Vaccination induces rapid protection against bacterial pneumonia via training alveolar macrophage in mice
Figures
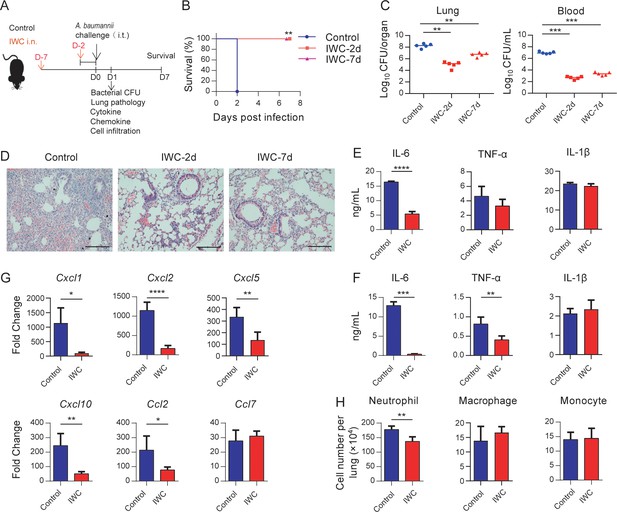
Rapid protection against A. baumannii pneumonia by a single intranasal vaccination.
(A) Schematic diagram of the experimental procedure. C57BL/6 mice were immunized intranasally (i.n.) with inactivated whole cell (IWC) of A. baumannii strain LAC-4 and challenged intratracheally (i.t.) with LAC-4 at day 2 (IWC-2d) or day 7 (IWC-7d) after immunization (n = 5/group). (B) Survival of mice was recorded for 7 days. **p<0.01 determined by log-rank test. (C) Bacterial burdens in lungs and blood at 24 hr post infection (hpi) were determined. Each plot represents one mouse. The line indicates the median of the data. **p<0.01, ***p<0.001 evaluated by ordinary one-way ANOVA followed by Tukey`s multiple comparisons test. (D) Representative histopathological images of lungs at 24 hpi. Scale bars: 100 μm. (E–G) IWC-immunized mice were challenged at day 7 and were sacrificed at 24 hpi. Levels of inflammatory cytokines in the lungs (E) and serum (F) were detected by ELISA. (G) Transcriptional levels of chemokines in the lungs were detected by real-time PCR. (H) Numbers of neutrophils in the lungs were detected by flow cytometry. Data are mean ± SD. n = 4–5 mice/group. For (E–H), *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001, determined by two-tailed unpaired t test. Data are representative of at least two independent experiments.
-
Figure 1—source data 1
Raw data for Figure 1.
- https://cdn.elifesciences.org/articles/69951/elife-69951-fig1-data1-v1.xlsx
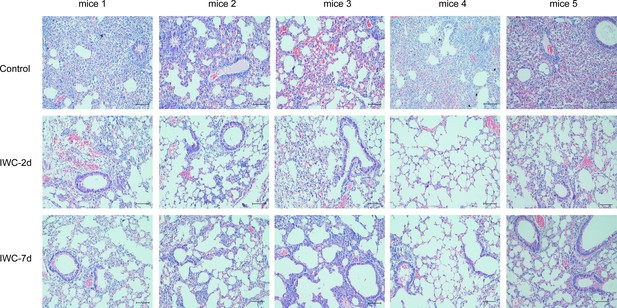
The histopathology images of lungs for each mouse in Figure 1.
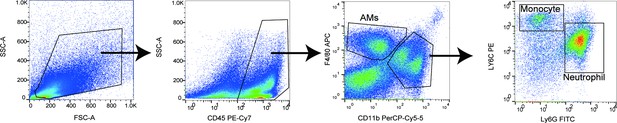
Gating strategy for detecting neutrophil, monocyte, and alveolar macrophages in lungs.
