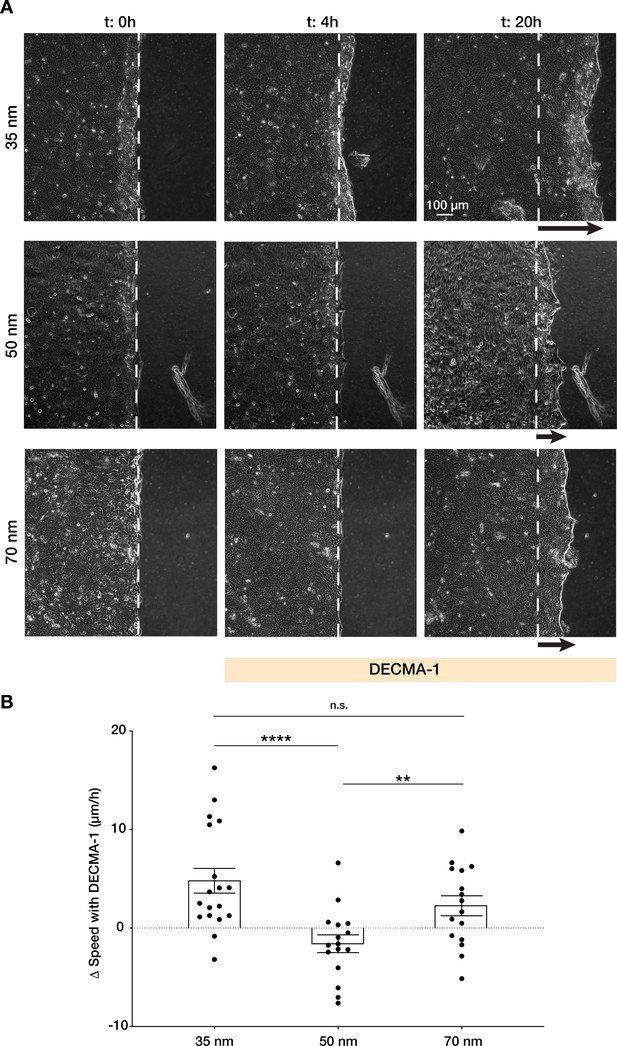
Integrin α5β1 spacing regulates intercellular force transmission via E-cadherin.
(A) Representative frames of keratinocyte sheet migration before (t = 0–4 hr) and after (t = 4–20 hr) the addition of E-cadherin blocking antibody (DECMA-1) in the medium. The arrows illustrate an approximation of the extent of sheet elongation. (B) Quantification of the change in migratory speed in the presence of DECMA-1 vs. control. Scatter plots show values and mean ± s.e.m. from at least three independent experiments. n.s. = not significant; **p < 0.01; ****p < 0.0001 using a Mann-Whitney test.
-
Figure 5—source data 1
Data points for graph in Figure 5.
- https://cdn.elifesciences.org/articles/69861/elife-69861-fig5-data1-v1.xlsx
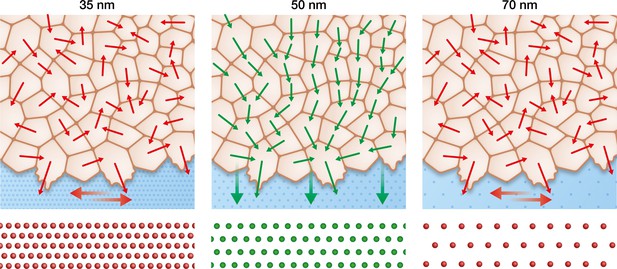
Keratinocytes require an optimum integrin α5β1 density to efficiently collectively migrate.
In contrast to the optimal integrin α5β1 lateral spacing (green), lower and higher spacings (red) lead to uncorrelated single-cell movement and stress propagation. This results in inefficient collective behaviour, significantly slowing keratinocyte sheet migration.
Videos
Keratinocyte sheet migration on 35, 50, and 70 nm integrin α5β1 ligand lateral spacing.
Time-lapse phase contrast imaging showing the migratory behaviour over 16 hr.
Focal adhesions dynamics on 35 nm integrin α5β1 ligand lateral spacing.
Representative time-lapse fluorescent imaging of a keratinocyte transfected with mCherry-α-paxillin to visualize focal adhesions dynamics during sheet migration.
Focal adhesions dynamics on 50 nm integrin α5β1 ligand lateral spacing.
Representative time-lapse fluorescent imaging of a keratinocyte transfected with mCherry-α-paxillin to visualize focal adhesions dynamics during sheet migration.
Focal adhesions dynamics on 70 nm integrin α5β1 ligand lateral spacing.
Representative time-lapse fluorescent imaging of a keratinocyte transfected with mCherry-α-paxillin to visualize focal adhesions dynamics during sheet migration.
Keratinocyte sheet migration on 35, 50, and 70 nm integrin α5β1 ligand lateral spacing is enhanced upon E-cadherin blocking.
Time-lapse phase contrast imaging showing the migratory behaviour before (1–230 min) and after (240–1240 min) E-cadherin blocking (DECMA-1).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (human) | HaCaT | Cell Lines Service – CLS, Germany | 300,493 | |
| Cell line (Homo-sapiens) | hKC | Sawant et al., 2018 | – | Gift from Dr Rudolf Leube |
| Cell line (Homo-sapiens) | nHEK | CellSystems | FC-0064 | |
| Recombinant DNA reagent | mCherry-α-Paxillin-C1 (plasmid) | Efimov et al., 2008 | Gift from Dr Irina Kaverina | |
| Antibody | Anti-human E-cadherin (DECMA-1, rat monoclonal) | Millipore | MABT26 | (10 µg/ml) |
| Peptide, recombinant protein | Thiolated integrin α5β1 peptidomimetic | Fraioli et al., 2015; Guasch et al., 2015; Neubauer et al., 2013 | – | – |
| Peptide, recombinant protein | Thiolated c(RGDfK) | PSL GmbH, Heidelberg, Germany | customized | – |
| Commercial assay or kit | Click-iT Plus EdU imaging kit | Invitrogen | C10640 | – |
| Software, algorithm | MATLAB | MathWorks | – | – |
| Software, algorithm | Imaris | Bitplane, Oxford Instrument | – | – |
| Software, algorithm | ImageJ | NIH | – | – |
| Other | DAPI stain | Invitrogen | D1306 | 1 µg/ml |