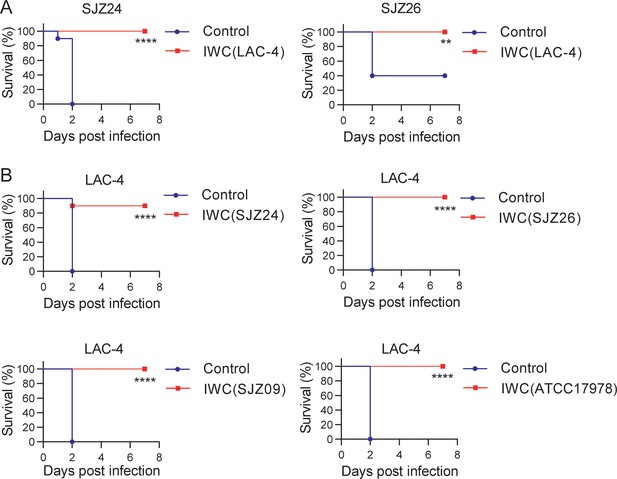
Broad protection against infection of clinical strains by intranasal immunization of inactivated whole cell (IWC) of A. baumannii.
(A) Mice were immunized intranasally (i.n.) with IWC of LAC-4 on day 0. On day 7, mice were infected intratracheally (i.t.) with lethal dose of clinical strains of A. baumannii SJZ24 or SJZ26 (n = 10). (B) Mice were immunized i.n. with IWC of SJZ24, SJZ26, SJZ09, or ATCC17978 and challenged with lethal dose of LAC-4 7 days later. Survival rate of mice was monitored for 7 days after challenge. n = 10. **p<0.01, ****p<0.0001 determined by log-rank test. These experiments were repeated at least twice.
-
Figure 2—source data 1
Raw data for the survival rate.
- https://cdn.elifesciences.org/articles/69951/elife-69951-fig2-data1-v1.xlsx
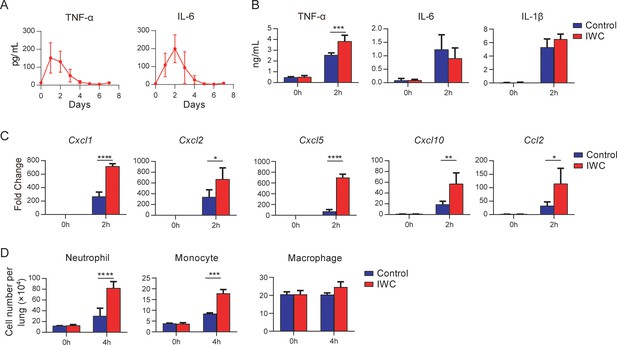
Rapid immune memory induced by a single intranasal vaccination.
(A) Dynamic responses of TNF-α and IL-6 in the lungs of A. baumannii inactivated whole cell (IWC) (LAC-4)-immunized mice (n = 3 per timepoint). (B–D) IWC (LAC-4)-immunized mice were challenged intratracheally (i.t.) with LAC-4 at day 7 after immunization. (B) Levels of TNF-α, IL-6, and IL-1β at 0 hr and 2 hr post infection (hpi) in the lungs were measured by ELISA. (C) Transcriptional levels of chemokines in the lungs at 0 hr and 2 hpi were assessed by real-time PCR. (D) Numbers of neutrophils, monocytes, and macrophages in the lungs of mice were determined by flow cytometry. Data are presented as mean ± SD. (n = 3–4 mice/group). *p<0.05; **p<0.01, ***p<0.001, ****p<0.0001, ordinary two-way ANOVA. Data are representative of two independent experiments.
-
Figure 3—source data 1
Raw data for Figure 3.
- https://cdn.elifesciences.org/articles/69951/elife-69951-fig3-data1-v1.xlsx
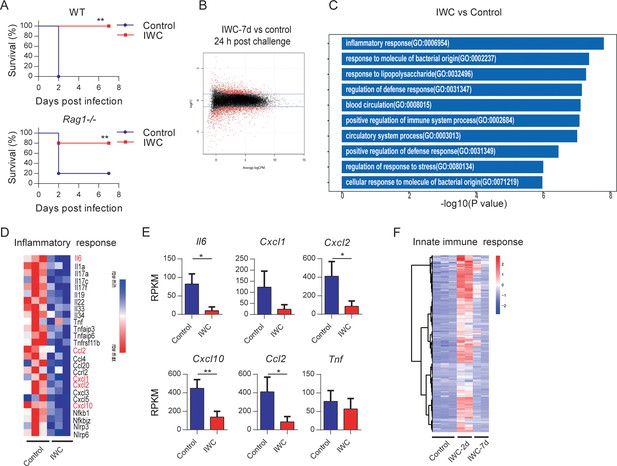
Trained innate immunity mediates vaccination-induced rapid protection.
(A) Survival of WT and Rag1-/- mice immunized intranasally (i.n.) with inactivated whole cell (IWC) (LAC-4) or PBS, challenged intratracheally (i.t.) by lethal LAC-4 7 days later (n = 5 for WT mice, n = 10 for Rag1-/- mice). **p<0.01 compared to control calculated by log-rank test. Data are representative of two independent experiments. (B–E) Rag1-/- mice were immunized with IWC (LAC-4) or PBS as control for 7 days and challenged with LAC-4. The lung tissue was collected to perform RNA-seq. (B) MA plot of the differentially expressed genes (DEGs) of IWC (LAC-4)-immunized lung (n = 3) vs. control lung of Rag1-/- mice (n = 3) at 24 hr post LAC-4 challenge. X-axis represents average counts per million (logCPM) and Y-axis represents log fold changes (logFC) in IWC-immunized mice vs. control mice. The blue line is the threshold, logFC >1 means upregulation and logFC < –1 means downregulation. (C) Top 10 Gene Ontology (GO) enrichment terms of downregulated DEGs in the IWC-immunized group at 24 hr post infection (hpi). (D) Heatmap of DEGs related to inflammatory response is shown. False discovery rate (FDR) < 0.05. (E) Reads per kilobase per million reads (RPKM) of inflammatory and chemokine genes. Data are mean ± SD. *p<0.05, **p<0.01 determined by two-tailed unpaired t test. (F) The heatmap of innate immune response-related genes (GO_0045087) of lung samples from control (n = 3), IWC (LAC-4)-immunized at day 2 (IWC-2d) (n = 2), and day 7 (IWC-7d) (n = 2) in Rag1-/- mice.
-
Figure 4—source data 1
Raw data for Figure 4A,E.
- https://cdn.elifesciences.org/articles/69951/elife-69951-fig4-data1-v1.xlsx
-
Figure 4—source data 2
RNA-seq results of reads per kilobase per million reads (RPKM) for all samples and all genes.
- https://cdn.elifesciences.org/articles/69951/elife-69951-fig4-data2-v1.xlsx
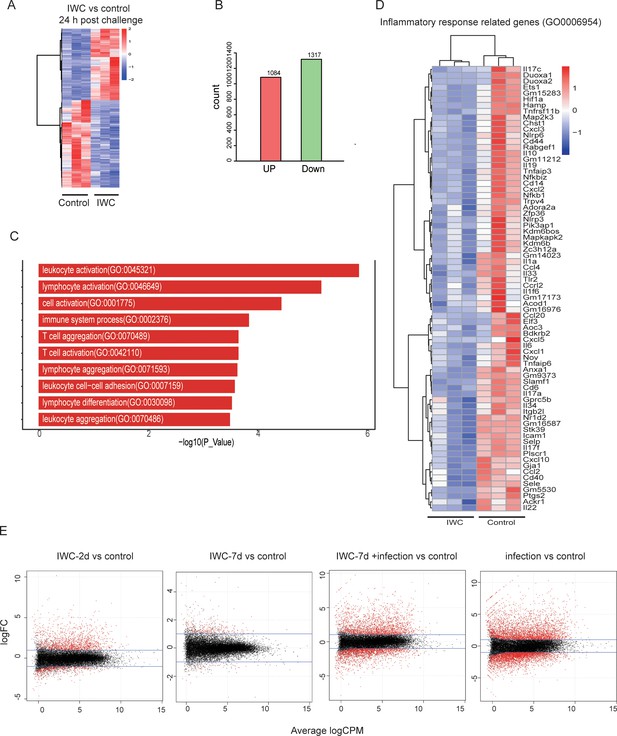
Supplementary RNA-seq results.
(A–C) RNA-seq data related to Figure 4B. (A) The heatmap of 2401 differentially expressed genes (DEGs) in lungs. (B) Numbers of upregulated and downregulated DEGs. (C) Top 10 Gene Ontology (GO) terms of upregulated DEGs. (D) Heatmap of DEGs related to inflammatory response (GO0006954). (E) Lung samples from control, inactivated whole cell (IWC)-immunized Rag1-/- mice at day 2 (IWC-2d), day 7 (IWC-7d), and IWC-immunized mice (7 day) at 24 hr after challenge with A. baumannii (IWC-7d+infection) and control mice at 24 hr after challenge (infection) were processed for RNA-seq. MA plot of DEGs in each treatment group compared with that in control group.
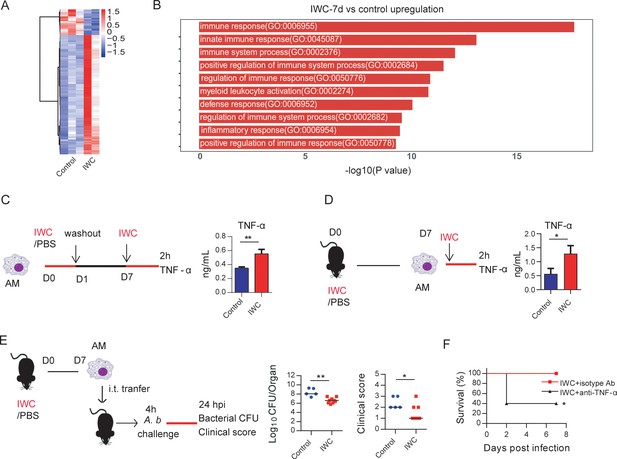
Trained immunity of alveolar macrophages (AMs) mediates rapid protection induced by vaccination.
(A) Heatmap of differentially expressed genes (DEGs) in lungs of inactivated whole cell (IWC) (LAC-4)-immunized and control Rag1-/- mice at day 7 after immunization. (B) Top 10 Gene Ontology (GO) terms of upregulated DEGs in IWC-immunized Rag1-/- mice at day 7. (C) In vitro model of IWC (LAC-4)-trained AMs. (D) C57BL/6 mice were immunized with IWC (LAC-4), and recall responses of trained AMs to IWC (LAC-4) were evaluated ex vivo by detection of TNF-α production at 2 hr after stimulation. For (C) and (D), data are mean ± SD. n = 3. *p<0.05, **p<0.01, two-tailed unpaired t test. (E) Schema of evaluating roles of AMs in BALF of PBS or IWC (LAC-4)-immunized C57BL/6 mice. Bacterial burdens of lung and clinical scores at 24 hr post infection (hpi) were measured (n = 5–9). Each dot means a mice, and the line represents the median. *p<0.05, **p<0.01 determined by Mann–Whitney U test. (F) WT mice were immunized with IWC (LAC-4) for 7 days. Mice were treated intraperitoneally with anti-TNF-α antibody or isotype control then were challenged with lethal LAC-4 1 hr later. The survival of mice was monitored (n = 5). *p<0.05, calculated by log-rank test. Data are representative of two independent experiments.
-
Figure 5—source data 1
Raw data for Figure 5.
- https://cdn.elifesciences.org/articles/69951/elife-69951-fig5-data1-v1.xlsx
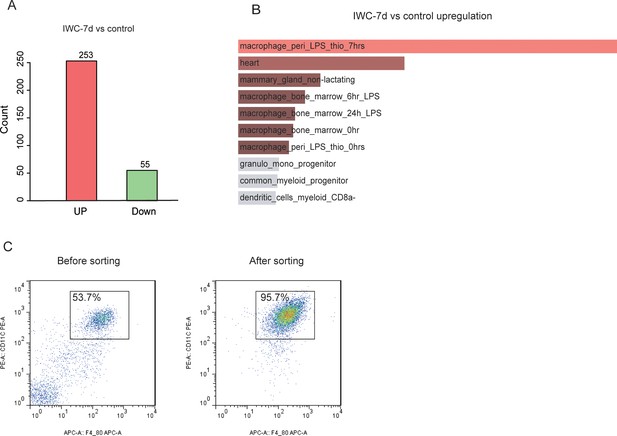
Transcriptional difference at day 7 after immunization of inactivated whole cell (IWC) (LAC-4) in Rag1-/- mice.
(A) Numbers of upregulated and downregulated differentially expressed genes (DEGs) of lungs in IWC-7d and control mice. (B) Top 10 mouse gene atlas terms of upregulated genes in lungs of IWC-7d vs. control mice analyzed by Enrichr. (C) The purity of alveolar macrophages (AMs) after magnetic-activated cell sorting (MACS) sorting is above 95%.
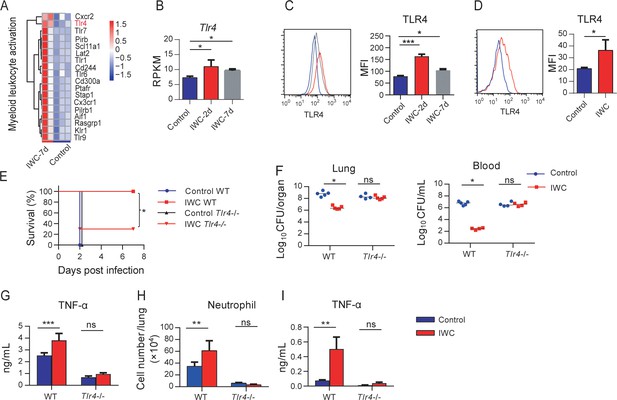
Higher TLR4 expression on inactivated whole cell (IWC)-trained alveolar macrophages (AMs) mediates rapid protection.
(A) Heatmap of differentially expressed genes (DEGs) associated with myeloid leukocyte activation at day 7 after A. baumannii IWC (LAC-4) immunization in Rag1-/- mice. (B) Reads per kilobase per million reads (RPKM) of Tlr4 in lungs on days 2 and 7 after IWC (LAC-4) immunization. (C) Representative histogram of TLR4 expression and mean fluorescence index (MFI) of TLR4 on AMs in BALF on day 2 (red line) and day 7 (gray line) after IWC (LAC-4) immunization or control (blue line). n = 3. For (B) and (C), *p<0.05, **p<0.01, evaluated by ordinary one-way ANOVA. (D) Representative histogram of TLR4 expression and MFI of TLR4 on AMs after IWC stimulation in vitro (red line) for 7 days. n = 3. Data are mean ± SD. *p<0.05, determined by two-tailed unpaired t test. (E–H) Tlr4-/- and WT mice were immunized intranasally (i.n.) with IWC (LAC-4) and challenged with LAC-4 7 days later. (E) Survival curve (n = 5 for WT mice, n = 10 for Tlr4-/- mice). (F) Bacterial burdens at 24 hr post infection (hpi). (G) TNF-α in lungs at 2 hpi. (H) Neutrophil infiltration in lungs at 4 hpi. n = 4–5 mice for (F–H). (I) TNF-α levels in 2 hr culture supernatants of ex vivo IWC (LAC-4)-stimulated AMs from 7-day-vaccinated WT or Tlr4-/- mice. n = 3. For survival, p-value was calculated by log-rank test. From (F) to (I), *p<0.05, **p<0.01, ***p<0.001, ns, not significant, compared by ordinary two-way ANOVA. In (F), each dot represents one mice and the line means median. Data are representative of at least two independent experiments.
-
Figure 6—source data 1
Raw data for Figure 6.
- https://cdn.elifesciences.org/articles/69951/elife-69951-fig6-data1-v1.xlsx
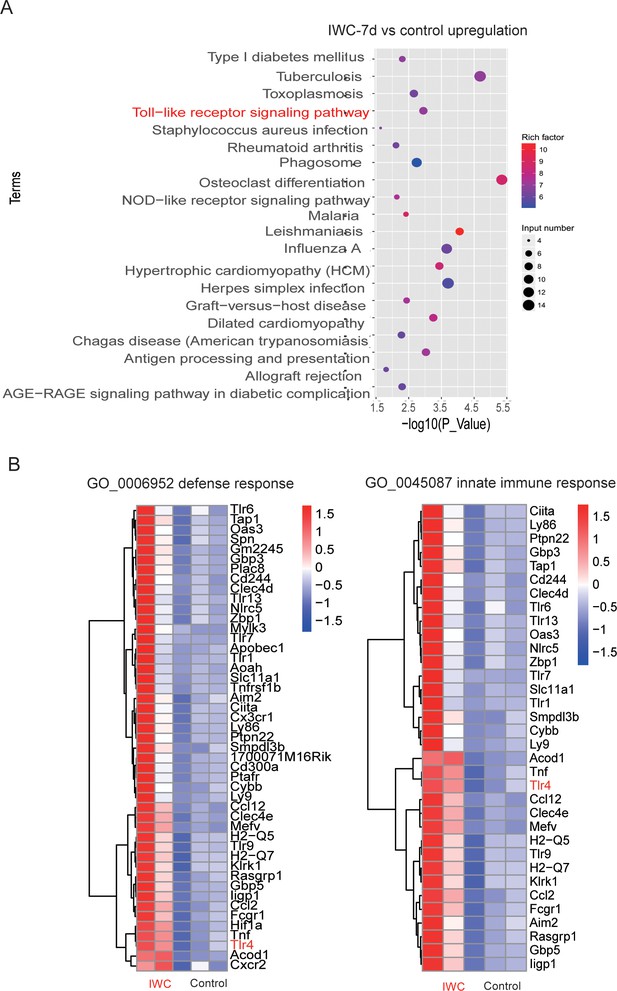
Upregulated differentially expressed genes (DEGs) at day 7 after inactivated whole cell (IWC) immunization in Rag1-/- mice.
(A) Top 20 KEGG terms of upregulated DEGs in IWC-7d vs. control mice. (B) Heatmap of DEGs related to defense response (GO: 0006952) and innate immune response (GO0045087).
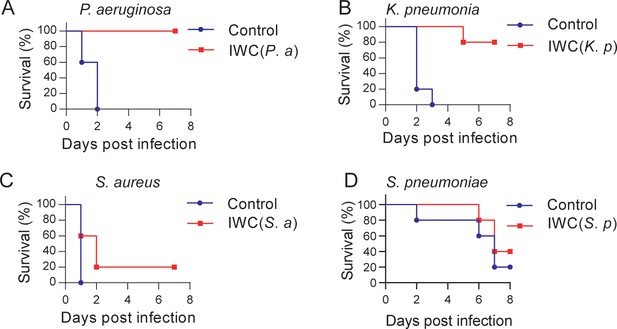
A rapid protection induced by intranasal vaccination against other bacteria.
C57BL/6 mice were immunized intranasally (i.n.) with inactivated whole cell (IWC) of (A) P. aeruginosa (IWC(P.a)), n = 5, (B) K. pneumoniae (IWC(K.p)), n = 5, (C) S. aureus (IWC(S.a)), n = 10, or (D) S. pneumoniae (IWC(S.p)), n = 5. On day 7, the immunized mice were challenged intratracheally (i.t.) with the same species. The survival rates were monitored for 7 days. Data are representative of at least two independent experiments.
-
Figure 7—source data 1
Raw data for Figure 7.
- https://cdn.elifesciences.org/articles/69951/elife-69951-fig7-data1-v1.xlsx
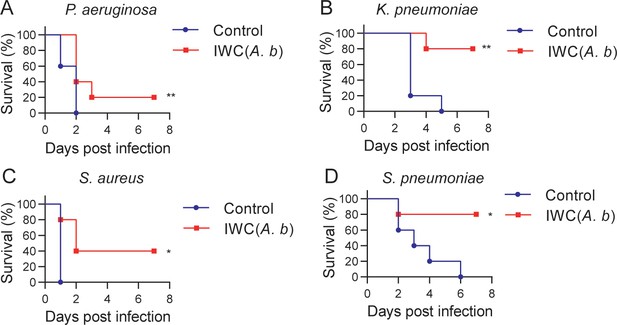
Cross-protection induced by intranasal vaccination of inactivated whole cell (IWC) of A. baumannii.
C57BL/6 mice were immunized intranasally (i.n.) with IWC of A. baumannii LAC-4 (IWC(A.b)) and challenged intratracheally (i.t.) with (A) P. aeruginosa (1 × 107 CFU, n = 10), (B) K. pneumoniae (1 × 107 CFU, n = 5), (C) S. aureus (5 × 107 CFU, n = 5), or (D) S. pneumoniae (2 × 107 CFU, n = 5) at day 7 after immunization. Survival was recorded for 7 days. *p<0.05, **p<0.01, determined by log-rank test. Data are representative of at least two independent experiments.
-
Figure 8—source data 1
Raw data for Figure 8.
- https://cdn.elifesciences.org/articles/69951/elife-69951-fig8-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Mus musculus) | C57BL/6J | Beijing HFK Bioscience Limited Company | N/A | |
| Genetic reagent (Mus musculus) | Rag1-/-: B6.129S7-Rag1tm1/Nju | Model Animal Research Center of Nanjing University | Stock no :N000088 | |
| Genetic reagent (Mus musculus) | Tlr4-/- mice: C57BL/10ScNJNju | Model Animal Research Center of Nanjing University | Stock no: N000192 | |
| Genetic reagent (Mus musculus) | C57BL/10JNju | Model Animal Research Center of Nanjing University | Stock no: N000023 | |
| Strain, strain background (Acinetobacter baumannii) | LAC-4 | Gift from Professor Chen, Harris et al., 2013 | N/A | |
| Strain, strain background (Acinetobacter baumannii) | ATCC-17978 | ATCC | ATCC-17978 | |
| Strain, strain background (Acinetobacter baumannii) | SJZ-09 | Clinical isolate, Zeng et al., 2019 | ||
| Strain, strain background (Acinetobacter baumannii) | SJZ-24 | Clinical isolate, Zeng et al., 2019 | ||
| Strain, strain background (Acinetobacter baumannii) | SJZ-26 | Clinical isolate, Zeng et al., 2019 | ||
| Strain, strain background (Pseudomonas aeruginosa) | XN-1 | Clinical isolate, Gao et al., 2020 | ||
| Strain, strain background (Klebsiella pneumoniae) | YBQ | Clinical isolate, Liu et al., 2020 | ||
| Strain, strain background (Staphylococcus aureus) | KM-22 | Clinical isolate, Zeng et al., 2020 | ||
| Strain, strain background (Streptococcus pneumoniae) | S. pneumoniae | Clinical isolate | ||
| Antibody | Rat monoclonal anti-mouse CD45-PE/Cy7 (clone 30-F11) | BioLegend | Cat# 103114 RRID:AB_312979 | (1:100) |
| Antibody | Rat monoclonal anti-mouse/human CD11b-PerCP/Cy5.5 (clone M1/70) | BioLegend | Cat# 101228 RRID:AB_893232 | (1:100) |
| Antibody | Rat monoclonal anti-mouse F4/80-APC (clone BM8) | BioLegend | Cat# 123116 RRID:AB_893481 | (1:100) |
| Antibody | Rat monoclonal anti-mouse Ly-6C-PE (clone HK1.4) | BioLegend | Cat# 128008 RRID:AB_1186132 | (1:100) |
| Antibody | Rat monoclonal anti-mouse Ly-6G-FITC (clone 1A8) | BioLegend | Cat# 127605 RRID:AB_1236488 | (1:100) |
| Antibody | Hamster monoclonal anti-mouse CD11c-PE (clone N418) | BioLegend | Cat# 117307 RRID:AB_313776 | (1:100) |
| Antibody | Rat monoclonal anti-mouse CD11c-FITC (clone N418) | BioLegend | Cat# 117305 RRID:AB_313774 | (1:100) |
| Antibody | Mouse monoclonal anti-mouse CD284(TLR4)-PE (clone UT41) | Thermo Fisher | Cat# 12-9041-80 RRID:AB_466236 | (1:100) |
| Antibody | Rat monoclonal rat IgG1 isotype control (clone HRPN) | Bio X Cell | Cat# BE0088, RRID:AB_1107775 | 200 μg/per mouse |
| Antibody | Rat monoclonal anti-mouse TNF-α (clone XT3.11) | Bio X Cell | Cat# BE0058, RRID:AB_1107764 | 200 μg/per mouse |
| Sequence-based reagent | Primers for qPCR | This paper | N/A | See Supplementary file 1 |
| Commercial assay or kit | RNAiso Plus | Takara | Cat# 9109 | |
| Commercial assay or kit | PrimeScript RT reagent kit | Takara | Cat# RR037A | |
| Commercial assay or kit | TB Green Premix Ex Taq II | Takara | Cat# RR820A | |
| Commercial assay or kit | IL-6 ELISA kit | BioLegend | Cat# 431304 | |
| Commercial assay or kit | TNF-α ELISA kit | BioLegend | Cat# 430904 | |
| Commercial assay or kit | IL-1β ELISA kit | BioLegend | Cat# 432604 | |
| Commercial assay or kit | Anti-mouse CD11c Microbeads kit | Miltenyi Biotec | Cat# 130-125-835 | |
| Software, algorithm | GraphPad Prism | GraphPad Software | RRID:SCR_002798 | |
| software, algorithm | FlowJo | FlowJo | RRID:SCR_008520 |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/69951/elife-69951-transrepform1-v1.docx
-
Supplementary file 1
Primers used in real-time PCR.
- https://cdn.elifesciences.org/articles/69951/elife-69951-supp1-v1.docx





